Abstract
MicroRNAs (miRNAs) are a novel class of small, non-coding RNAs that play a significant role in both inflammatory and cardiovascular diseases. Immune cells, especially T helper (Th) cells, are critical in the development of atherosclerosis and the onset of acute coronary syndrome (ACS). To assess whether inflammation-related miRNAs (such as miR-155, 146a, 21, 125a-5p, 125b, 31) are involved in the imbalance of Th cell subsets in patients with ACS, we measured the expression of related miRNAs in patients with acute myocardial infarction (AMI), unstable angina (UA), stable angina (SA) and chest pain syndrome (CPS); analyzed the relationship between miRNA expression and the frequency of Th cell subsets; and observed the co-expression of miR-155 and IL-17A in peripheral blood mononuclear cells (PBMCs) of patients with ACS. The results showed that the expression of miR-155 in the PBMCs of patients with ACS was decreased by approximately 60%, while the expression of both miR-21 and miR-146a was increased by approximately twofold. The expression patterns of miRNAs in plasma correlated with those in PBMCs, except for miR-21, which was increased by approximately sixfold in the AMI group and showed no significant difference between the UA group and the CPS group. We also found that the expression of miR-155 inversely correlated with the frequency of Th17 cells (r=−0.896, P<0.01) and that miR-155 was co-expressed with IL-17A in patients with ACS. In conclusion, our study revealed the expression patterns of inflammation-related miRNAs in patients with ACS and found that miR-155 may be associated with Th17 cell differentiation.
Keywords: acute coronary syndrome, microRNA-155, Th17
Introduction
MicroRNAs (miRNAs, miRs) are a novel class of 19- to 23-nucleotide, untranslated, small RNAs that post-transcriptionally regulate 30% of the genes in eukaryotic organisms.1 To date, about 1000 miRNAs have been found in humans. Many miRNAs have been shown to contribute to various physiological and pathological conditions, including some cardiovascular diseases.2 Using the conditional deletion of Dicer or Argonaute 2, two key enzymes that control the production of mature miRNAs, the importance of miRNAs in the development of the immune system and the regulation of immune responses was discovered.3, 4 Several miRNAs have been found to be involved in the activation, differentiation and function of immune cells by affecting key transcripts.5 The aberrant expression of miRNAs in some inflammatory diseases has also been investigated. For example, in rheumatoid arthritis, both miR-146a and miR-155 were upregulated.6 In psoriasis, miR-21 and miR-146a were upregulated, while miR-125b was downregulated.7
Atherosclerosis is a chronic inflammatory disease that is regulated by immune cells, especially T helper (Th) cells. The imbalance of both Th1/Th2 cells and Th17/CD4+CD25+Foxp3+ regulatory T (Treg) cells plays a critical role in the pathogenesis of atherosclerosis.8, 9 According to previous studies, Th1 cells induce plaque rupture and the onset of acute coronary syndrome (ACS), and Th17 cells contribute to vascular and systemic inflammation and may modulate plaque stability, whereas Treg cells play the opposite role.10, 11 However, the underlying molecular mechanisms of the imbalance of Th cells in atherosclerosis have not been thoroughly clarified. Recently, miRNAs (such as miR-155 and miR-146a) were determined to be key molecules in modulating the differentiation of Th cells.12, 13
Based on the relationships among miRNAs, inflammation and atherosclerosis, we hypothesized that inflammation-related miRNAs (such as miR-155, 146a, 21, 125a-5p, 125b and 31) might play a role in the development of atherosclerosis and the onset of ACS. Moreover, the expression of these miRNAs and their relationship with Th cell subsets in patients with acute myocardial infarction (AMI), unstable angina (UA) and stable angina (SA) remain to be determined. In the present study, we aimed to detect the relative expression levels of inflammation-related miRNAs in patients with AMI, UA and SA compared to expression levels in patients with chest pain syndrome (CPS) as well as to determine the relationship between the frequencies of Th cell subsets and the expression of these miRNAs.
Materials and methods
Study population
The study was guided by the Declaration of Helsinki and its amendments and also conformed to the approved institutional guidelines. Informed consent was obtained from each patient.
All 62 subjects—including patients with AMI,14 UA,15 SA15 and CPS14—enrolled in this study were patients in the Cardiology Department of Union Hospital, Wuhan, China, from October 2009 to October 2010. The AMI group was composed of 12 men and 4 women with a mean age of 58±7 years. Inclusion criteria were as follows: chest pain lasting >30 min within 24 h before enrollment and myocardial infarction confirmed by significant rise of creatine kinase MB and troponin I levels. The UA group was composed of 12 men and 3 women with a mean age of 60±9 years. Inclusion criteria were as follows: chest pain with an accelerating pattern or prolonged duration (>20 min) or recurrent episodes at rest or with minimal effort with documented transient ST-segment elevation or ST-segment depression of 0.1 mV in at least two contiguous electrocardiograph leads. The SA group was composed of 11 men and 4 women with a mean age of 59±10 years. Inclusion criteria were as follows: chest discomfort, including spreading to the left shoulder and arm, which could be relieved with nitroglycerin or rest. These patients also had a downsloping or horizontal ST-segment depression <1 mm in an exercise test. The CPS group was composed of 12 men and 4 women with a mean age of 60±10 years. Inclusion criteria were as follows: chest pain with no electrocardiographic changes and coronary spasm or coronary artery stenosis when acetylcholine was injected into coronary artery during arteriography. Exclusion criteria were as follows: patients who were treated with anti-inflammatory drugs such as aspirin or dexamethasone or patients who had collagen disease, thromboembolism, renal failure, advanced liver disease, malignant disease or any other inflammatory disease (such as psoriasis, septicemia or rheumatoid arthritis).
Sample preparation
One 8-ml blood sample from each subject was collected in EDTA-treated tubes. Plasma was collected after centrifugation, and then peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll-Hypaque density gradient centrifugation within 1 h.
RNA isolation
Total RNA was extracted from both freshly isolated PBMCs and plasma by TRIzol and TRIzol LS (Invitrogen, Carlsbad, CA, USA), respectively, according to the manufacturer's protocols, respectively. The concentration and purity of the RNA samples were measured by a spectrophotometer, and samples were used only if the ratio of the absorbance at 260 and 280 nm (A260/280) was between 1.8 and 2.0. RNA samples were diluted with DEPC-treated water to ensure a constant starting concentration of 0.05 µg/µl for each reverse transcription (RT) reaction.
RT and quantitative real-time polymerase chain reaction (qRT-PCR)
RT was carried out according to the manual for the Reverse Transcription System, which was purchased from Promega (#A3500). The reagent mixes were incubated at 16 °C for 30 min, 42 °C for 42 min and 95 °C for 5 min. cDNA samples were stored at −20 °C until used for qRT-PCR. qRT-PCR was performed using the SYBR Green PCR Master Mix Kit (#4309155; Applied Biosystems, Foster City, CA, USA) and was processed on a 7300HT analyzer (Applied Biosystems) in 48-well PCR plates. In the first cycle, reagent mixes were incubated at 95 °C for 10 min, then 95 °C for 15 s and finally 60 °C for 60 s. This cycle was repeated 40 times. miRNA levels were normalized to U6 small nuclear RNA levels. RT and PCR specific primers for U6 and miRNAs were synthesized using Invitrogen (Table 1).
Table 1. Sequences of RT and PCR primers for U6 and miRNAs.
| miRNA | RT primer | PCR primer |
|---|---|---|
| U6 | CGCTTCACGAATTTGCGTGTCAT | F: GCTTCGGCAGCACATATACTAAAATR: CGCTTCACGAATTTGCGTGTCAT |
| miR-21 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCAACA | F: GCCGCTAGCTTATCAGACTGATGTR: GTGCAGGGTCCGAGGT |
| miR-31 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGCTAT | F: GGAGAGGCAAGATGCTGGCAR: GTGCAGGGTCCGAGGT |
| miR-125a-5p | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCACAGG | F: TCCCTGAGACCCTTTAACCTGTGR: GGGTCCGAGGTATTCGCACT |
| miR-125b | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCACAA | F: CGTCCCTGAGACCCTAACTTGTR: GTGCAGGGTCCGAGGT |
| miR-155 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCCCTA | F: GGAGGTTAATGCTAATCGTGATAGR: GTGCAGGGTCCGAGGT |
| miR-146a | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAACCC | F: AGCAGTGAGAACTGAATTCCATR: GTGCAGGGTCCGAGGT |
Abbreviations: PCR, polymerase chain reaction; RT, reverse transcription.
Flow cytometric analysis
Freshly isolated PBMCs (2×106) from each subject were stimulated for 4 h with 25 ng/ml of phorbol myristate acetate and 1 µg/ml of ionomycin in the presence of 2 µM of monensin. Cells were incubated at 37 °C under a 5% CO2 environment in a 24-well culture plate. The cells were then stained with fluorescein isothiocyanate-labeled anti-human CD4, and some were then Allophycocyanin-labeled anti-human CD25 monoclonal antibodies (moAbs) for 30 min at 4 °C. After washing, the cells were fixed and permeabilized according to the manufacturer's protocol (ebioscience, Foxp3 Staining Buffer Set, #005523). For intracellular staining, cells were continually stained with phycoerythrin-labeled anti-human Foxp3 or phycoerythrin-Cy7-labeled anti-human IL-17A and PerCP/Cy5.5-labeled anti-human interferon (IFN)-γ and APC-labeled anti-human IL-4 moAbs. Finally, cells were resuspended in 200 µl of washing buffer and analyzed using flow cytometry with FACSCalibur (BD Biosciences, San Jose, CA, USA). Isotype-matched antibody controls were used to confirm antibody specificity. All reagents used for flow cytometric analysis were purchased from eBioscience (San Diego, CA, USA).
Dual miR-155 fluorescent in situ hybridization and IL-17A immunohistochemical detection
First, PBMCs from patients with AMI and UA were stimulated for 3 days with 5 µg/ml of anti-CD3 and 2 µg/ml of anti-CD28 moAbs (eBioscience) in the presence of 20 ng/ml of IL-6 and 2 ng/ml of TGF-β1 (PeproTech, Rocky Hill, NJ, USA), as well as 10 µg/ml of anti-IFN-γ and anti-IL-4 moAbs (eBioscience). Then, cells were stimulated with phorbol myristate acetate, ionomycin and monensin for 4 h (as described above) and seeded on polyane-covered slides at 5×108 cells/ml.
For miR-155 fluorescent in situ hybridization (FISH), slides were fixed with 4% paraformaldehyde for 15 min at room temperature, digested with proteinase K for 5 min at 37 °C, washed with phosphate-buffered saline (PBS) twice and hydrated in ethanol solutions (sequentially hydrated in 70%, 96% and 99.9% ethanol). After air-drying the slides, hybridization mix with double-DIG LNA miR-155 probe, positive control (U6) or negative control (scramble-miR) (Exiqon, Woburn, MA, USA) was applied and allowed to hybridize for 1 h at 54 °C in the wet chamber. Low-stringency post-hybridization washes were performed with ×2 SSC, ×1 SSC and ×0.2 SSC, and slides were washed with PBS twice and incubated in PBS-BB (PBS containing 1.0% bovine serum albumin, 0.2% powdered skim milk and 0.3% Triton X-100) for 15 min. Finally, slides were incubated for at least 14 h at 4 °C with sheep anti-digoxigenin moAbs (Roche Molecular; Indianapolis, IN, USA) diluted 1∶1000 in PBS-BB, followed by PBS washes and TSA Plus Direct-Green deposition according to the manufacturer's protocol (PerkinElmer, Waltham, MA, USA).
For IL-17A immunohistochemistry, sections were blocked in PBS-BB for 30 min. The sections were incubated in the dark with mouse anti-human IL-17A moAbs (eBioscience) for 12–24 h at 4 °C and then washed with PBS. Next, TSA Plus Direct-Cyanine 3 was applied according to the manufacturer's protocol (PerkinElmer Life and Analytical Sciences, Shelton, CT, USA). After washing with PBS, sections were incubated with DAPI for 5 min and covered with coverglass. Finally, sections were analyzed using laser scanning confocal microscopy.
Statistical analysis
Values were expressed as mean±s.d. To determine the overall differences between different independent groups, one-way ANOVA was used. The statistical significance was evaluated using the Bonferroni/Dunn post hoc test. Spearman correlation analysis was used to test for correlation between the variables. In all cases, a probability (P) value of <0.05 was considered statistically significant and P<0.01 was considered highly statistically significant.
Results
Clinical characteristics of patients
There were no significant differences in age, gender or risk factors except for hyperlipidemia among patients with AMI, UA, SA and CPS. However, the number of diseased vessels and the incidence rate of hyperlipidemia in patients with AMI, UA and SA were significantly higher than in patients with CPS (Table 2).
Table 2. Clinical characteristics of patients with AMI, UA, SA and CPS.
| Characteristics | AMI (n=16) | UA (n=15) | SA (n=15) | CPS (n=16) |
|---|---|---|---|---|
| Age (years) | 58±7 | 60±9 | 59±10 | 60±10 |
| Sex (male/female) | 12/4 | 12/3 | 11/4 | 12/4 |
| No. of diseased vessels | ||||
| 1.7±1.1* | 2.1±0.7* | 1.3±1.0* | 0.0±0.0 | |
| Risk factors | ||||
| Hypertension, n (%) | 8 (50) | 7 (42) | 8 (53) | 3 (19) |
| Diabetes mellitus, n (%) | 4 (25) | 4 (27) | 3 (20) | 3 (19) |
| Smoking, n (%) | 4 (25) | 4 (27) | 4 (27) | 4 (27) |
| Obesity, n (%) | 5 (31) | 4 (27) | 5 (33) | 2 (13) |
| Hyperlipidemia, n (%) | 10 (63)** | 9 (60)** | 10 (67)** | 3 (19) |
Abbreviations: AMI, acute myocardial infarction; CPS, chest pain syndrome; SA, stable angina; UA, unstable angina.
Values are expressed as mean±s.d. or number.
*P<0.01 vs. CPS;
**P<0.05 vs. CPS.
The altered expression of miRNAs in patients with ACS
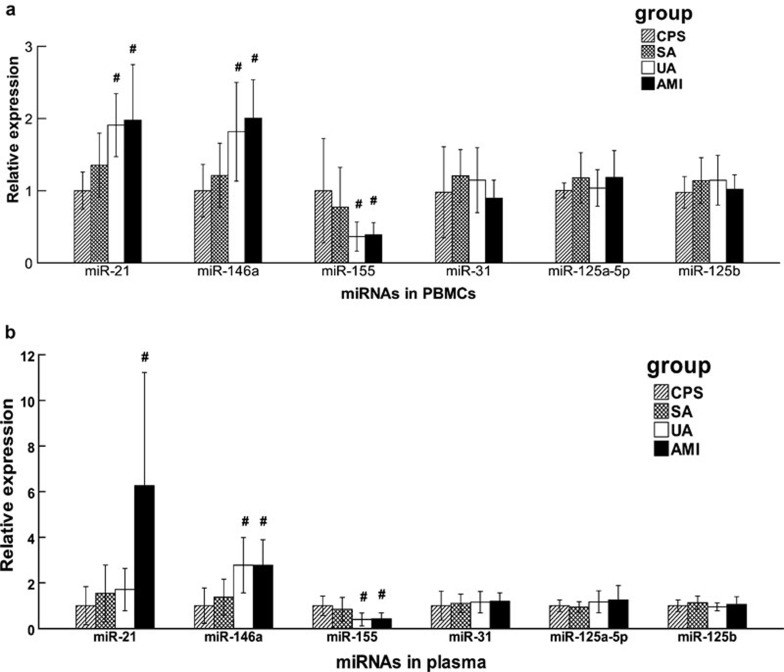
In PBMCs, miR-21 and miR-146a levels were higher and the level of miR-155 was lower in patients with UA and AMI than in patients with CPS, while no statistically significant differences were observed between patients with SA and CPS. Additionally, there were no significant differences in the expression of miR-31,125a-5p and 125b among patients with AMI, UA, SA and CPS (Figure 1a). In plasma, expression patterns of miRNAs were in accordance with the patterns in PBMCs, except for miR-21. miR-21 levels in plasma were remarkably high in patients with AMI, but there was no significant difference between patients with UA and patients with CPS, which was strikingly different from the levels measured in PBMCs (Figure 1b).
Figure 1.
Relative expression of miRNAs in patients with SA, UA and AMI compared with patients with CPS. miRNA levels were measured with qRT-PCR. The expression of miRNAs in PBMCs (a) and in plasma (b) from patients with SA, UA and AMI is shown as relative levels compared with CPS. Data are shown as mean±s.d. *P<0.05 vs. CPS; #P<0.01 vs. CPS. AMI, acute myocardial infarction; CPS, chest pain syndrome; PBMCs, peripheral blood mononuclear cells; qRT-PCR, quantitative real-time polymerase chain reaction; SA, stable angina; UA, unstable angina.
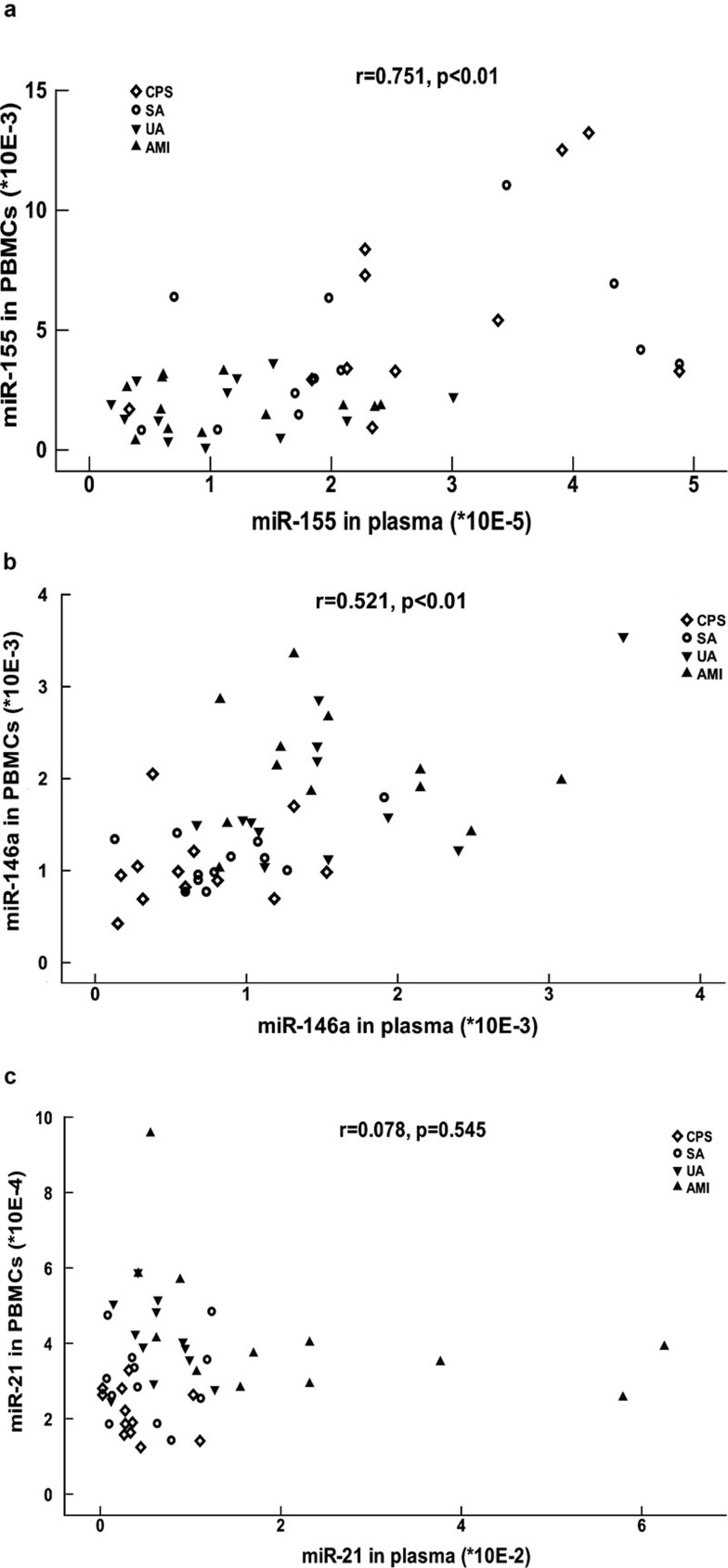
We also analyzed the relationship between the expression of miRNAs in plasma and in PBMCs by Spearman's correlation analysis. This analysis showed that the expression of miR-155 and miR-146a in plasma positively correlated with the expression in PBMCs (Figure 2a and b). However, the expression of miR-21 in PBMCs did not correlate with the expression in plasma (Figure 2c).
Figure 2.
Correlation between the expression of miRNAs in plasma and in PBMCs. miR-155 (a), miR-146a (b) and miR-21 (c) levels in plasma were plotted against their levels in PBMCs. Different shapes were used to distinguish between patients with CPS, SA, UA and AMI. The correlation between miRNAs levels in plasma and PBMCs was calculated using the Spearman correlation analysis, and the r and P values are shown. AMI, acute myocardial infarction; CPS, chest pain syndrome; PBMCs, peripheral blood mononuclear cells; SA, stable angina; UA, unstable angina.
The imbalance of Th cell subsets in patients with ACS
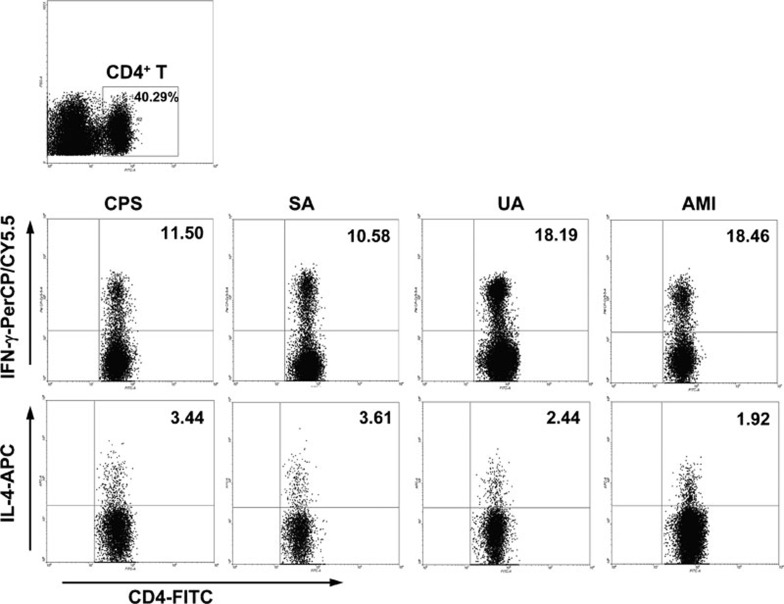
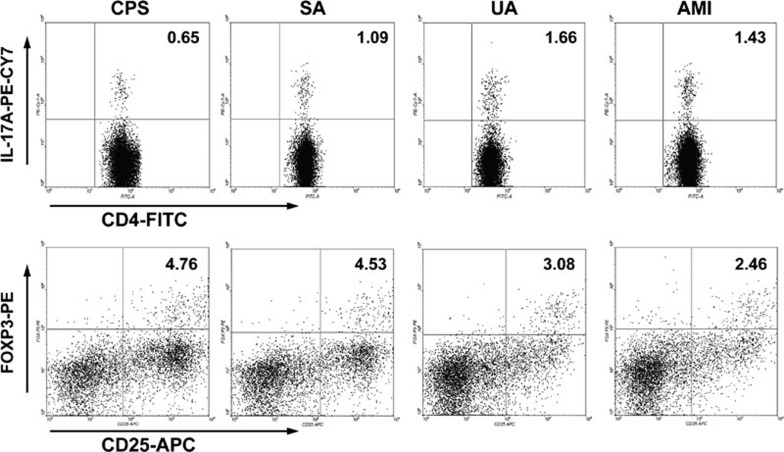
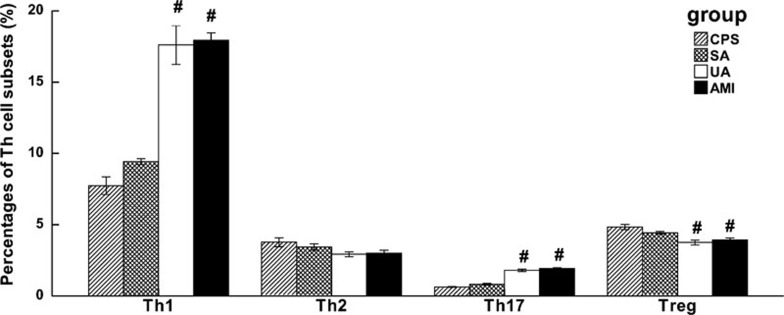
As shown in Figures 3–5, the frequencies of Th1 and Th17 cells were increased in patients with UA and AMI, whereas the frequency of Treg cells was decreased. The frequency of Th2 cells showed no significant difference among different patient groups. Also, there were no significant differences in the frequencies of Th cell subsets (including Th1, Th2, Th17 and Treg cells) between patients with SA and CPS.
Figure 3.
Flow cytometric analysis of the frequencies of Th1 and Th2 cells in the PBMCs of patients with CPS, SA, UA and AMI. First, CD4+ Th cells were gated. Then, Th1 and Th2 cell subsets were gated with CD4+IFN-γ+ and CD4+IL-4+, respectively. Representative FACS plots from a single patient from the CPS, SA, UA and AMI groups are shown, and the percentage of positive cells is shown in each panel. AMI, acute myocardial infarction; CPS, chest pain syndrome; FACS, fluorescence-activated cell sorting; PBMCs, peripheral blood mononuclear cells; SA, stable angina; Th, T helper cells; UA, unstable angina.
Figure 4.
Flow cytometric analysis of the frequencies of Th17 and Treg cells in the PBMCs of patients with CPS, SA, UA and AMI. First, CD4+ Th cells were gated. Then, Th17 and Treg cell subsets were gated with CD4+IL-17+ and CD4+CD25+Foxp3+, respectively. Representative FACS plots from a single patient from the CPS, SA, UA and AMI groups are shown, and the percentage of positive cells is shown in each panel. AMI, acute myocardial infarction; CPS, chest pain syndrome; FACS, fluorescence-activated cell sorting; PBMCs, peripheral blood mononuclear cells; SA, stable angina; Th, T helper cells; Treg, CD4+CD25+Foxp3+ regulatory T cells; UA, unstable angina.
Figure 5.
Collective analysis of the frequencies of Th cell subsets in the PBMCs of patients with CPS, SA, UA and AMI. The frequencies of Th cell subsets (Th1, Th2, Th17 and Treg) in the PBMCs of all patients with CPS, SA, UA and AMI were collectively analyzed, and the results are shown as mean±s.d.. *P<0.05 vs. CPS, #P<0.01 vs. CPS. AMI, acute myocardial infarction; CPS, chest pain syndrome; PBMCs, peripheral blood mononuclear cells; SA, stable angina; Th, T helper cells; Treg, CD4+CD25+Foxp3+ regulatory T cells; UA, unstable angina.
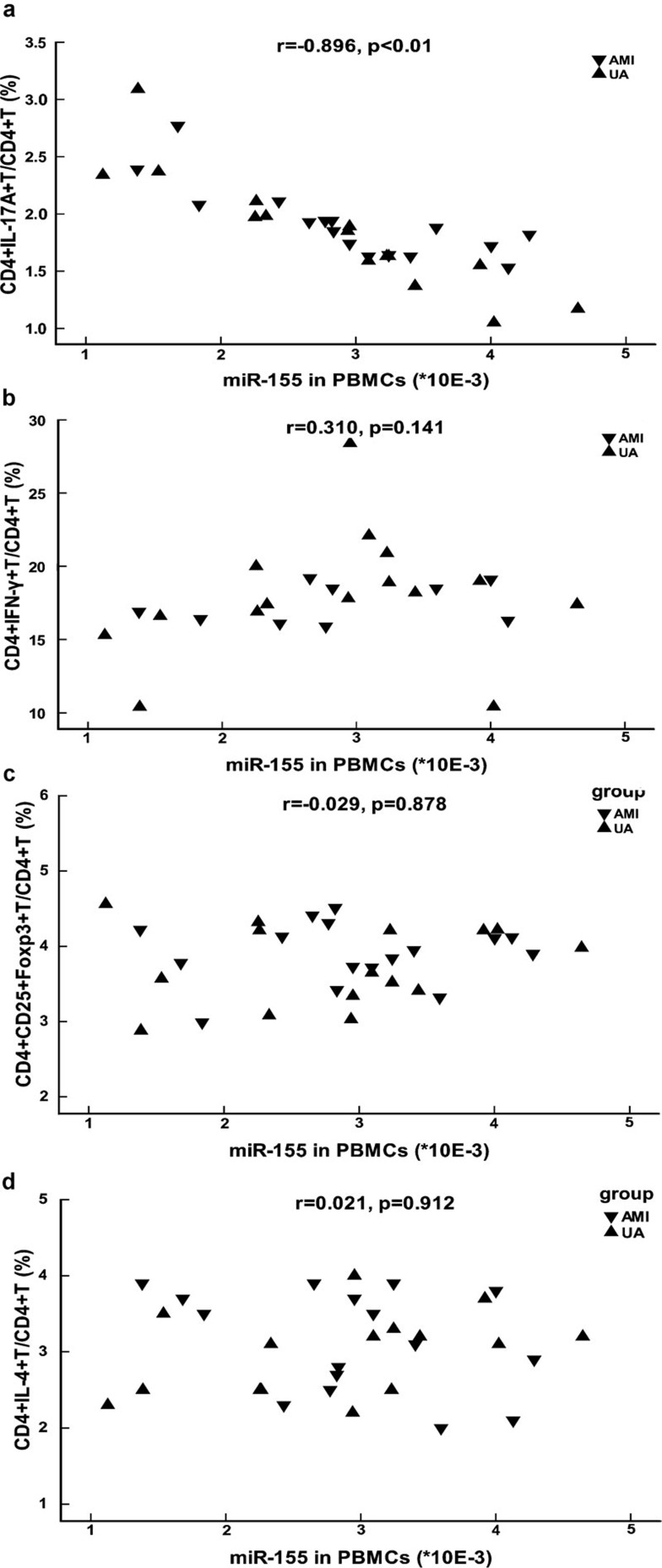
The expression of miR-155 inversely correlated with the frequency of Th17 cells in patients with ACS
To investigate whether differentially expressed miRNAs correlated with the imbalance of Th cell subsets in patients with ACS, we performed Spearman's correlation analysis examining the relationship between the expression of miRNAs and the frequencies of Th cell subsets. The results showed that the expression of miR-155 in PBMCs was highly inversely correlated with the frequency of Th17 cells in patients with AMI and UA (Figure 6a), but did not correlate with the frequency of Th1, Treg or Th2 cells (Figure 6b–d). No significant correlation was found between the expression of other miRNAs and the frequencies of Th cell subsets (data not shown).
Figure 6.
Correlation between the expression of miR-155 in PBMCs and the frequencies of Th cell subsets in patients with ACS. miR-155 levels in the PBMCs of patients with ACS were plotted against the frequencies of Th17 (a), Th1 (b), Treg (c) and Th2 (d) cell subsets. Th17, Th1, Treg and Th2 cells were gated with CD4+IL-17+, CD4+IFN-γ+, CD4+CD25+Foxp3+ and CD4+IL-4+, respectively. Different shapes were used to distinguish between patients with UA and patients with AMI. The correlation between the expression of miR-155 in PBMCs and the frequency of Th cell subsets was calculated by using the Spearman's correlation analysis, and the r and P values are shown in the figure. ACS, acute coronary syndrome; AMI, acute myocardial infarction; PBMCs, peripheral blood mononuclear cells; Th, T helper cells; Treg, CD4+CD25+Foxp3+ regulatory T cells; UA, unstable angina.
The co-expression of miR-155 and IL-17A in PBMCs of patients with ACS
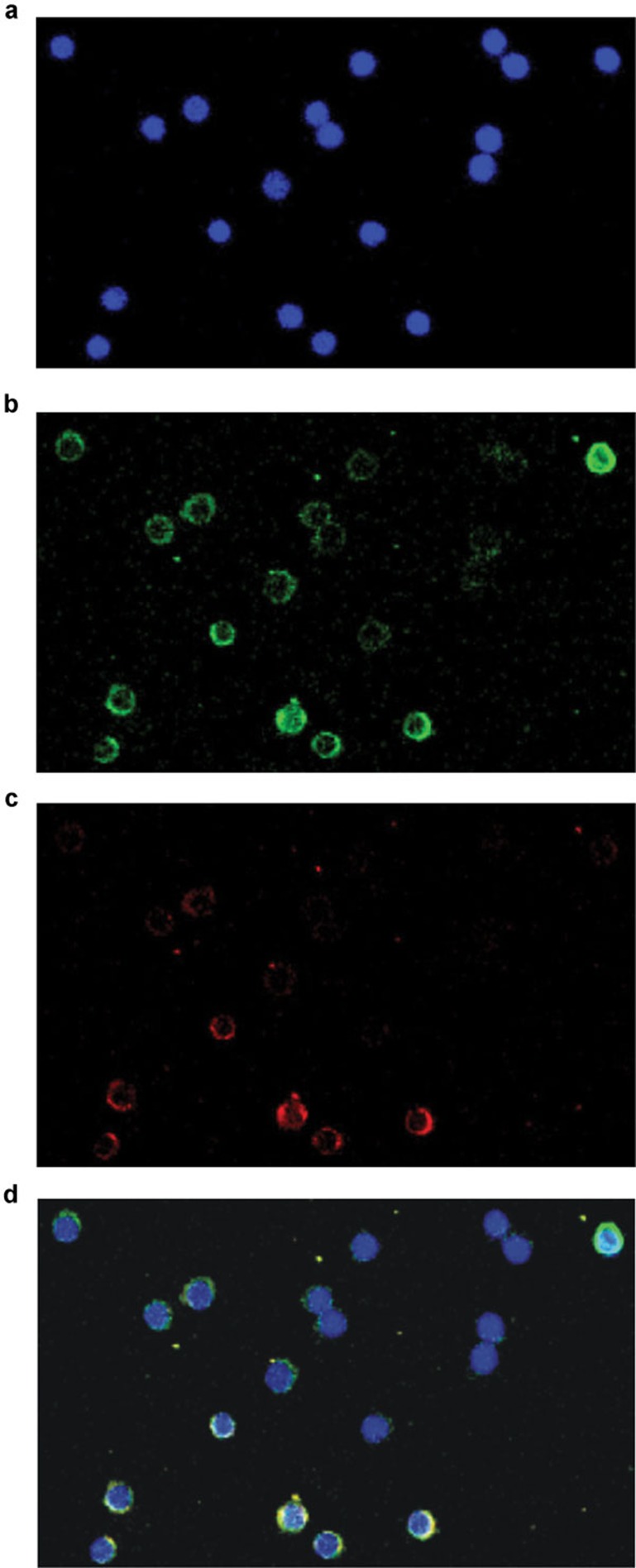
To further examine the relationship between the expression of miR-155 and the differentiation of Th17 cells, we performed sequential miR-155 FISH and IL-17A immunohistochemistry using TSA Plus Direct-Green and TSA Plus Direct-Cyanine 3. Both miR-155 (Figure 7b) and IL-17A (Figure 7c) were observed in a subset of PBMCs in patients with UA and AMI. Virtually all IL-17A-positive cells possessed miR-155 (Figure 7d).
Figure 7.
Co-expression of miR-155 and IL-17A in patients with ACS. The expression of miR-155 and IL-17A in the PBMCs of patients with ACS was detected sequentially. A typical ×40 image of the slides from a single patient is shown under different wavelengths of excitation. First, the nuclei of PBMCs were measured (blue, DAPI) (a). Then, the expression of miR-155 was detected by FISH using TSA Plus Direct-Green (b), and the expression of IL-17A was detected by IHC using TSA Plus Direct-Cyanine 3 (c). Finally, colocalization of the nuclei of PBMCs, the expression of miR-155 and the expression of IL-17A were shown together by overlaying the images (d). n=2 experiments performed in triplicate in both UA and AMI patients. ACS, acute coronary syndrome; AMI, acute myocardial infarction; FISH, fluorescent in situ hybridization; IHC, immunohistochemistry; PBMCs, peripheral blood mononuclear cells; UA, unstable angina.
Discussion
Inflammation contributes to the initiation and progression of atherosclerosis. In recent years, studies have indicated that a variety of miRNAs (miRNA-155, 146a, 21, 125a-5p, 125b and 31) targeted key transcripts of immune cells and were involved in inflammatory diseases. To assess whether these miRNAs were involved in the development of atherosclerosis, we studied the relative expression of these miRNAs in both PBMCs and plasma of patients with AMI, UA, SA and CPS. The results demonstrated that miR-155 was downregulated, while miR-21 and miR-146a were upregulated in PBMCs of patients with UA and AMI. Additionally, the expression patterns of miRNAs in patient plasma were in accordance with the pattern in PBMCs, except for miR-21. miR-21 was remarkably upregulated in the plasma of patients with AMI but showed no important change in patients with UA. Next, we performed a correlation analysis of miRNAs in plasma and miRNAs in PBMCs to determine whether the miRNAs in plasma were derived from the immune system. The results showed that both miR-155 and miR-146a in plasma positively correlated with the expression levels in PBMCs, but miR-21 levels in the plasma did not correlate to levels found in PBMCs. As miR-21 was highly expressed in injured cardiac myocytes as well as activated immune cells,14, 15 we speculated that the upregulation of miR-21 in patients with ACS may be due not only to overactivation of immune cells but also to injury of cardiac myocytes.
Both miR-146a and miR-155 can regulate immune responses, but they have been shown to have a particularly strong effect on inflammatory processes. miR-146a negatively regulates activated macrophages by directly inhibiting interleukin-1 receptor-associated kinase-1 and tumor necrosis factor receptor-associated factor 6, which are key molecules in the Toll-like receptor 4 signaling pathway.16 It also indirectly limits overactivation of Th1 cells by regulating Treg cell function.13 miR-155 is upregulated in activated immune cells17 and modulates immune responses by regulating cell differentiation (such as Th1 and Treg) and cytokine secretion (such as tumor necrosis factor-alpha) and IL-6).18, 19, 20 However, we found that miR-155 was downregulated in patients with ACS, which was consistent with what Fichtlscherer et al.21 found in patients with coronary artery disease. Two mechanisms may explain these findings. First, downregulation of miR-155 could be a feedback mechanism that controls the overactivation of immune cells. miR-155 can be activated by the upstream activation of key transcription factors in traditional signaling pathways, like nuclear factor-κB and c-Jun N-terminal kinase; however, it may be regulated by related factors in other signaling pathways, such as Toll-like receptor, B-cell receptor and T-cell receptor signaling pathways as well as cytokines, such as IL-10.22, 23 Second, the reduction of circulating miR-155 in patients with ACS might be explained by the uptake of miR-155 by atherosclerotic lesions.21
Immune cells, especially Th cells, are key factors that induce the rupture of atheroma. In addition to the observation of altered expression levels of miRNAs, we also confirmed the imbalance of Th1/Th2 and Th17/Treg in patients with ACS. In our study, Th1 and Th17 cells were upregulated, whereas Treg cells were downregulated in patients with UA and AMI, which was in accordance with previous studies.9, 24, 25 Th1 and Th17 cells are pro-inflammatory Th cell subsets that can induce the rupture of atheroma by secreting pro-inflammatory cytokines, such as IFN-γ and IL-17A.26, 27, 28, 29 Th2 cells, which antagonize the effects of Th1 cells, are considered an anti-atherosclerotic Th cell subset. Treg cells suppress inflammatory cells by directly contacting or secreting suppressive cytokines, and they play a protective role in the pathogenesis of atherosclerosis.11 The functional imbalance of Th17/Treg cells was detected by our group in both ACS patients and ApoE−/− mice with atherosclerosis lesions.9, 30 The relationship between the imbalance of Treg/Th17 cells and the progression of atherosclerosis is under investigation by our group.
Several studies investigated the influence of miR-155 on Th cell differentiation. In vitro, bic/miR-155−/− naive T cells were biased toward the Th2 cell subset, with increased production of Th2 cytokines, including IL-4, IL-5 and IL-10, and decreased production of IL-2 and IFN-γ.12, 31 In experimental autoimmune encephalomyelitis, Th1 and Th17 cells were found to be defective in miR-155−/− mice.32 Furthermore, the proportion and the absolute number of Treg cells were also smaller in miR-155-deficient mice.19, 33 In summary, these studies indicated that miR-155 is essential for the differentiation of Th cell subsets. However, different transcripts were found to be targeted by miR-155 in different Th cell subsets, and additional targets still need to be confirmed. Moreover, only certain transcripts were inhibited by miR-155 in specific cellular phases and microenvironments.22 Therefore, in different physiological and pathological conditions, miR-155 might only directly inhibit certain transcripts and could play quite a different role in the differentiation of each Th cell subset. Sma- and Mad-related protein 2 and suppressor of cytokine signaling 1, which are confirmed targets of miR-155,34, 35 may regulate Th17 cell differentiation in the opposite way. Sma- and Mad-related protein 2, which is essential for TGF-β signaling, induces the generation of Th17 cells,36 whereas suppressor of cytokine signaling 1 negatively regulates Th17 cell differentiation by inhibiting the IL-6/STAT3 signaling pathway.37 In our study, the expression of miR-155 negatively correlated with the frequency of Th17 cells, and miR-155 was co-expressed with IL-17A in PBMCs, which suggests that miR-155 might negatively regulate Th17 cells in patients with ACS. O'Connell et al.32 presumed that miR-155 positively regulated Th17 cell differentiation in experimental autoimmune encephalomyelitis. More studies are needed to clarify whether alternative transcripts are inhibited by miR-155 during the pathogenesis of ACS and experimental autoimmune encephalomyelitis. However, as mentioned above, it is also possible that downregulation of miR-155 may be explained by a feedback mechanism to limit overactivated inflammatory responses, including Th17 cell differentiation.
In summary, our study found that miR-155 was obviously downregulated and that miR-21 and miR-146a were upregulated in patients with ACS. The expression of miR-155 closely correlated with the frequency of Th17 cells, and miR-155 was co-expressed with IL-17A in the PBMCs of patients with ACS. Based on previous studies of the relationship between miR-155 expression and Th cell differentiation, our study suggests that miR-155 might be involved in the imbalance of Th cell subsets in the development of atherosclerosis, especially the upregulation of Th17 cells. However, our study involved a limited number of subjects and methods, and the relationship between miR-155 expression and Th cell differentiation in patients with ACS may not be completely resolved. More studies are necessary to clarify the exact relationship between miR-155 expression and Th cell differentiation and the mechanisms of miR-155 modulation of Th cell differentiation in patients with ACS.
Acknowledgments
The work described in this article was supported by the National Natural Science Foundation of China (no. 81000085) and the Doctor and Novo-teacher Foundation of Chinese National Ministry of Education (no. 20090142120049).
References
- Zhang B, Pan X, Wang Q, Cobb GP, Anderson TA. Computational identification of microRNAs and their targets. Comput Biol Chem. 2006;30:395–407. doi: 10.1016/j.compbiolchem.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res. 2008;79:581–588. doi: 10.1093/cvr/cvn156. [DOI] [PubMed] [Google Scholar]
- Cobb BS, Nesterova TB, Thompson E, Hertweck A, O'Connor E, Godwin J, et al. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med. 2005;201:1367–1373. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Carroll D, Mecklenbrauker I, Das PP, Santana A, Koenig U, Enright AJ, et al. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev. 2007;21:1999–2004. doi: 10.1101/gad.1565607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay MA. microRNAs and the immune response. Trends Immunol. 2008;29:343–351. doi: 10.1016/j.it.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Stanczyk J, Pedrioli DM, Brentano F, Sanchez-Pernaute O, Kolling C, Gay RE, et al. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008;58:1001–1009. doi: 10.1002/art.23386. [DOI] [PubMed] [Google Scholar]
- Bostjancic E, Glavac D. Importance of microRNAs in skin morphogenesis and diseases. Acta Dermatovenerol Alp Panonica Adriat. 2008;17:95–102. [PubMed] [Google Scholar]
- Laurat E, Poirier B, Tupin E, Caligiuri G, Hansson GK, Bariéty J, et al. In vivo downregulation of T helper cell 1 immune responses reduces atherogenesis in apolipoprotein E-knockout mice. Circulation. 2001;104:197–202. doi: 10.1161/01.cir.104.2.197. [DOI] [PubMed] [Google Scholar]
- Cheng X, Yu X, Ding YJ, Fu QQ, Xie JJ, Tang TT, et al. The Th17/Treg imbalance in patients with acute coronary syndrome. Clin Immunol. 2008;127:89–97. doi: 10.1016/j.clim.2008.01.009. [DOI] [PubMed] [Google Scholar]
- Madhur MS, Funt SA, Li L, Vinh A, Chen W, Lob HE, et al. Role of interleukin 17 in inflammation, atherosclerosis, and vascular function in apolipoprotein E-deficient mice. Arterioscler Thromb Vasc Biol. 2011;31:01–09. doi: 10.1161/ATVBAHA.111.227629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taleb S, Tedgui A, Mallat Z. Adaptive T cell immune responses and atherogenesis. Curr Opin Pharmacol. 2010;10:197–202. doi: 10.1016/j.coph.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu LF, Boldin MP, Chaudhry A, Lin LL, Taganov KD, Hanada T, et al. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2010;142:914–929. doi: 10.1016/j.cell.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O'Leary JJ, Ruan Q, et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. 2010;11:141–147. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Liu X, Zhang S, Lin Y, Yang J, Zhang C. MicroRNA-21 protects against the H2O2-induced injury on cardiac myocytes via its target gene PDCD4. J Mol Cell Cardiol. 2009;47:5–14. doi: 10.1016/j.yjmcc.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl HF, Fauti T, Ullrich N, Bopp T, Kubach J, Rust W, et al. miR-155 inhibition sensitizes CD4+ Th cells for TREG mediated suppression. PLoS One. 2009;4:e7158. doi: 10.1371/journal.pone.0007158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, Schambach F, DeJong CS, Hammond SM, Reiner SL. Micro-RNA-155 inhibits IFN-gamma signaling in CD4+ T cells. Eur J Immunol. 2010;40:225–231. doi: 10.1002/eji.200939381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu LF, Thai TH, Calado DP, Chaudhry A, Kubo M, Tanaka K, et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RS, Hu GQ, Lin B, Lin ZY, Sun CC. MicroRNA-155 silencing enhances inflammatory response and lipid uptake in oxidized low-density lipoprotein-stimulated human THP-1 macrophages. J Investig Med. 2010;58:961–967. doi: 10.231/JIM.0b013e3181ff46d7. [DOI] [PubMed] [Google Scholar]
- Fichtlscherer S, de Rosa S, Fox H, Schwietz T, Fischer A, Liebetrau C, et al. Circulating MicroRNAs in patients with coronary artery disease. Circ Res. 2010;107:677–684. doi: 10.1161/CIRCRESAHA.109.215566. [DOI] [PubMed] [Google Scholar]
- Tili E, Croce CM, Michaille JJ. miR-155: on the crosstalk between inflammation and cancer. Int Rev Immunol. 2009;28:264–284. doi: 10.1080/08830180903093796. [DOI] [PubMed] [Google Scholar]
- McCoy CE, Sheedy FJ, Qualls JE, Doyle SL, Quinn SR, Murray PJ, et al. IL-10 inhibits miR-155 induction by Toll-like receptors. J Biol Chem. 2010;285:20492–20498. doi: 10.1074/jbc.M110.102111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methe H, Brunner S, Wiegand D, Nabauer M, Koglin J, Edelman ER. Enhanced T-helper-1 lymphocyte activation patterns in acute coronary syndromes. J Am Coll Cardiol. 2005;45:1939–1945. doi: 10.1016/j.jacc.2005.03.040. [DOI] [PubMed] [Google Scholar]
- Mor A, Luboshits G, Planer D, Keren G, George J. Altered status of CD4+CD25+ regulatory T cells in patients with acute coronary syndromes. Eur Heart J. 2006;27:2530–2537. doi: 10.1093/eurheartj/ehl222. [DOI] [PubMed] [Google Scholar]
- Libby P, Sukhova G, Lee RT, Galis ZS. Cytokines regulate vascular functions related to stability of the atherosclerotic plaque. J Cardiovasc Pharmacol. 1995;25:S9–S12. doi: 10.1097/00005344-199500252-00003. [DOI] [PubMed] [Google Scholar]
- Rabbani R, Topol EJ. Strategies to achieve coronary arterial plaque stabilization. Cardiovasc Res. 1999;41:402–417. doi: 10.1016/s0008-6363(98)00279-x. [DOI] [PubMed] [Google Scholar]
- Smith E, Prasad KM, Butcher M, Dobrian A, Kolls JK, Ley K, et al. Blockade of interleukin-17A results in reduced atherosclerosis in apolipoprotein E-deficient mice. Circulation. 2010;121:1746–1755. doi: 10.1161/CIRCULATIONAHA.109.924886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid RE, Rao DA, Zhou J, Lo SF, Ranjbaran H, Gallo A, et al. Interleukin-17 and interferon-gamma are produced concomitantly by human coronary artery-infiltrating T cells and act synergistically on vascular smooth muscle cells. Circulation. 2009;119:1424–1432. doi: 10.1161/CIRCULATIONAHA.108.827618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie JJ, Wang J, Tang TT, Chen J, Gao XL, Yuan J, et al. The Th17/Treg functional imbalance during atherogenesis in ApoE−/− mice. Cytokine. 2010;49:185–193. doi: 10.1016/j.cyto.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, et al. Regulation of the germinal center response by MicroRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- O'Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, et al. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–619. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlhaas S, Garden OA, Scudamore C, Turner M, Okkenhaug K, Vigorito E. Cutting edge: the Foxp3 target miR-155 contributes to the development of regulatory T cells. J Immunol. 2009;182:2578–2582. doi: 10.4049/jimmunol.0803162. [DOI] [PubMed] [Google Scholar]
- Louafi F, Martinez-Nunez RT, Sanchez-Elsner T. Microrna-155 (miR-155) targets SMAD2 and modulates the response of macrophages to transforming growth factor-β (TGF-β) J Biol Chem. 2010;285:41328–41336. doi: 10.1074/jbc.M110.146852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Zhang HW, Lu MH, He XH, Li Y, Gu H, et al. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 2010;70:3119–3127. doi: 10.1158/0008-5472.CAN-09-4250. [DOI] [PubMed] [Google Scholar]
- Martinez GJ, Zhang Z, Reynolds JM, Tanaka S, Chung Y, Liu T, et al. Smad2 positively regulates the generation of Th17 cells. J Biol Chem. 2010;285:29039–29043. doi: 10.1074/jbc.C110.155820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa H, Matsubara K, Qian GS, Jackson P, Groopman JD, Manning JE, et al. SOCS-1, a negative regulator of the JAK/STAT pathway, is silenced by methylation in human hepatocellular carcinoma and shows growth-suppression activity. Nat Genet. 2001;28:29–35. doi: 10.1038/ng0501-29. [DOI] [PubMed] [Google Scholar]