Abstract
The chlamydial glycolipid exoantigen (GLXA), a glycolipid antigen derived from Chlamydia muridarum, has been implicated in chlamydial–host cell interaction. Although glycolipid antigens from Sphingomonas and related bacteria have been shown to activate invariant natural killer T (iNKT) cells, it is not yet known whether GLXA can activate these cells. In this study, we have for the first time investigated the role of GLXA in iNKT cell activation using in vitro as well as in vivo settings. First, we examined the effect of GLXA on iNKT cell activation in a cell-free antigen-presentation assay, and found that GLXA specifically stimulated iNKT1.4 hybridoma cell produce enhanced amounts of IL-2. Next, we analyzed the effect of pharmacological activation of iNKT cells by GLXA using iNKT cell-deficient (iNKT knockout (KO)) mice and bone marrow-derived dendritic cell (BMDC)–liver mononuclear cell (LMC) coculture system. On stimulation with GLXA, iNKT cells produced higher quantities of cytokines in a CD1d-dependent fashion. More importantly, iNKT cells from GLXA-treated, but not from cell mock-treated, mice showed higher expression of activation marker, CD69, and enhanced production of interferon (IFN)-γ and IL-4 in vivo. Cumulatively, these data provide evidence on the pharmacological ability of GLXA in specifically activating iNKT cells.
Keywords: CD1d, Chlamydia, GLXA, iNKT
Introduction
Chlamydiae are obligate intracellular bacterial pathogens. The genus Chlamydia includes two species, Chlamydia trachomatis and Chlamydia pneumoniae, which cause various human diseases. C. trachomatis is a major cause of respiratory, ocular and sexually transmitted diseases.1 In addition, C. trachomatis can also act as a precipitating factor in pathogenesis of human immunodeficiency virus and human papilloma virus infections.2,3 Further, C. pneumoniae causes respiratory diseases like bronchitis, sinusitis and pneumonia. Recently, C. pneumoniae has also been implicated in the pathogenesis of atherosclerosis, Alzheimer's disease and multiple sclerosis.4,5,6 To date, there is no effective vaccine available against human chlamydial diseases. Considering the public health significance of chlamydial diseases, it is highly desirable to have an effective and safe chlamydial vaccine. However, one of the main constraints in the way of vaccine development is poor understanding of the role of chlamydial components in host–chlamydial interactions.7
Chlamydial glycolipid exoantigen (GLXA) is a glycolipid component of the chlamydial membrane and intracellular inclusion bodies, which has been implicated in chlamydial–host cell interaction, and can also be found in the host cell cytoplasm and micromilieu of infected cells.8,9,10,11,12,13 It is different from chlamydial lipopolysaccharide and possesses a polysaccharide epitope having weak immunogenicity.8,9,11 Moreover, GLXA has been exploited for its use as a vaccine candidate against the infection.14,15 Due to poor immunogenicity of the epitope, a monoclonal anti-idiotypic antibody (mAb2) to the epitope has been generated, which mimics the biological features of the epitope.14 On immunization with mAb2, mice developed an anti-mAb2 response that recognized purified GLXA.15 Further studies showed that immunization with mAb2 induced significant protective immune responses that reduced C. trachomatis infectivity.14,15
Invariant natural killer T (iNKT) cells represent an innate subset of T lymphocytes expressing the markers of both αβ T cells and natural killer cells. They are the most widely studied class of NKT cells that express the invariant T-cell receptor (TCR).16 In contrast to αβ T cells, iNKT cells recognize glycolipid antigens presented by CD1d, which is a non-classical MHC class I molecule expressed on antigen presenting cells (APC), such as DC.17 However, they can be activated by CD1d-independent stimulation as well.18 CD1d-restricted glycolipid Ag, α-galactosylceramide (α-GalCer), originally extracted from the marine sponge, Agelasmauritianus, is the prototypic glycolipid antigen for iNKT cells.19,20 Further, iNKT cells are unique in their ability to secrete large quantities of cytokines such as interferon (IFN)-γ and IL-4, and possess the unique characteristics of rapid activation, effector functions and modulation of other immune cells.21,22 Owing to these characteristics, iNKT cells have been shown to play a regulatory role in immune responses to autoimmunity, tumor rejection and various infections.23,24,25,26
Previous studies have shown that iNKT cells can respond to the natural glycolipid antigens from certain microbes such as Sphingomonas sps and Borrelia burgdorferi.27,28,29 However, it remains to be determined that whether iNKT cells can recognize glycolipid antigens from other taxa of microbes. Further, we have previously reported that iNKT cells are activated in vivo and play an important role in immune responses to chlamydial infections.30,31 Taking account of these facts, we hypothesized that GLXA might specifically activate iNKT cells. To test the hypothesis, we investigated the role of GLXA derived from Chlamydia muridarum, a mouse biovar of C. trachomatis (also known as mouse pneumonitis) in iNKT cell activation using in vitro as well as in vivo settings. First, we investigated whether GLXA could directly stimulate iNKT cells in a cell-free antigen-presentation assay using an iNKT hybridoma cell. Further, we analyzed the ability of GLXA in activating iNKT cells using a combination of bone marrow derived dendritic cells (BMDC)–liver mononuclear cell (LMC) coculture system and iNKT knockout (KO) mice. Furthermore, we analyzed the effect of GLXA on iNKT cells in vivo. Overall, the data show that GLXA plays an important role in iNKT cell activation.
Materials and methods
Extraction and identification of GLXA
GLXA was purified from C. muridarum as described previously.32 Briefly, C. muridarum organisms (Nigg strain) were grown in Hep-2 cells and the GLXA-containing culture supernatant centrifuged (8000g) to remove any cellular debris after 96 h post-infection. Supernatants harvested from uninfected cell monolayers were prepared in an identical manner and used as cell mock control. Then the supernatant was subjected to ultracentrifugation at 180 000g for 3 h at 4 °C. The pellets were resuspended in 1 ml phosphate-buffered saline (PBS) and sequentially digested for a minimum of 2 h at 37 °C with DNase (50 µg/ml), RNase (50 µg/ml) and Proteinase K (100 µg/ml) in the presence of 4.2 mM MgCl2 and 1 mM CaCl2 (Sigma, St Louis, MO, USA). The solution was incubated at 85 °C for 2 h to remove Proteinase K activity, followed by dialysis (15000 MWCO) against 0.075 M PBS containing 0.01% Sodium Azide overnight at 4 °C. This purified product was used at a 1∶100 dilution in all subsequent experiments. SDS–PAGE and western blot analysis were done to identify GLXA as described previously.8 Briefly, GLXA and cell mock were subjected to SDS–PAGE and transferred to a polyvinylidene difluoride membrane. The following antibodies were used: rabbit polyclonal antibody to Chlamydia and goat anti-rabbit IgG horseradish peroxidase. (Abcam Company, Cambridge, MA, USA).
CD1d fusion protein iNKT cell culture
CD1d fusion protein and iNKT cell stimulation assay was performed as described previously.33 Briefly, 96-well plates (Costar, Corning Life Sciences, Union City, CA, USA) were incubated with mouse CD1d fusion protein (BD Biosciences, San Diego, CA, USA) at a concentration of 0.5 µg/well in coating buffer (eBioscience, San Diego, CA, USA) at 4 °C for overnight. After the incubation, plates were washed thrice with PBS. α-Galcer, GLXA, cell mock or PBS at different doses (20 or 40 µl) was added to each well of the plate and incubated at 37 °C for 4 h. The concentrations of α-Galcer in the culture were 0.05 or 0.1 µg/ml, while the concentrations of GLXA were 2.5 or 5 µg/ml. iNKT1.4 hybridoma cell (kindly provided by Dr Mitchell Kronenberg, La Jolla Institute for Allergy and Immunology, La Jolla, CA, USA) cells were added to each well (5×104 cells per well) and incubated at 37 °C for 24 h. The culture supernatant was measured for IL-2 by ELISA.
Mice
C57BL/6 mice were bred at the facility of the University of Manitoba animal care facility. Breeding pairs of Jα18 KO (iNKT KO) mice with B6 background were kindly provided by Dr Masaru Taniguchi (RIKEN Research Center for Allergy and Immunology, Yokohama, Japan). iNKT KO mice were generated by specific deletion of the Jα18 gene segment using homologous recombination and aggregation chimera techniques.34 All mice used in this study were males of 6–8 weeks of age, and bred and maintained at a pathogen-free animal care facility at the University of Manitoba. All experiments were done in compliance with the guidelines issued by the Canadian Council of Animal Care, and the animal protocol was approved by the institutional ethical committee.
BMDC generation
Dendritic cells (DCs) were generated in vitro from bone marrow cells as described previously.35 In brief, bone marrow cells were isolated from the femurs of mice and cultured in complete medium RPMI 1640 (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (Gibco), 10−5 M 2-ME (Sigma Chemicals, St. Louis, MO, USA), and 2 mM l-glutamine (Gibco) in a six-well plate (Costar) at a concentration of 106 cells/ml in the presence of granulocyte-macrophage colony-stimulating factor (20 ng/ml) and IL-4 (1 ng/ml) for 6 days.
Spleen and liver cell isolation
Spleens were harvested from the animals at specified time and digested in 2 mg/ml collagenase D (Roche Diagnostics, Meylan, France) in RPMI 1640 for 30 min at 37 °C. EDTA (5 mM) was added and the cell suspension was pipetted up and down and filtered. The single-cell suspension so prepared was then subjected to red blood cell lysis by ACK lysis buffer (150 mM NH4Cl, 10 mM KHCO3 and 0.1 mM EDTA). The cells were washed and resuspended in RPMI 1640 for further applications. For liver single-cell preparation, livers were minced into small pieces and pipetted thoroughly in RPMI 1640. The cell suspension was passed through a 70 µm cell filter and resuspended in 40 ml PBS. The cells were centrifuged at 700g for 1 min and then centrifuged at 1200g for 10 min at 4 °C. The cell pellet was resuspended in 8 ml 35% (v/v) Percoll (Pharmacia, Uppsala, Sweden) and centrifuged at 2400g for 12 min at room temperature. Red blood cells were lysed with ACK lysis buffer followed by two washes in RPMI 1640 with 5% fetal calf serum and resuspended in the media according to different applications.
LMC–BMDC coculture
2BMDC were cultured at 1×106 cells in 1 ml culture medium per well in 24-well plates in the presence of α-Galcer, GLXA, cell mock or PBS for 24 h. The concentrations of α-Galcer and GLXA in the culture were 0.1 and 5 µg/ml, respectively. After 24 h of the incubation, the treated BMDC were cocultured with LMC (2×105 cells/well) in RPMI 1640 complete media in 96-well plates for 48 h. The levels of IFN-γ and IL-4 in the culture supernatants were measured by ELISA using unlabeled (capture) and biotinylated (detection) antibodies (BD Pharmingen, San Diego, CA, USA) as described previously.36
Serum cytokine measurements in vivo
C57BL/6 (wild-type (WT)) and iNKT KO mice were intravenously treated with α-Galcer (1 µg in 200 µl PBS), GLXA (10 µg/ml in 200 µl) or cell mock (200 µl). The serum levels of IL-4 and IFN-γ were measured at 2 and 12 h respectively after the treatment. The measurement of the cytokine levels was done by ELISA.
GLXA administration in vivo
Male C57BL/6 mice between 6 and 8 weeks of age were injected with α-Galcer (2 µg in 400 µl PBS), GLXA (10 µg/ml of 400 µl) or cell mock (400 µl) intraperitoneally. Mice receiving cell mock were considered as controls. After 72 h, spleens and livers were harvested from the animals, and processed into single-cell suspensions. The cells were then stained with the appropriate antibodies for flow cytometric analysis.
Flow cytometry
The staining for iNKT cells was done by using PE-mCD1d/PBS57 ligand tetramer (provided by the National Institute of Allergy and Infectious Disease MHC Tetramer Core Facility, Atlanta, GA, USA). For iNKT cell surface marker staining, freshly isolated splenic cells and hepatic cells were stained using fluorescent-labeled anti-CD3e-FITC, anti-CD69–APC or with their isotype controls (eBioscience) in the staining buffer. The analysis for IFN-γ and IL-4 production by iNKT cells was done by intracellular cytokine staining as described previously.31 Briefly, the cells were stimulated with phorbol myristate acetate (50 ng/ml) and ionomycin (1 µg/ml), and incubated at a concentration of 7.5×106 cells/ml at 37 °C in complete RPMI 1640 medium. After 2 h, brefeldin A was added to the culture, and the cells were cultured for another 4 h to accumulate cytokines intracellularly. The cells were subsequently washed and blocked for 20 min with anti-CD16/CD32 antibodies in FACS buffer and then surface-stained with the appropriate antibodies. Cells were fixed in fixation buffer, and then washed in permeabilization buffer. They were subsequently stained with anti-IFN-γ–APC or anti-IL-4–APC for 30 min. Sample data were collected using a LSR II flow cytometer (BD Biosciences) and analyzed using FACS express software.
Statistical analysis
Data were analyzed using unpaired, two-tailed Student's t-test (SPSS 11.0; SPSS Inc., Chicago, IL, USA). A P value <0.05 was considered significant.
Results
Preparation of GLXA antigen
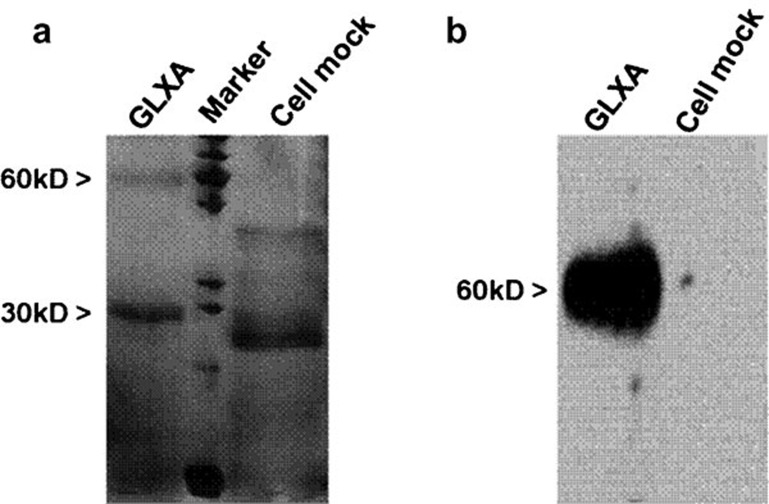
We prepared GLXA antigen from the supernatant of C. muridarum-infected cell culture as described.32 To confirm that the purified antigen was GLXA, we examined the antigen by SDS–PAGE and western blot analysis. SDS–PAGE and silver staining of the antigen sample demonstrated a banding pattern similar to GLXA as described previously,8 cell mock preparation failed to show the bands although (Figure 1a). Further, in a western blot, the major bands of GLXA, but not cell mock, produce a banding pattern that was apparently similar to GLXA (Figure 1b), indicating that the prepared antigen was GLXA.
Figure 1.
SDS–PAGE and western blot analysis for analysis of purified GLXA from C. muridarum-infected cell culture. (a) SDS–PAGE and silver staining demonstrating the banding pattern of purified GLXA, molecular weight standards and cell mock control. (b) Western blot. Immnoidentification of GLXA and cell mock bands transferred from the SDS–PAGE system. GLXA, glycolipid exoantigen.
GLXA specifically stimulates iNKT cells
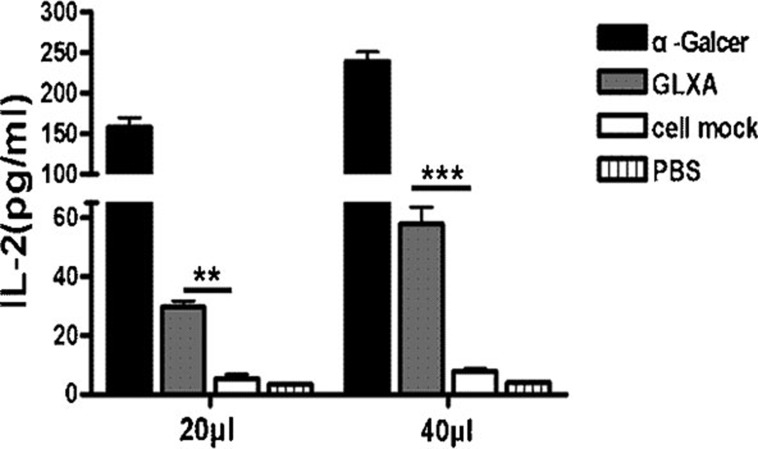
iNKT cells recognize glycolipid antigens presented to them by CD1d molecules. To determine whether the GLXA preparation from C. muridarum can activate iNKT cells, we incubated iNKT hybridoma cell with plate-bound CD1d in the presence of GLXA, α-Galcer, cell mock or PBS in a cell-free antigen-presentation assay. On stimulation with GLXA-treated CD1d, in contrast to the cell mock or PBS, iNKT cells produced IL-2 in a dose-dependent manner, suggesting that GLXA can specifically stimulate iNKT cells (Figure 2).
Figure 2.
Recognition of GLXA by iNKT hybridoma cells using cell-free antigen-presentation assay. iNKT hybridoma cells (5×104 cells per well) were added and cultured in CD1-coated 96-well plates in the presence of α-Galcer, GLXA, cell mock or PBS at different doses (20 or 40 µl) for 24 h. The concentrations of α-Galcer in the culture were 0.05 or 0.1 µg/ml, while the concentrations of GLXA were 2.5 or 5 µg/ml. Concentration of IL-2 in supernatant was determined by ELISA. One representative experiment of three independent experiments is shown. The results are shown as mean±s.d. of each group (**P<0.01 and ***P<0.001). α-Galcer, α-galactosylceramide; GLXA, glycolipid exoantigen; iNKT, invariant natural killer T; PBS, phosphate-buffered saline.
GLXA induces increased cytokine production by iNKT cells in vitro, and the cytokine production is CD1d-dependent
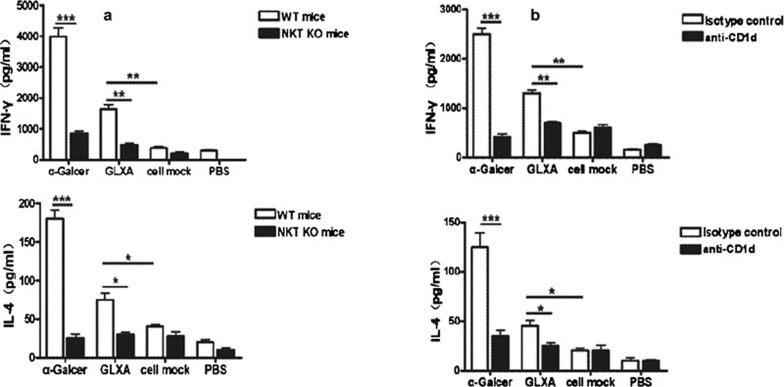
To confirm the finding in the cell-free system that GLXA can stimulate iNKT cells, we further tested the capacity of GLXA to activate iNKT cells for cytokine production using a BMDC–LMC coculture system. We treated BMDC with GLXA, α-Galcer, cell mock or PBS, and then cocultured the treated BMDC with LMC, and examined the cytokine levels of the culture supernatants. We found that GLXA stimulated LMC from WT mice to produce higher quantities of IFN-γ and IL-4 compared with cell mock. To confirm that the cytokines produced in the coculture were secreted by iNKT cells, we isolated the LMC from iNKT KO mice and cocultured it with BMDC. The data showed that the LMC from iNKT KO mice produced significantly lower quantities of IFN-γ and IL-4 compared with those from WT mice (Figure 3a), indicating that the cytokines were mainly produced by iNKT cells. Since iNKT cells can be activated via CD1d-dependent and/or CD1d-independent pathways, we further examined whether the cytokine production by the iNKT cells was CD1d-dependent by blocking CD1d using anti-CD1d antibodies in the BMDC–LMC coculture system. On addition of the antibodies, the GLXA-treated coculture showed a dramatic decrease in IFN-γ and IL-4 production compared with the controls, demonstrating the dependence of iNKT cell activation by GLXA on CD1d molecules (Figure 3b).
Figure 3.
Enhanced cytokine production by iNKT cells after GLXA treatment in BMDC–LMC coculture system. BMDC were cultured at 1×106 cells in 1 ml culture medium in 24-well plates in the presence of α-Galcer, GLXA, cell mock or PBS for 24 h, and then cocultured with LMC from NKT KO or WT mice for 48 h. The concentrations of α-Galcer and GLXA used in the culture were 0.1 and 5 µg/ml respectively. (a) Concentrations of IFN-γ and IL-4 in the culture supernatants were determined by ELISA. (b) Blocking of CD1d in LMC–BMDC coculture. BMDC–LMC coculture was treated with anti-CD1d antibodies or isotype control antibodies. Concentrations of IL-4 and IFN-γ in supernatants were determined by ELISA. The results are shown as mean ± SD. One of the three similar experiments is shown (*P<0.05, **P<0.01 and ***P<0.001). α-Galcer, α-galactosylceramide; BMDC, bone marrow-derived dendritic cell; GLXA, glycolipid exoantigen; IFN, interferon; iNKT, invariant natural killer T; KO, knockout; LMC, liver mononuclear cell; PBS, phosphate-buffered saline; WT, wild-type.
GLXA contributes to the cytokine production by iNKT cells in vivo
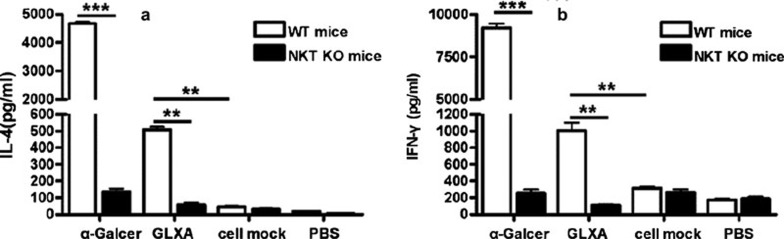
To examine the function of GLXA on cytokine production by iNKT cells in vivo, we injected GLXA in iNKT KO and WT mice intravenously, and measured the cytokine levels in sera at appropriate time points. Our data showed that GLXA stimulated WT mice to produce higher quantities of IFN-γ and IL-4 compared with cell mock, whereas there was no significant difference in the cytokine production between GLXA and cell mock in iNKT KO mice (Figure 4). These data suggested that the cytokines produced in the serum were largely secreted by iNKT cells, and that iNKT cells were activated by GLXA in this process.
Figure 4.
GLXA contributes to the cytokine production by iNKT cells in vivo. Both WT mice and iNKT KO mice were intravenously injected with α-Galcer (1 µg in 200 µl PBS), GLXA (10 µg/ml of 200 µl) or cell mock (200 µl), and the sera were collected at appropriate times for cytokine measurement by ELISA. (a) IFN-γ levels at 12 h after injections; (b) IL-4 levels at 2 h after injections. The results are shown as mean±s.d. (**P<0.01 and ***P<0.001). α-Galcer, α-galactosylceramide; GLXA, glycolipid exoantigen; IFN, interferon; iNKT, invariant natural killer T; KO, knockout; PBS, phosphate-buffered saline; WT, wild-type.
Enhanced iNKT cell activation in vivo by GLXA treatment
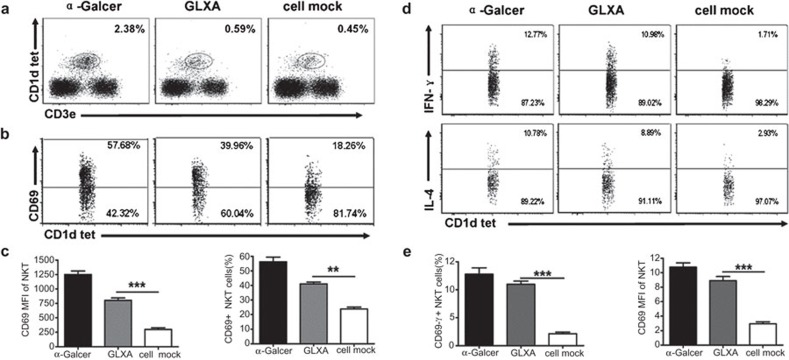
To confirm the in vitro and in vivo data from the present study that GLXA induces iNKT cell activation, we examined the role of GLXA in iNKT cell activation in vivo in terms of cytokine production as well as activation marker expression. We immunized the mice with GLXA, α-Galcer or cell mock and analyzed the splenic iNKT cells for activation marker, CD69, expression and intracellular cytokine production. CD1d tetramer+CD3e+ cells were gated and analyzed as iNKT cells by flow cytometry. Our data showed that the percentage of iNKT cells in GLXA group was a little higher than those in cell mock group, although there was no statistically significant difference between them (Figure 5a). Compared with those from the cell mock-treated, iNKT cells from GLXA-treated mice showed significantly higher expression of CD69 (Figure 5b and c). On flow cytometric analysis of the pattern of intracellular cytokine production by iNKT cells in vivo, we found that iNKT cells from the GLXA-treated mice produced large amounts of IFN-γ and IL-4 compared with the cell mock-treated mice (Figure 5d and e). Similarly, analysis of hepatic iNKT cells showed that the GLXA group exhibited higher iNKT cells, CD69 expression (37% versus 21%), IFN-γ production (13% versus 3%) and IL-4 production (14% versus 4%) when compared to control group. These data show that GLXA stimulates iNKT cells in vivo.
Figure 5.
Effect of GLXA treatment on iNKT cell activation in vivo in terms of surface marker expression and cytokine production. C57BL/6 mice (four mice per group) were injected intraperitoneally with α-Galcer (2 µg in 400 µl PBS), GLXA (10 µg/ml of 400 µl) or cell mock (400 µl). All the mice were killed at 72 h after GLXA administration. Spleens were aseptically removed and processed into single-cell suspensions and then stained with appropriate antibodies for flow cytometry analysis. (a) Flow cytometry. Dotplot demonstrating the gating strategy for iNKT cells. CD1d tetramer+CD3e+ cells were analyzed as iNKT cells. (b) Representative flow cytometry images showing analyses performed on iNKT cells for CD69 expression from each group. (c) Graphs summarizing flow cytometry data showing MFI and percentage of CD69+ iNKT cells. (d) Cells from different treatment groups of mice were cultured and stained for intracellular cytokine production as described in the section on ‘Materials and methods'. Representative flow cytometry images demonstrating IFN-γ and IL-4 production by iNKT cells. (e) Graphs represent the summary of percentage of IFN-γ+ and IL-4+ iNKT cells in each group. Data are shown as mean±s.d. Results of one representative of three experiments are shown (**P<0.01 and ***P<0.001). α-Galcer, α-galactosylceramide; GLXA, glycolipid exoantigen; IFN, interferon; iNKT, invariant natural killer T; MFI, median fluorescence intensity; PBS, phosphate-buffered saline.
Discussion
The present study demonstrates that GLXA, a glycolipid antigen derived from Chlamydia-infected cell culture supernatants, can activate iNKT cells in vitro and in vivo. Specifically, the data showed that GLXA can enhance iNKT cell activation marker and induce iNKT cell cytokine production. GLXA specifically stimulated iNKT cells in a cell-free antigen-presentation assay. More importantly, when exposed to GLXA-pulsed DC in a coculture system, iNKT cells produced large amounts of IFN-γ and IL-4 in a CD1d-dependent manner shown by antibody blocking. The in vitro results were further supported by the findings from experiments using the in vivo administration of GLXA in mice. The systemic administration of GLXA resulted in iNKT cell activation in which the cells showed higher expression of activation marker, CD69, and produced enhanced quantities of a mixture of IFN-γ and IL-4.
The data enhance our understanding on the molecular basis by which iNKT cells are activated in chlamydial infection. iNKT cells express a highly conserved invariant TCR that recognizes a prototypic glycolipid antigen extracted from marine sponges, α-Galcer. Emerging evidence demonstrates that iNKT cells can also recognize glycolipid antigens from certain microorganisms, although activated DC may activate iNKT cells without presenting microbial antigens to iNKT cells.27,28,29,38 So far, the identified microbial glycolipid antigens of microbes are still limited, much less than the microbes which have been shown to be able to activate iNKT cells in vitro and in vivo studies. In our previous study, we showed that iNKT plays an important role in immune responses to chlamydial infections.30,31 Interestingly, our previous studies have shown that iNKT cells are activated in vivo and produce IFN-γ and IL-4. C. pneumoniae infection in mice showed enhanced Th1 cytokine, IFN-γ, production by iNKT cells, whereas C. muridarum increased Th2 response reflected by enhanced IL-4 and IL-5 production.30,31 In this study, we further explored the molecular nature of chlamydial antigen which activates iNKT cells. Our data from the study showed that GLXA, a previously reported chlamydial glycolipid, can stimulate iNKT hybridoma cell in either cell-free antigen-presentation assay or a DC based assay. More importantly, in vivo experiments also confirmed the activation of iNKT cells by GLXA based on activation marker expression and cytokine production. Notably, our data from the present study show that GLXA treatment stimulated the production of the mixture of Th1 and Th2 cytokines, i.e. IFN-γ and IL-4 by iNKT cells, and that there is no preference for either Th1 or Th2 cytokines. The data have certain discrepancy with our previous data by testing live C. muridarum infection,30 showing preferential IL-4 production. At least two reasons may be related to this inconsistency. First, it is possible that GLXA is not the only iNKT-specific lipid antigen in chlamydial cells. There may be other chlamydial antigens that act independently or in coordination with GLXA to skew the cytokine response of iNKT cells to either IFN-γ or IL-4 polarization; Second, and probably more likely, the cytokine pattern of iNKT cells especially in population level may be determined by the live process of the infection rather than particular lipid antigen. For example, the process and status of infection may influence the function of DC which may modulate the cytokine production of iNKT cells when they present chlamydial glycolipid antigen to the iNKT cells. Therefore, further study to explore other chlamydial glycolipid antigens and the influence of infection process on iNKT cell cytokine patterns is needed.
It remains unknown how chlamydial GLXA fits into CD1d molecule during antigen presentation. So far, no data are available on the molecular structure of GLXA. A commonly accepted view on the topology of the interaction of α-GalCer, the prototypic antigen for iNKT cells, and CD1d is that the lipid chains of α-GalCer is placed into the hydrophobic groove of CD1d and the α-linked sugar moiety interacts with the TCR of iNKT cells.37 Since GLXA consists of C17 and C18:1 fatty acids and a carbohydrate moiety of polyglucose,9 we speculate that the fatty acid chains of the GLXA might bind to the floor of the hydrophobic cleft of CD1d, whereas the carbohydrate moiety is likely to interact with iNKT TCR. However, further study is required to test this possibility.
In conclusion, the findings from the present study show that GLXA, a glycolipid antigen derived from the chlamydia, can activate iNKT cells, and this activated function depend on CD1d molecules. This finding provides significant new knowledge on the molecular and cellular mechanisms of immune response to chlamydial infection.
Acknowledgments
This work was supported by grants from National Natural Sciences Foundation of China (nos. 30770110 and 30811120425) to WZ and a grant from Canadian Institutes of Health Research (CCI 92213) to XY. YP and LZ were supported by PhD Studentships for Oversea Study from Shandong University.
References
- Schachter, J Chlamydial infections (first of three parts) N Engl J Med. 1978;298:428–435. doi: 10.1056/NEJM197802232980805. [DOI] [PubMed] [Google Scholar]
- Darville T, Hiltke TJ. Pathogenesis of genital tract disease due to Chlamydia trachomatis. . J Infect Dis. 2010;201 Suppl 2:S114–S125. doi: 10.1086/652397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samoff E, Koumans EH, Markowitz LE, Sternberg M, Sawyer MK, Swan D, et al. Association of Chlamydia trachomatis with persistence of high-risk types of human papillomavirus in a cohort of female adolescents. Am J Epidemiol. 2005;162:668–75. doi: 10.1093/aje/kwi262. [DOI] [PubMed] [Google Scholar]
- Johnston SC, Messina LM, Browner WS, Lawton MT, Morris C, Dean D, et al. C-reactive protein levels and viable I in carotid artery atherosclerosis. Stroke. 2001;32:2748–52. doi: 10.1161/hs1201.099631. [DOI] [PubMed] [Google Scholar]
- Balin BJ, Gérard HC, Arking EJ, Appelt DM, Branigan PJ, Abrams JT, et al. Identification and localization of Chlamydia pneumoniae in the Alzheimer's brain. Med Microbiol Immunol. 1998;187:23–42. doi: 10.1007/s004300050071. [DOI] [PubMed] [Google Scholar]
- Grimaldi LM, Pincherle A, Martinelli-Boneschi F, Fillippi M, Patti F, Reggio A, et al. An MRI study of Chlamydia pneumoniae infection in Italian multiple sclerosis patients. Mult Scler. 2003;9:467–471. doi: 10.1191/1352458503ms944oa. [DOI] [PubMed] [Google Scholar]
- Brunham RC, Rey-Ladino J. Immunology of Chlamydia infection: implications for a Chlamydia trachomatis vaccine. Nat Rev Immunol. 2005;5:149–161. doi: 10.1038/nri1551. [DOI] [PubMed] [Google Scholar]
- Stuart ES, Macdonald AB. Some characteristics of a secreted chlamydial antigen recognized by IgG from C. trachomatis patient sera. Immunology. 1989;68:469–473. [PMC free article] [PubMed] [Google Scholar]
- Stuart ES, Tirrell SM, MacDonald AB. Characterization of an antigen secreted by Chlamydia-infected cell culture. Immunology. 1987;61:527–533. [PMC free article] [PubMed] [Google Scholar]
- Stuart ES, Wyrick PB, Choong J, Stoler SB, MacDonald AB. Examination of chlamydial glycolipid with monoclonal antibodies: cellular distribution and epitope binding. Immunology. 1991;74:740–747. [PMC free article] [PubMed] [Google Scholar]
- Webley WC, Vora GJ, Stuart ES. Cell surface display of the chlamydial glycolipid exoantigen (GLXA) demonstrated by antibody-dependent complement-mediated cytotoxicity. Curr Microbiol. 2004;49:13–21. doi: 10.1007/s00284-003-4181-7. [DOI] [PubMed] [Google Scholar]
- Vora GJ, Stuart ES. A role for the glycolipid exoantigen (GLXA) in chlamydial infectivity. Curr Microbiol. 2003;46:217–223. doi: 10.1007/s00284-002-3843-1. [DOI] [PubMed] [Google Scholar]
- Wyrick PB, Davis CH, Raulston JE, Knight ST, Choong J. Effect of clinically relevant culture conditions on antimicrobial susceptibility of Chlamydia trachomatis. . Clin Infect Dis. 1994;19:931–936. doi: 10.1093/clinids/19.5.931. [DOI] [PubMed] [Google Scholar]
- An LL, Hudson AP, Prendergast RA, O'Brien TP, Stuart ES, Whittum-Hudson JA, et al. Biochemical and functional antigenic mimicry by a polyclonal anti-idiotypic antibody for chlamydial exoglycolipid antigen. Pathobiology. 1997;65:229–240. doi: 10.1159/000164134. [DOI] [PubMed] [Google Scholar]
- Whittum-Hudson JA, An LL, Saltzman WM, Prendergast RA, MacDonald AB. Oral immunization with an anti-idiotypic antibody to the exoglycolipid antigen protects against experimental Chlamydia trachomatis infection. Nat Med. 1996;2:1116–1121. doi: 10.1038/nm1096-1116. [DOI] [PubMed] [Google Scholar]
- Godfrey DI, McCluskey J, Rossjohn J. CD1d antigen presentation: treats for NKT cells. Nat Immunol. 2005;6:754–756. doi: 10.1038/ni0805-754. [DOI] [PubMed] [Google Scholar]
- Kronenberg, M Toward an understanding of NKT cell biology: progress and paradoxes. Annu Rev Immunol. 2005;23:877–900. doi: 10.1146/annurev.immunol.23.021704.115742. [DOI] [PubMed] [Google Scholar]
- Godfrey DI, MacDonald HR, Kronenberg M, Smyth MJ, van Kaer L. NKT cells: what's in a name. Nat Rev Immunol. 2004;4:231–237. doi: 10.1038/nri1309. [DOI] [PubMed] [Google Scholar]
- Brossay L, Chioda M, Burdin N, Koezuka Y, Casorati G, Dellabona P, et al. CD1d-mediated recognition of an alpha-galactosylceramide by natural killer T cells is highly conserved through mammalian evolution. J Exp Med. 1998;188:1521–1528. doi: 10.1084/jem.188.8.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, et al. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- Taniguchi M, Harada M, Kojo S, Nakayama T, Wakao H. The regulatory role of Valpha14 NKT cells in innate and acquired immune response. Annu Rev Immunol. 2003;21:483–513. doi: 10.1146/annurev.immunol.21.120601.141057. [DOI] [PubMed] [Google Scholar]
- Carnaud C, Lee D, Donnars O, Park SH, Beavis A, Koezuka Y, et al. Cutting edge: cross-talk between cells of the innate immune system: NKT cells rapidly activate NK cells. J Immunol. 1999;163:4647–4650. [PubMed] [Google Scholar]
- Skold M, Behar SM. Role of CD1d-restricted NKT cells in microbial immunity. Infect Immun. 2003;71:5447–5455. doi: 10.1128/IAI.71.10.5447-5455.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Bao M, Yoon JW. Intrinsic defects in the T-cell lineage results in natural killer T-cell deficiency and the development of diabetes in the nonobese diabetic mouse. Diabetes. 2001;50:2691–2699. doi: 10.2337/diabetes.50.12.2691. [DOI] [PubMed] [Google Scholar]
- Smyth MJ, Thia KY, Street SE, Cretney E, Trapani JA, Taniguchi M, et al. Differential tumor surveillance by natural killer (NK) and NKT cells. J Exp Med. 2000;191:661–668. doi: 10.1084/jem.191.4.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukreja A, Maclaren NK. NKT cells and type-1 diabetes and the “hygiene hypothesis” to explain the rising incidence rates. Diabetes Technol Ther. 2002;4:323–333. doi: 10.1089/152091502760098465. [DOI] [PubMed] [Google Scholar]
- Mattner J, Debord KL, Ismail N, Goff RD, Cantu C 3 rd, Zhou D, et al. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 2005;434:525–529. doi: 10.1038/nature03408. [DOI] [PubMed] [Google Scholar]
- Kinjo Y, Wu D, Kim G, Xing GW, Poles MA, Ho DD, et al. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature. 2005;434:520–525. doi: 10.1038/nature03407. [DOI] [PubMed] [Google Scholar]
- Kinjo Y, Tupin E, Wu D, Fujio M, Garcia-Navarro R, Benhnia MR, et al. Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nat Immunol. 2006;7:978–986. doi: 10.1038/ni1380. [DOI] [PubMed] [Google Scholar]
- Bilenki L, Wang S, Yang J, Fan Y, Joyee AG, Yang X. NK T cell activation promotes Chlamydia trachomatis infection in vivo. . J Immunol. 2005;175:3197–3206. doi: 10.4049/jimmunol.175.5.3197. [DOI] [PubMed] [Google Scholar]
- Joyee AG, Qiu H, Wang S, Fan Y, Bilenki L, Yang X. Distinct NKT cell subsets are induced by different Chlamydia species leading to differential adaptive immunity and host resistance to the infections. J Immunol. 2007;178:1048–1058. doi: 10.4049/jimmunol.178.2.1048. [DOI] [PubMed] [Google Scholar]
- Whittum-Hudson JA, Rudy D, Gèrard H, Vora G, Davis E, Haller PK, et al. The anti-idiotypic antibody to chlamydial glycolipid exoantigen (GLXA) protects mice against genital infection with a human biovar of Chlamydia trachomatis. . Vaccine. 2001;19:4061–4071. doi: 10.1016/s0264-410x(01)00117-7. [DOI] [PubMed] [Google Scholar]
- Chiba A, Cohen N, Brigl M, Brennan PJ, Besra GS, Brenner MB. Rapid and reliable generation of invariant natural killer T-cell lines in vitro. . Immunology. 2009;128:324–333. doi: 10.1111/j.1365-2567.2009.03130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Shin T, Kawano T, Sato H, Kondo E, Toura I, et al. Requirement for Valpha14 NKT cells in IL-12-mediated rejection of tumors. Science. 1997;278:1623–1626. doi: 10.1126/science.278.5343.1623. [DOI] [PubMed] [Google Scholar]
- Inaba K, Inaba M, Romani N, Aya H, Deguchi M, Ikehara S, et al. Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/macrophage colony-stimulating factor. J Exp Med. 1992;176:1693–1702. doi: 10.1084/jem.176.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, HayGlass KT, Brunham RC. Genetically determined differences in IL-10 and IFN-gamma responses correlate with clearance of Chlamydia trachomatis mouse pneumonitis infection. J Immunol. 1996;156:4338–4344. [PubMed] [Google Scholar]
- McCarthy C, Shepherd D, Fleire S, Stronge VS, Koch M, Illarionov PA, et al. The length of lipids bound to human CD1d molecules modulates the affinity of NKT cell TCR and the threshold of NKT cell activation. J Exp Med. 2007;204:1131–1144. doi: 10.1084/jem.20062342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriram V, Du W, Gervay-Hague J, Brutkiewicz RR. Cell wall glycosphingolipids of Sphingomonas paucimobilis are CD1d-specific ligands for NKT cells. Eur J Immunol. 2005;35:1692–1701. doi: 10.1002/eji.200526157. [DOI] [PubMed] [Google Scholar]