Abstract
HMBOX1 is a new member of the homeobox family. Homeobox members have been reported to participate in embryonic development and systemic metabolism, but the function of HMBOX1 remains unclear, especially in the hematopoietic system. Here, we show that HMBOX1 is expressed at a high level in primary human NK cells but is expressed at much lower levels in NK cell lines. Overexpression of HMBOX1 significantly inhibited NK cell activities, including natural cytotoxicity against tumor cells, the level of CD107a (a marker protein for degranulation) and the production of cytolytic proteins (perforin and granzymes). More interestingly, HMBOX1 negatively regulated the expression of NKG2D and the activation of the NKG2D/DAP10 signaling pathway in NK cells. This effect was reversed by knocking down HMBOX1. Taken together, these findings demonstrate that HMBOX1 may act as a negative regulator of NK cell functions via suppressing the NKG2D/DAP10 signaling pathway.
Keywords: HMBOX1, NK cells, NKG2D
Introduction
HMBOX1, a novel homeobox gene, was first isolated from a pancreatic cDNA library in 2006.1 Homeobox genes constitute a large gene family with approximately 300 members. Homeobox proteins are characterized by their typical DNA binding homeodomain, which contains about 60 amino acids and is encoded by a conserved DNA motif (180 bp).2 Most homeobox genes are transcription factors that regulate the transcription of genes associated with embryonic development and cell differentiation. For example, the HOX genes regulate normal hematopoiesis and are usually involved in leukemic transformation.3 The HMBOX1 protein contains an atypical homeodomain and a putative HNF1_N domain and was classified into the HNF homeobox class of the Hmbox family.2 Recently, a novel splicing variant of HMBOX1, HMBOX1b, was identified.4 HMBOX1 is expressed in many human tissues, such as the cerebrum, intestines, stomach, liver, pancreas, lungs, thyroid gland, cardiac muscle, testis and prostate.1, 5 Chen et al. speculated that HMBOX1 may be a ubiquitous transcription repressor that is expressed in most human tissues.1 Recently, Su et al. reported that HMBOX1 is a key factor in the differentiation of bone marrow stromal cells into vascular endothelial cells.6 However, the expression of HMBOX1 and its function in the hematopoietic system are still unclear.
Natural killer (NK) cells are critical components of the immune system; they not only play important roles in immune surveillance but also regulate the adaptive immune response, making NK cells a bridge between the innate and adaptive arms of the immune system.7 The particular role of NK cells is elaborately regulated by the microenvironment and is controlled by a series of transcription factors, including T-bet, ETS1, MEF, IRF2, MITF and GATA3.8, 9 These transcription factors are transcriptional activators involved in NK cell development, maturation, cytotoxicity generation and perforin or IFN-γ production.9 Even though much is known about NK cells, there are few reports about transcription factors that negatively regulate the function of NK cells.
In this study, we observed that the expression level of HMBOX1 in primary NK cells purified from human peripheral blood mononuclear cells (PBMCs) was much higher than that in NK cell lines. Then we found that overexpression of HMBOX1 significantly inhibited the cytotoxicity of NK cells in addition to decreasing the expression levels of perforin and granzyme. Furthermore, we observed decreased expression of NKG2D and its adaptor molecule DAP10, as well as reduced activity of the NKG2D/DAP10 signaling proteins. Knockdown of HMBOX1 expression by RNA interference reversed these effects. Therefore, HMBOX1 may act as a negative regulator of NK cell function.
Materials and methods
Cell lines and cell culture
Natural killer lymphoma (NKL) cells were cultured in RPMI-1640 (GIBICO, Grand Island, NY, USA) containing 10% fetal bovine serum and 100 U/ml rhIL-2. NK-92 cells were cultured in α-MEM (GIBICO/BRL) containing 12.5% horse serum (GIBICO), 12.5% fetal bovine serum (Sijiqing, Hangzhou, China), 100 U/ml rhIL-2 (ChangSheng, Changchun, China), and 0.1 mM β-mercaptoethanol. K562 cells were cultured in RPMI-1640 containing 10% fetal bovine serum. All cell lines were maintained in our lab.
Isolation and stimulation of primary NK cells
PBMCs were obtained from a healthy human donor and isolated from human peripheral blood by Ficoll gradient centrifugation separation. NK cells were purified by negative selection using a human NK cell isolation kit (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer's instructions. As determined by flow cytometry with anti-CD3 and anti-CD56 antibodies (BD Pharmingen, San Jose, CA, USA), more than 98% of the cells were CD3−CD56+.
Cytotoxicity assay
The cytotoxicity of NK cells was detected using an MTT assay. K562 cells, which were used as the target cells, were placed in a 96-well plate at 1×104 cells per well. NK cells were added at an effector-to-target ratio of 10∶1 or 5∶1. After incubation at 37 °C in 5% CO2 for 6 h, 20 µl (5 mg/ml) of MTT was added, and the cells were incubated for another 4 h. The absorbance at 570/630 nm was determined using a microplate autoreader (Bio-Rad, Hercules, CA, USA). The percentage of cytotoxicity was calculated using the following formula: specific lysis percent (%)=1−(ODE+T−ODE)/ODT×100% (ODE+T: OD value of the effector cell and target cell group; ODE: OD value of the effector cell group; ODT: OD value of the target cell group).
NK cell degranulation assay
A degranulation assay was performed as previously described.10 Briefly, NK cells were stimulated with K562 cells at an effector-to-target ratio of 10∶1, and CD107a monoclonal antibody (mAb) and monensin (6 µg/ml) were added directly into the medium 1 h later. After another 3 h of incubation at 37 °C and 5% CO2, the cells were harvested, washed with phosphate-buffered saline and stained with anti-CD56 mAb (NK-92 cells)/anti-CD94 mAb (NKL cells). Next, flow cytometric analysis was performed. Cells stimulated with PMA (2.5 µg/ml) and ionomycin (0.5 µg/ml) were used as a positive control, and non-stimulated cells were used as a negative control.
Antibodies
Phycoerythrin (PE)-labeled mouse anti-human CD314 (NKG2D), PE-labeled anti-human NKp46, PE-labeled mouse IgG1κ, PE-cy5-labeled anti-human CD54, PE-cy5-labeled mouse IgG1κ isotype control, Alexa Fluor 488-labeled mouse anti-human CD56, Alexa Fluor 488-labeled mouse IgG1κ isotype control, CD3-PE-cy5, PE-cy5 mouse IgG1κ isotype control (BD Pharmingen), PE-conjugated anti-human NKG2A, PE-labeled mouse IgG2a isotype control (R&D Systems, Minneapolis MN, USA), PE-conjugated anti-human TRAIL and PE-conjugated anti-human Fas ligand (CD178) (eBioscience, San Diego, CA, USA) were used for phenotype analyses. PE-labeled mouse anti-human CD107a (BD Pharmingen) was used for the NK cell degranulation assay. PE-conjugated mouse anti-human perforin and PE-conjugated mouse IgG2bκ isotype control were used to detect intracellular proteins. Mouse β-actin mAb, goat anti-mouse immunoglobulin mAb/HRP, rabbit polyclonal antibody DAP10 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), PI3K p85 rabbit mAb, phospho-PI3K p85(Tyr458)/p55(Tyr199) rabbit Ab, ERK1/2 rabbit mAb, phospho-ERK1/2(Thr202/Tyr204) rabbit mAb, PLCγ2 antibody, phospho-PLCγ2(Tyr759) antibody, Myc-tag mAb and anti-rabbit IgG HRP-linked antibody (Cell Signaling Technology, Irvine, CA, USA) were used for western blot analysis. The anti-HMBOX1 monoclonal antibody used for western blot analysis was prepared by our lab.5 Monoclonal mouse anti-human NKG2D mAb (R&D Systems) was used to stimulate NK cells.
Flow cytometry
For the phenotype assay, cells were harvested, washed with phosphate-buffered saline, stained with antibodies and incubated for 30 min at 4 °C. After another wash, flow cytometric analysis was performed using FACSCalibur system (BD Biosciences, San Jose, CA, USA). For intracellular staining, NK cells were stimulated with K562 cells for 1 h, and then brefeldin A (10 µg/ml) and monensin (6 µg/ml) were added. Cells were harvested 2 h later for intracellular staining of perforin. FITC-labeled anti-CD94 was used to specifically label NKL cells. Data were analyzed with WinMDI 2.9.
RNA isolation and real-time PCR
Total cellular RNA was extracted using TRIzol (Invitrogen, Carlsbad, CA, USA). cDNA was synthesized using M-MLV reverse transcriptase (Invitrogen). Real-time PCR was performed using a TransStart SYBR qPCR Kit (TransGen, Ltd, Beijing, China) and a MyiQ thermocycler (Bio-Rad). The relative expression levels were calculated using the algorithm. The primers used are listed in Table 1.
Table 1. Primer sequences used in the real-time PCR assays.
| Genes | Primer sequences | Product size (bp) (ref.) |
|---|---|---|
| GAPDH | F: GAAGGTGAAGGTCGGAGT | 155 |
| R: CATGGGTGGAATCATATTGGAA | ||
| HMBOX1 | F: AACCCTGGCGCTACACTAAG | 140 |
| R: TCCTTTCTCCAGGTAAATCGAC | ||
| Perforin | F: AGTACAGCTTCAGCACTGACA | 175 |
| R: ATGAAGTGGGTGCCGTAGTT | ||
| Granzyme A | F: TCAGGTTGATTGATGTGGGACAG | 163 34 |
| R: GACCATGTAGGGTCTTGAATGAGGA | ||
| Granzyme B | F: GCGGTGGCTTCCTGATACAAG | 82 34 |
| R: CCCCCAAGGTGACATTTATGG | ||
| Granzyme H | F: TGGCGGCATCCTAGTGAGAA | 81 34 |
| R: GCCCCCAAGGTGACATTTATG | ||
| Granzyme K | F: ATCAACACATTTCATCTGGGCTTC | 197 34 |
| R: AAACGTGATGTCCGCCATACTG | ||
| Granzyme M | F: GGACACCCGCATGTGTAACAAC | 187 34 |
| R: GATGTCAGTGCAGACCCTGGAG |
HMBOX1 shRNA plasmid construction and transfection
The lentivirus-derived pGCSIL-HMBOX1 RNAi system was constructed by Shanghai GeneChem Co., Ltd (Shanghai, China). The target sequence was GCCTTGCACTACCAATCAA. NKL cells were infected with the lentivirus or the control virus (i.e., the lentivirus containing a negative control sequence, TTCTCCGAACGTGTCACGT, which is not complementary to any human gene) at 100 IU/ml and harvested 48–96 h later for functional analysis.
NK cell activation and western blot analysis
NK cells were incubated with 10 µg/ml anti-NKG2D mAb (R&D Systems) for 40 min on ice and then washed with ice-cold phosphate-buffered saline. These NK cells were suspended in complete medium and incubated at 37 °C for 10 min. The total protein of the NK cells was isolated, and the phosphorylation levels of PI3K (p85 and p55), ERK1/2 and PLCγ2 were detected using western blotting.
Statistical analysis
Data are presented as mean±s.e.m. of three independent experiments. Statistical significance was determined using Student's t-test (one-tail) (*P<0.05; **P<0.01).
Results
HMBOX1 is highly expressed in resting human primary NK cells
To investigate the function of HMBOX1 in NK cells, we determined the expression level of HMBOX1 in human primary NK cells. The resting NK cells were enriched and purified (98% pure) from human PBMCs, and the HMBOX1 mRNA level was measured by real-time PCR. As shown in Figure 1a, the expression level of HMBOX1 in resting human primary NK cells was significantly higher than that in human PBMCs and non-NK cells. This high expression of HMBOX1 implied that this protein has a special status in the regulation of NK cell function. Because NK cell lines are expedient cell models for the investigation of NK cell-associated genes, we also analyzed the expression levels of HMBOX1 in human NK cell lines such as NK-92 and NKL. As shown in Figure 1b, both of these cell lines expressed the HMBOX1 gene at a level similar to that in PBMCs, indicating that the expression of HMBOX1 in primary NK cells is much higher than that in cultured NK cell lines. Because NK-92 and NKL cells are routinely cultured in a IL-2-containing medium and possess the characteristics of activated NK cells,11 these results indicate that the expression level of HMBOX1 in human NK cells is dependent on the status of cell activation, i.e., HMBOX1 is highly expressed in resting primary NK cells, while only a low level of HMBOX1 is detected in activated NK cells. We also compared the expression of HMBOX1 in NK cell lines (NK-92 and NKL) and in primary NK cells cultured in IL-2 (100 U) for 6 h. As shown in Figure 1c, stimulation with IL-2 clearly reduced the expression of HMBOX1 in primary NK cells and made it closer to that in NK cell lines. Additionally, we observed a slight increase in HMBOX1 expression in NK-92 cells that were starved of IL-2 for 24 h (Figure 1d). These results further suggest that HMBOX1 is associated with NK cell activation, which may affect the functions of NK cells.
Figure 1.
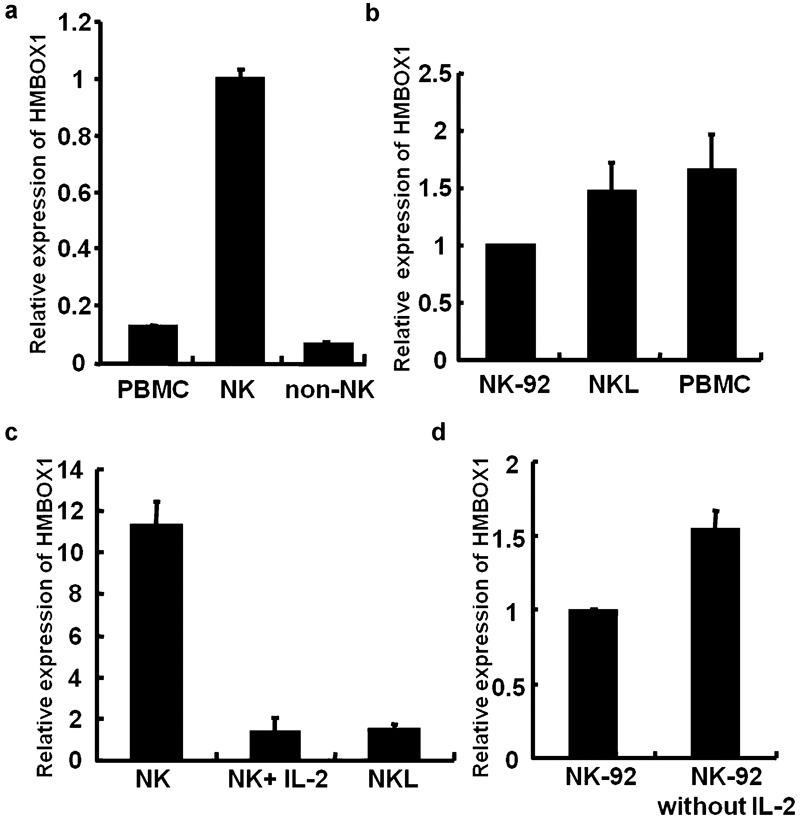
Expression of HMBOX1 in primary NK cells and NK cell lines. (a) HMBOX1 is expressed in primary NK cells isolated from PBMCs. Primary NK cells were separated from PBMCs by negative selection as described in the section on ‘Materials and methods', and the purity of the NK cells was determined by FACS. Total RNA from PBMCs and primary NK cells was extracted, and the mRNA expression level of HMBOX1 was measured using real-time PCR. (b) HMBOX1 is expressed in several NK cell lines. Total RNA from NK-92 cells, NKL cells and PBMCs isolated from human peripheral blood were extracted, and the mRNA expression levels of the HMBOX1 gene were measured using real-time PCR. (c) IL-2 decreased the expression of HMBOX1 in primary NK cells. Primary NK cells were stimulated with IL-2 (100 U/ml) for 6 h. (d) The expression of HMBOX1 increased in NK cells without IL-2. NK-92 cells were cultured in medium with or without IL-2 for 24 h and then harvested for mRNA analysis. Results are expressed as mean±s.e.m. of three independent experiments. NK, natural killer; NKL, natural killer lymphoma; PBMC, peripheral blood mononuclear cell.
HMBOX1 inhibited NK cell cytolytic activity
To investigate whether HMBOX1 is involved in the regulation of NK cell functions, we constructed a HMBOX1 overexpression vector, pcDNA3.1-HMBOX1-Myc/his, and then electroporated it into the human NK cell lines NK-92 and NKL. After selection with G418, the stable transfectants NK-92-vector, NK-92-HMBOX1, NKL-vector and NKL-HMBOX1 were obtained. The effect of HMBOX1 on NK cytotoxicity against tumor cells was examined using these HMBOX1-modified stable NK cell lines. K562, human leukemia cells, were used as the target cells. The results showed that the cytotoxicities of both NK-92 and NKL cells were inhibited by approximately 15%–30% upon overexpression of HMBOX1 compared with controls (Figure 2a). At the same time, the expression levels of CD107a, a marker for the cytotoxic activity of NK cells, were also significantly reduced in HMBOX1-overexpressing NK cells compared with that in control cells in response to stimulation with K562 cells (Figure 2b). These results indicate that HMBOX1 could suppress NK cell cytotoxicity. Subsequently, we knocked down HMBOX1 expression in NKL-HMBOX1 cells with lentivirus-delivered shRNA, and we observed a recovery of the NK cell' cytolytic activity (Figure 2c). The upregulation of cytolytic function was also observed when HMBOX1 was knocked down in the parent NKL cells (Figure 2d). These results indicate that HMBOX1 negatively regulates the cytolytic function of NK cells.
Figure 2.
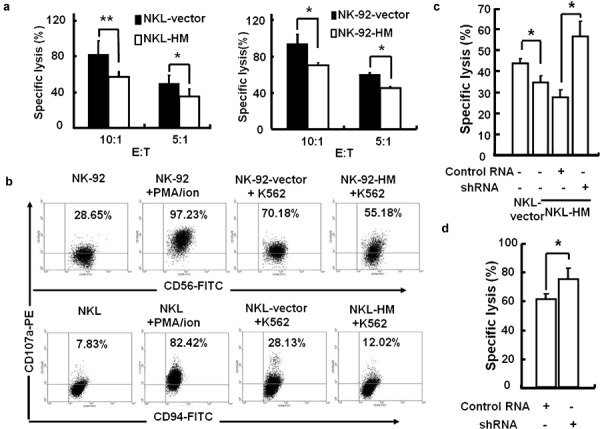
Transgenic expression of HMBOX1 inhibited the cellular cytotoxicity of human NK cells. (a) Overexpression of HMBOX1 decreased the cytotoxicity of NK-92 and NKL cells against K562 leukemia cells. NK cells were incubated with K562 cells at a ratio of 10∶1 or 5∶1 for 6 h, and the cytotoxicity was analyzed using the MTT assay. (b) Degranulation was decreased in NK-92 and NKL cells overexpressing HMBOX1. NK cells were stimulated with K562 cells, and the expression of the lysosomal marker CD107a was analyzed by flow cytometry as described in the section on ‘Materials and methods'. (c, d). RNA interference of HMBOX1 expression increased the cytolysis of NK cells. HMBOX1-overexpressing NKL cells (NKL-HMBOX1) (c) or wild-type NKL (d) cells were treated with HMBOX1 shRNA or control RNA. The specific cytolysis of NK cells against K562 cells was tested at 96 h after treatment. Results are expressed as mean±s.e.m. of three independent experiments. *P<0.05; **P<0.01. NK, natural killer; NKL, natural killer lymphoma.
Production of anti-tumor cytolytic granules by NK cells was inhibited by HMBOX1 overexpression
The cytotoxicity of NK cells is always accompanied with the production of cytolytic granules, such as perforin and granzyme family proteins (GZM A, B, M, H and K), which are the key mediators and effectors of NK cell cytotoxicity.12, 13 Therefore, we determined whether HMBOX1 overexpression could influence the expression of these molecules. As shown in Figure 3a, the mRNA levels of perforin, GZM K and GZM H were clearly reduced in NKL-HMBOX1 cells, whereas the others showed no significant changes. At the same time, the protein expression levels of intracellular perforin in NKL-vector and NKL-HMBOX1 cells stimulated with K562 target cells were analyzed by flow cytometry, and we observed an obvious decrease in the perforin level (Figure 3b). These results demonstrate that HMBOX1 suppressed NK cell cytolytic activity and reduced the production of cytolytic granules by human NK cells.
Figure 3.
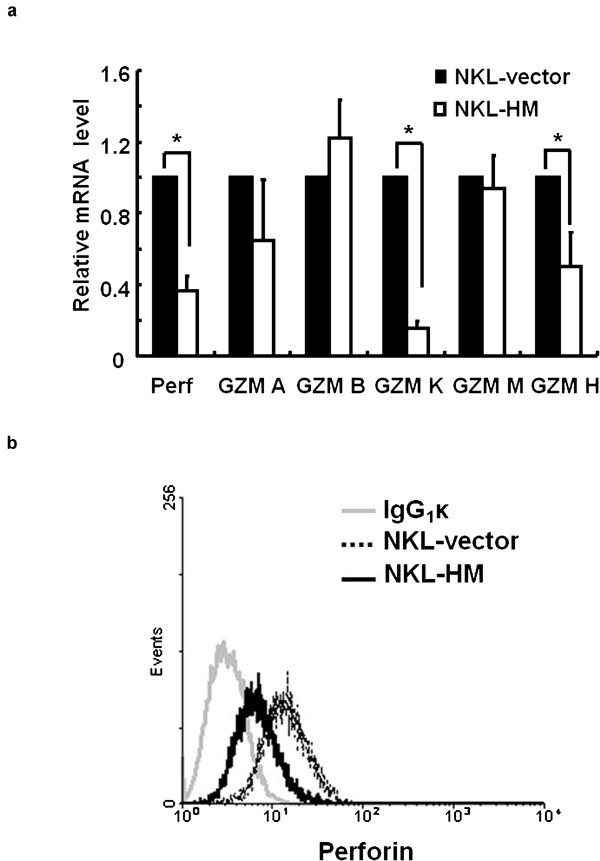
HMBOX1 inhibited the production of cytolytic granules by NKL cells. (a) The mRNA levels of perforin and granzyme family proteins (GZM A, B, M, K and H) were analyzed using real-time PCR. Data are expressed as mean±s.e.m. of three independent experiments. *P<0.05. (b) HMBOX1 inhibited the expression of perforin. NKL cells were stimulated with K562 cells, and intracellular levels of perforin were analyzed by FACS. NKL, natural killer lymphoma.
HMBOX1 negatively regulates NK cell functions via NKG2D/DAP10 signaling pathway
NK cell function is strictly regulated by a balance between positive and negative signals provided by a diverse array of activating and inhibitory receptors, among which NKG2D and NKG2A are two critical players.14 We performed a screen to investigate whether HMBOX1 overexpression could impair the expression balance between NKG2D and NKG2A, as well as the expression of molecules associated with the cytotoxicity of NK cells, including NKp46, TRAIL, Fas ligand and CD54. We found that the expression of NKG2D on the NKL-HMBOX1 cell surface decreased significantly compared with that of control cells, while no significant change in NKG2A was observed (Figure 4a). Because there are many factors that may influence the expression of NKG2D on the NK cell surface, the total amount of NKG2D (both surface and intracellular) expressed by NK cells was detected to exclude other potential influential factors. Coinciding with the expression of NKG2D on the cell surface (Figure 4a), a decrease in the total NKG2D level was also observed in NKL-HMBOX1 cells (Figure 4b). No significant changes in the expression levels of NKp46, TRAIL, Fas ligand or CD54 were detected by flow cytometry (Figure 4c). To further confirm the negative regulatory effects of HMBOX1 on NKG2D expression, the HMBOX1 gene was knocked down in both NKL-HMBOX1 cells (Figure 4d and e) and the parent NKL cells (Figure 4f and g). The knockdown of HMBOX1 expression resulted in a significant elevation in the level of NKG2D on NK cells.
Figure 4.

HMBOX1 negatively regulated NKG2D expression. (a) Flow cytometric analysis of NKG2D and NKG2A on NKL cells. (b) The surface and total NKG2D (surface and intracellular) levels expressed by NKL and NKL-HMBOX1 cells were analyzed by FACS. The MFI of NKG2D was normalized to the NKL-vector group. Data are expressed as mean±s.e.m. of three independent experiments. *P<0.05. (c) Flow cytometric analysis of other NK receptors on NKL cells, including CD54, NKp46, Fas ligand and TRAIL. (d–g). RNA interference of HMBOX1 expression increased NKG2D expression. HMBOX1-overexpressing NKL cells (NKL-HMBOX1) (d, e) or wild-type NKL cells (f, g) were treated with HMBOX1 shRNA or control RNA. At 72 h after treatment, the levels of NKG2D were analyzed by FACS (d, f, histogram map; e and g, statistics of relative NKG2D expression level. The MFI of NKG2D was normalized to the NKL-vector/control group). Data are expressed as mean±s.e.m. of three independent experiments. *P<0.05. NK, natural killer; NKL, natural killer lymphoma.
Because NKG2D expression and signal transmission rely on the adapter molecules DAP10 in humans,15 which is also considered important for NKG2D cell surface expression,16 we analyzed the influence of HMBOX1 on the expression of DAP10. As shown in Figure 5a, in accordance with NKG2D, the expression of DAP10 was negatively correlated with the expression of HMBOX1. These findings indicate that HMBOX1 regulates the expression of both NKG2D and DAP10.
Figure 5.
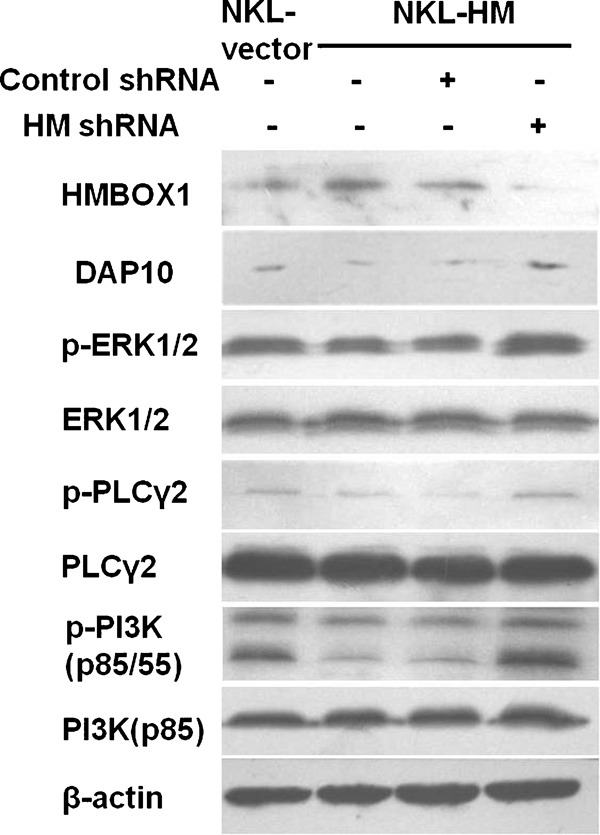
HMBOX1 suppressed NK cell function via negatively regulating the NKG2D/DAP10 signaling pathways. NKL-vector and NKL-HM cells were treated with shRNA or control RNA for 96 h, and then the expression levels of HMBOX1 and DAP10 were detected by western blotting. NK cells treated as above were stimulated with anti-NKG2D mAb as described in the section on ‘Materials and methods', and then the levels of phosphorylated PI3K, ERK1/2 and PLCγ2 were detected by western blotting. mAb, monoclonal antibody; NK, natural killer; NKL, natural killer lymphoma.
The results above imply that HMBOX1 may suppress NK cell cytolytic function by negatively regulating the NKG2D/DAP10 signaling pathway. Several proteins in this signaling pathway are generally acknowledged to play important roles in granule release and the cytotoxicity of NK cells after the formation of NKG2D/DAP10 complexes; these proteins include PI3K, ERK1/2, Grb2/vav1 and PLCγ2.17, 18 To further determine whether HMBOX1 can impair NKG2D-mediated signaling transduction, four groups of NKL cells (NKL-vector, NKL-HM, NKL-HM-negative control and NKL-HM-shRNA) were activated by cross-linking with an anti-NKG2D mAb for 10 min. The expression and phosphorylation levels of PI3K (p85 and p55), ERK1/2 and PLCγ2 were then assayed by western blotting. As shown in Figure 5a, the phosphorylated levels of PI3K, ERK1/2 and PLCγ2 were obviously decreased in HMBOX1-overexpressing NKL cells and could be recovered by the use of HMBOX1 RNA interference. Thus, the inactivation of the NKG2D/DAP10 signaling pathway may mediate the negative regulation NK cell functions by HMBOX1.
Discussion
The expression of HMBOX1 has been reported in many human tissues, such as the cerebrum, intestines, stomach, liver, pancreas, lungs, thyroid gland, cardiac muscle, testis and prostate.1 Here, we observed a high expression level of HMBOX1 in resting primary NK cells. To explore the role of HMBOX1 in the regulation of NK cell functions, we investigated the effect of expression of HMBOX1 on NK cell cytolytic activity. As presented in the results (Figure 2), the cytotoxicity of NK cells against tumor cells was significantly inhibited by HMBOX1 overexpression, and the inhibition was reversed by knockdown of HMBOX1 in NK cells. Because the key effector of NK cell cytolytic functions is the degranulation of cytolytic granules,13 we subsequently detected the expression and production of perforin and granzyme family proteins (GZM A, B, M, H and K), and we observed the suppressive effect of HMBOX1 on perforin and granzyme expression. The above evidence indicates that HMBOX1 functions as a novel negative modulator of NK cell cytotoxicity.
The functions of NK cells are tightly regulated by signals derived from multiple activating and inhibitory receptors.19, 20 There are many activating receptors on NK cells, such as NKG2D, NKp46, NKp30, NKp44, CD16 and 2B4, among which NKG2D plays an extremely important role in the cytolytic activity of NK cells.19 It has been reported that the activation of signaling in NKL cells related to killing K562 tumor cells mainly originates from NKG2D, while this signaling in NK-92 cells originates from both NKG2D and NKp30.21, 22 In the present study, we investigated the changes in the above NK cell receptors and found that NKG2D was the only receptor that underwent obvious changes in expression. The expression levels of NKG2D exhibited a negative correlation with HMBOX1 expression. Furthermore, the downstream signaling pathways of NKG2D in NK cells were analyzed. We found that the expression of NKG2D and its adaptor protein DAP10, as well as the activation of related downstream signaling proteins (PI3K, ERK1/2 and PLCγ2) were reduced significantly by the overexpression of HMBOX1, and knockdown of HMBOX1 by RNA interference clearly reversed the negative regulatory functions of HMBOX1. However, the expression of HMBOX1 did not significantly affect NKp46, TRAIL, Fas ligand or CD54 (Figure 4c). Therefore, HMBOX1 negatively regulated NK cell functions by repressing NKG2D/DAP10 expression and the activation of the associated signaling pathway.
A variety of factors have been shown to influence the expression of NKG2D. Cytokines, such as IL-2,23 IL-12,24 IL-1525 and IFN-α,26 can promote NKG2D expression; IL-21,27 TGF-β28 and IFN-γ29 have a repressive effect. In addition, the expression of NKG2D adapter proteins DAP10 or DAP1230 and chronic exposure to soluble or membrane-bound NKG2D ligands such as major histocompatibility complex class I-related chain A (MICA) could also influence the expression of NKG2D on NK cells.31 However, the transcriptional factor that influences NKG2D expression remains unknown. Additionally, we could not define the direct relationship between HMBOX1 and NKG2D or DAP10 based on our present results. Therefore, the molecular regulation mechanism is worth exploring further.
Many transcription factors are involved in the development and maturation of NK cells.8, 9 In addition to controlling the development and maturation of NK cells, these transcription factors also participate in the regulation of the expression of cytolytic effector molecules. HMBOX1 belongs to a family of transcription factors, many members of which, such as HOXA and HOXB, regulate the development of hematopoietic stem cells.32 It has been reported that another member of the homeobox family, Distal-less, regulates the development and maturation of NK cells.33 In our study, HMBOX1 showed its ability to regulate the cytotoxicity of NK cells by negatively controlling the production of cytolytic molecules, but whether HMBOX1 participates in the development of NK cells requires further investigation.
In summary, we observed a higher level of expression of HMBOX1 in primary NK cells than in non-NK cells. In addition, HMBOX1 negatively regulates the cytotoxicity of NK cells by suppressing the NKG2D/DAP10 signaling pathway. These findings suggest that HMBOX1 may be a novel negative regulator of NK cell functions.
Acknowledgments
This work was supported by the Natural Science Foundation of China (30901307 and 30671901) and the Ministry of Science and Technology of China (2007AA021000, 2007AA021109 and 2006CB504303).
References
- Chen S, Saiyin H, Zeng X, Xi J, Liu X, Li X, et al. Isolation and functional analysis of human HMBOX1, a homeobox containing protein with transcriptional repressor activity. Cytogenet Genome Res. 2006;114:131–136. doi: 10.1159/000093328. [DOI] [PubMed] [Google Scholar]
- Holland PW, Booth HA, Bruford EA. Classification and nomenclature of all human homeobox genes. BMC Biol. 2007;5:47. doi: 10.1186/1741-7007-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramovich C, Humphries RK. Hox regulation of normal and leukemic hematopoietic stem cells. Curr Opin Hematol. 2005;12:210–216. doi: 10.1097/01.moh.0000160737.52349.aa. [DOI] [PubMed] [Google Scholar]
- Zhang M, Chen S, Li Q, Ling Y, Zhang J, Yu L. Characterization of a novel human HMBOX1 splicing variant lacking the homeodomain and with attenuated transcription repressor activity. Mol Biol Rep. 2010;37:2767–2772. doi: 10.1007/s11033-009-9815-9. [DOI] [PubMed] [Google Scholar]
- Dai J, Wu L, Zhang C, Zheng X, Tian Z, Zhang J. Recombinant expression of a novel human transcriptional repressor HMBOX1 and preparation of anti-HMBOX1 monoclonal antibody. Cell Mol Immunol. 2009;6:261–268. doi: 10.1038/cmi.2009.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L, Zhao HL, Sun CH, Zhao BX, Zhao J, Zhang SL, et al. Role of Hmbox1 in endothelial differentiation of bone-marrow stromal cells by a small molecule. ACS Chemical Biology. 2010;5:1035–43. doi: 10.1021/cb100153r. [DOI] [PubMed] [Google Scholar]
- Raulet DH. Interplay of natural killer cells and their receptors with the adaptive immune response. Nat Immunol. 2004;5:996–1002. doi: 10.1038/ni1114. [DOI] [PubMed] [Google Scholar]
- Di Santo JP. Natural killer cell developmental pathways: a question of balance. Annu Rev Immunol. 2006;24:257–286. doi: 10.1146/annurev.immunol.24.021605.090700. [DOI] [PubMed] [Google Scholar]
- Glimcher LH, Townsend MJ, Sullivan BM, Lord GM. Recent developments in the transcriptional regulation of cytolytic effector cells. Nat Rev Immunol. 2004;4:900–911. doi: 10.1038/nri1490. [DOI] [PubMed] [Google Scholar]
- Alter G, Malenfant JM, Altfeld M. CD107a as a functional marker for the identification of natural killer cell activity. J Immunol Methods. 2004;294:15–22. doi: 10.1016/j.jim.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Gong JH, Maki G, Klingemann HG. Characterization of a human cell line (NK-92) with phenotypical and functional characteristics of activated natural killer cells. Leukemia. 1994;8:652–658. [PubMed] [Google Scholar]
- Pardo J, Balkow S, Anel A, Simon MM. Granzymes are essential for natural killer cell-mediated and perf-facilitated tumor control. Eur J Immunol. 2002;32:2881–2887. doi: 10.1002/1521-4141(2002010)32:10<2881::AID-IMMU2881>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Fehniger TA, Cai SF, Cao X, Bredemeyer AJ, Presti RM, French AR, et al. Acquisition of murine NK cell cytotoxicity requires the translation of a pre-existing pool of granzyme B and perforin mRNAs. Immunity. 2007;26:798–811. doi: 10.1016/j.immuni.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Raulet DH, Guerra N. Oncogenic stress sensed by the immune system: role of natural killer cell receptors. Nat Rev Immunol. 2009;9:568–580. doi: 10.1038/nri2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess SJ, Marusina AI, Pathmanathan I, Borrego F, Coligan JE. IL-21 down-regulates NKG2D/DAP10 expression on human NK and CD8+ T cells. J Immunol. 2006;176:1490–1497. doi: 10.4049/jimmunol.176.3.1490. [DOI] [PubMed] [Google Scholar]
- Wu J, Song Y, Bakker AB, Bauer S, Spies T, Lanier LL, et al. An activating immunoreceptor complex formed by NKG2D and DAP10. Science. 1999;285:730–732. doi: 10.1126/science.285.5428.730. [DOI] [PubMed] [Google Scholar]
- Upshaw JL, Leibson PJ. NKG2D-mediated activation of cytotoxic lymphocytes: unique signaling pathways and distinct functional outcomes. Semin Immunol. 2006;18:167–175. doi: 10.1016/j.smim.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Awasthi A, Samarakoon A, Dai X, Wen R, Wang D, Malarkannan S. Deletion of PI3K-p85alpha gene impairs lineage commitment, terminal maturation, cytokine generation and cytotoxicity of NK cells. Genes Immun. 2008;9:522–535. doi: 10.1038/gene.2008.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9:503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R, DeStephan CM, Madge LA, May MJ, Orange JS. NKp30 ligation induces rapid activation of the canonical NF-kappaB pathway in NK cells. J Immunol. 2007;179:7385–7396. doi: 10.4049/jimmunol.179.11.7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Trivedi PP, Ge B, Krzewski K, Strominger JL. Many NK cell receptors activate ERK2 and JNK1 to trigger microtubule organizing center and granule polarization and cytotoxicity. Proc Natl Acad Sci USA. 2007;104:6329–6334. doi: 10.1073/pnas.0611655104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maasho K, Opoku-Anane J, Marusina AI, Coligan JE, Borrego F. NKG2D is a costimulatory receptor for human naive CD8+ T cells. J Immunol. 2005;174:4480–4484. doi: 10.4049/jimmunol.174.8.4480. [DOI] [PubMed] [Google Scholar]
- Zhang C, Zhang J, Niu J, Zhou Z, Zhang J, Tian Z. Interleukin-12 improves cytotoxicity of natural killer cells via upregulated expression of NKG2D. Hum Immunol. 2008;69:490–500. doi: 10.1016/j.humimm.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Zhang C, Zhang J, Niu J, Zhang J, Tian Z. Interleukin-15 improves cytotoxicity of natural killer cells via up-regulating NKG2D and cytotoxic effector molecule expression as well as STAT1 and ERK1/2 phosphorylation. Cytokine. 2008;42:128–136. doi: 10.1016/j.cyto.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Zhang C, Zhang J, Sun R, Feng J, Wei H, Tian Z. Opposing effect of IFNgamma and IFNalpha on expression of NKG2 receptors: negative regulation of IFNgamma on NK cells. Int Immunopharmacol. 2005;5:1057–1067. doi: 10.1016/j.intimp.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Burgess SJ, Marusina AI, Pathmanathan I, Borrego F, Coligan JE. IL-21 down-regulates NKG2D/DAP10 expression on human NK and CD8+ T cells. J Immunol. 2006;176:1490–1497. doi: 10.4049/jimmunol.176.3.1490. [DOI] [PubMed] [Google Scholar]
- Castriconi R, Cantoni C, Della CM, Vitale M, Marcenaro E, Conte R, et al. Transforming growth factor beta 1 inhibits expression of NKp30 and NKG2D receptors: consequences for the NK-mediated killing of dendritic cells. Proc Natl Acad Sci USA. 2003;100:4120–4125. doi: 10.1073/pnas.0730640100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Tian ZG, Zhang J, Feng JB, Zhang JH, Xu XQ. The negative regulatory effect of IFN-gamma on cognitive function of human natural killer cells. Zhonghua Zhong Liu Za Zhi. 2004;26:324–327. [PubMed] [Google Scholar]
- Karimi M, Cao TM, Baker JA, Verneris MR, Soares L, Negrin RS. Silencing human NKG2D, DAP10, and DAP12 reduces cytotoxicity of activated CD8+ T cells and NK cells. J Immunol. 2005;175:7819–7828. doi: 10.4049/jimmunol.175.12.7819. [DOI] [PubMed] [Google Scholar]
- Burgess SJ, Maasho K, Masilamani M, Narayanan S, Borrego F, Coligan JE. The NKG2D receptor: immunobiology and clinical implications. Immunol Res. 2008;40:18–34. doi: 10.1007/s12026-007-0060-9. [DOI] [PubMed] [Google Scholar]
- Lawrence HJ, Sauvageau G, Humphries RK, Largman C. The role of HOX homeobox genes in normal and leukemic hematopoiesis. Stem Cells. 1996;14:281–291. doi: 10.1002/stem.140281. [DOI] [PubMed] [Google Scholar]
- Sunwoo JB, Kim S, Yang L, Naik T, Higuchi DA, Rubenstein JL, et al. Distal-less homeobox transcription factors regulate development and maturation of natural killer cells. Proc Natl Acad Sci USA. 2008;105:10877–10882. doi: 10.1073/pnas.0805205105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Zhang J, Tian Z. Comparison in the effects of IL-2, IL-12, IL-15 and IFNalpha on gene regulation of granzymes of human NK cell line NK-92. Int Immunopharmacol. 2008;8:989–996. doi: 10.1016/j.intimp.2008.03.001. [DOI] [PubMed] [Google Scholar]


