The human immune system is equipped with vast numbers of lymphocytes possessing distinct antigen specificities. The enormous repertoire and heterogeneity of lymphocytes ensures their ability to effectively fight against external microbial invaders, as well as internal aberrant cells. However, the understanding of the specific phenotypes and functions of lymphocytes is hindered by the lack of tools that can be used to visualize this complexity. Fluorescent dye-based flow cytometry, which can simultaneously analyze 8–10 parameters of a single cell, is commonly used in immunological research, but further expansion of the number of parameters (up to 10–18) is limited by the overlapping excitation and emission spectra of different fluorescent dyes. Recently, Bendall et al.1 used transition element isotopes as chelated antibody tags for the atomic mass spectrometric analysis of single cells, called cytometry by time-of-flight (CyTOF), which expanded the analysis to 34 or more parameters simultaneously. In addition to greatly increasing the number of the parameters that can be evaluated, these isotopes have very little crosstalk and no need for background adjustment, thus eliminating two other common problems associated with fluorescent dyes.
Using this newly developed CyTOF technology, Newell et al.2 analyzed the phenotypic and functional characteristics of human CD8+ T cells by measuring 17 cell surface markers, six cytokines, two cytotoxic granule components and three viral antigen peptide-MHC tetramers on human CD8+ T cells. These parameters covered nine functional attributes and distinguished all reported CD8+ T-cell subsets, such as naive, central memory, effector memory, terminal effector, long-lived memory precursor effector cells and short-lived effector cells. The authors used principal component analysis (PCA) to analyze the large CyTOF datasets and presented their results either in two-dimensional PCA or in combination with a protein structure program (PyMOL) as three-dimensional PCA.
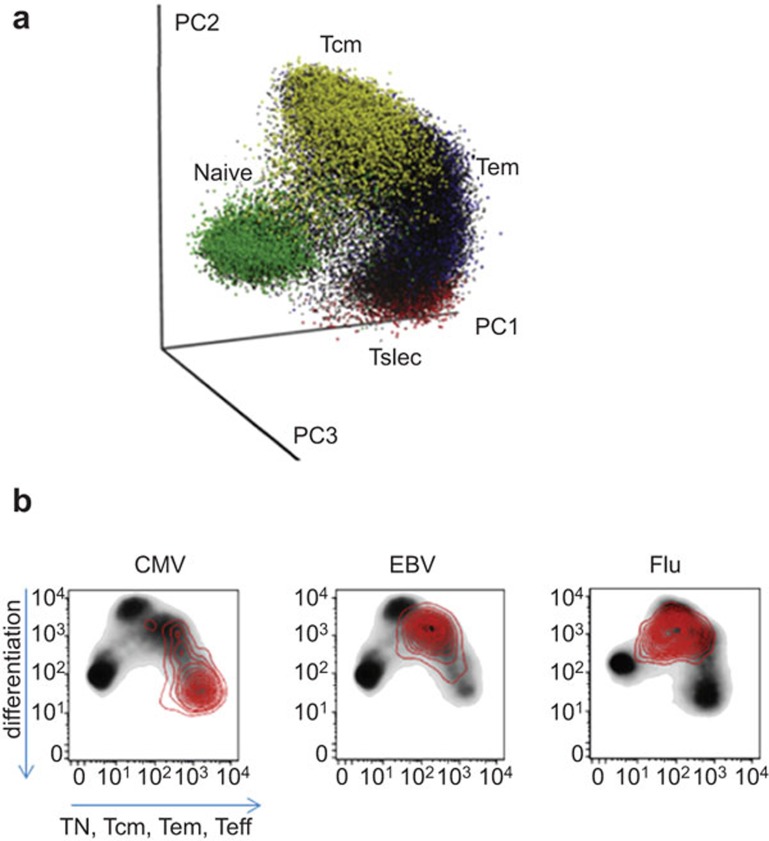
CD8+ T-cell subsets visualized by three-dimensional PCA show a uniform ‘folded Y' pattern. The previously defined subsets are grouped in distinct locations, and the interrelatedness of these subsets can be observed clearly. For example, naive T cells are present at the base of the Y, and short-lived effector cells and central memory cells form distinct nodes at the tips of the Y pattern (Figure 1a). Furthermore, a graded progression of some markers could be observed between the subsets, especially within the memory compartment, generating a continuum of T-cell phenotypes. This continuity reveals the stochastic nature of the expression of CD8+ T-cell progression markers, and the pattern might be useful for defining the differentiation state of a given cell or identifying correlates of effective versus non-effective CD8+ T cell-mediated immune responses. It could also be used for the classification of additional subsets of CD8+ T cells, such as exhausted T cells and the recently described memory T cells with stem cell-like properties.3
Figure 1.
Visualization of CD8+ T-cell subsets identified by CyTOF. (a) A uniform ‘folded Y'-shaped pattern was established using CyTOF PCA. (b) The distinct niche occupation (red) of different viral specific CD8+ T cells was demonstrated by CyTOF two-dimensional PCA. CMV, cytomegalovirus; CyTOF, cytometry by time-of-flight; EBV, Epstein–Barr virus; PCA, principal component analysis; Tcm, central memory T cell; Teff, terminal effector T cell; Tem, effector memory T cell; Tn, naive T cell; Tslec, short-lived effector T cell.
One major contribution of this study is its illustration of the functional complexity of CD8+ T cells. Newell et al.2 identified over 200 functional phenotypes of CD8+ T cells defined by the expression status of cytokines, cytotoxic granule components and CD107, a degranulation marker. A shift in the proportions of cells with each phenotype was identified after in vitro stimulation with either PMA/ionomycin or anti-CD3±anti-CD28. PMA/ionomycin induced a greater number of distinct functional types of CD8+ T cells than did anti-CD3±anti-CD28. However, because these studies were performed at the same timepoint after stimulation, it is not clear whether the observed difference in the number of functional types of CD8+ T cells between these two stimuli is unique to the stimulators or reflects the difference in cell response kinetics to different stimulators, or possibly represents a combination of these factors. Nevertheless, the functional heterogeneity of CD8+ T cells can now be visualized by the combinatorial diversity of the expression status of large numbers of effector molecules. It is certain that more types of functional CD8+ T cells will be uncovered as more effector molecules are included in the analysis.
Further extending this analysis to different viral-specific CD8+ T cells, the authors made a very intriguing finding, i.e., different viral-specific CD8+ T cells have distinct niches in the CyTOF three-dimensional PCA. Three virus-specific cells were inspected: two cause chronic infection (Epstein–Barr virus and cytomegalovirus), and one causes episodic acute infection influenza virus (Flu). Epstein–Barr virus-specific CD8+ T cells had effector memory T cell-like phenotypes, whereas cytomegalovirus-specific CD8+ T cells had the properties of both late-stage effectors and effector memory cell T cells. In contrast, Flu-specific CD8+ T cells showed central memory T cell-like phenotypes (Figure 1b). Furthermore, they found that none of these viral antigen-specific CD8+ T cells expressed CD49d (α4 integrin, VLA-4), which suggests that these cells may share a cell trafficking/migration pattern that does not include the participation of CD49d. However, the cause of these distinct characteristic responses of CD8+ T cells to different viruses is currently unknown. The authors suggest that virus-specific antigen exposure and epitope placement within a dominance hierarchy may provide some explanation. Further study will be needed to elucidate the distinct and shared characteristics of these viral-specific CD8+ T cells.
Despite the weaknesses of the CyTOF technique, which include: (i) the lower signal from metal-labeled probes compared with fluorescent probes; (ii) its inability to be used for cell sorting; and (iii) the requirement for specific instruments, CyTOF technology can detect more than twice the maximum number of tags available for fluorescence-based flow cytometry, and it offers an unprecedented depth for analyzing the complexity of the phenotypes and functions of lymphocytes, as well as other types of cells. It will likely become an essential tool in immunology and in other fields that currently use flow cytometry. In CD8+ T cells, this study has revealed the scale of phenotypic and functional complexity of viral specific CD8+ T cells. Further characterization of these different CD8+ T cells will provide new insights into their relative contributions to an effective immune response.
References
- Bendall SC, Simonds EF, Qiu P, Amir E, Krutzik PO, Finck R, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell EW, Sigal N, Bendall SC, Nolan GP, Davis MM. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity. 2012;36:142–152. doi: 10.1016/j.immuni.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattinoni L, Lugli E, Ji Y, Pos Z, Paulos CM, Quigley MF, et al. A human memory T cell subset with stem cell-like properties. Nat Med. 2011;17:1290–1297. doi: 10.1038/nm.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]