The authors reviewed the excision repair cross-complementation group 1 (ERCC1) expression results from three different laboratories. None of the laboratories’ assays could predict resistance to platinum-based therapies in non-small cell lung cancer with a specificity of greater than 50% and only 4 of 18 tumors examined in round-robin analysis were fully concordant. The authors conclude that commercial laboratories should not offer ERCC1 testing until assays are better standardized and results are more thoroughly validated.
Keywords: ERCC1, Tumor predictive biomarker, Platinum sensitivity, Lung cancer
Abstract
Introduction.
Excision repair cross-complementation group 1 (ERCC1) expression by non-small cell lung cancer (NSCLC) has been reported to predict resistance to platinum-based therapies. On this basis, several commercial laboratories have offered ERCC1 testing to facilitate clinical decision making, but the reliability of such assays has recently been called into question.
Methods.
First, three large commercial laboratories were queried for their cumulative ERCC1 test results in NSCLC patients to compare their independent rates of ERCC1 expression. Second, identical tumor blocks from individual NSCLC patients underwent round-robin analysis to evaluate interlaboratory concordance for ERCC1 expression. Third, a retrospective review of medical records from NSCLC patients identified those who were both highly responsive and resistant to platinum-based chemotherapies. Tumor blocks from these patients were then used in a gold standard analysis to determine individual laboratory sensitivity and specificity for ERCC1 results.
Results.
Significant differences were observed in independent laboratory ERRC1 expression rates (Clarient 70% vs. Genzyme 60% vs. Third Laboratory 44%, p < .0001 for all two-way comparisons). Only 4 of 18 tumors examined in round-robin analysis were fully concordant (κ ≤ 0.222 for all two-way comparisons). In preselected platinum responsive and resistant specimens, none of these three commercially marketed laboratory assays achieved a specificity of greater than 50%.
Conclusion.
The results of commercial laboratory testing for ERCC1 are inconsistent and unreliable. Better validation and postmarketing surveillance should be mandated before tumor biomarker assays are allowed to enter the clinical arena.
Implications for Practice:
Prior reports have suggested that clinical benefit from platinum-based chemotherapy may be predicted by determining tumor expression levels of the excision repair cross-complementation group 1 (ERCC1) enzyme. On this basis, ERCC1 testing has been recognized in consensus guidelines and offered by commercial laboratories. In this study, we compared ERCC1 expression levels on identical tumor specimens, as determined by three different commercial laboratories. We also evaluated each laboratory’s ERCC1 assay for its ability to correctly predict platinum resistance or sensitivity in tumor specimens that were preselected on the basis of clinically observed platinum responsiveness. ERCC1 testing was found to be both highly discordant and uniformly unreliable at all three laboratories. We conclude that these ERCC1 assays should not be used in routine clinical practice or recommended in current practice guidelines. Our experience also demonstrates how postmarketing surveillance may help to ensure the technical reliability and clinical utility of predictive tumor biomarker assays.
Introduction
Platinum-based chemotherapies represent the first-line metastatic treatment of choice [1, 2] and the only effective adjuvant option for most patients with non-small cell lung cancer (NSCLC) [3–5]. Even so, these therapies have modest benefits that must be weighed against significant treatment-related side effects [6]. Predictive markers are much needed to allow selective treatment of the subpopulation of NSCLC patients who are most likely to benefit from platinum therapies while sparing the remainder from treatment-associated morbidity. Excision repair cross-complementation group 1 (ERCC1) is one of the critical proteins involved in the process whereby cells normally repair platinated DNA and circumvent treatment-induced cytotoxicity [7–11]. Consequently, the upregulation of ERCC1 by tumors represents a plausible mechanism for their resistance to platinum-based chemotherapies [11–13].
Numerous prior reports have described an association between the tumor expression of ERCC1 and lack of clinical benefits following platinum-based chemotherapy [14–20]. In the adjuvant setting, Olaussen et al. reported that early-stage NSCLC patients whose tumors were “positive” for high levels of ERCC1 protein expression derived no benefit from platinum-based therapies (hazard ratio for death, 1.14, p = .40), whereas ERCC1 “negative” patients realized important platinum attributable benefit (hazard ratio for death, 0.65, p = .002) [18]. In the metastatic setting, Cobo et al. prospectively randomized NSCLC patients to standard platinum-based chemotherapy (control group) versus ERCC1-directed therapy (experimental group). Patients in the experimental group, who received platinum therapy only if their tumors demonstrated low ERCC1 mRNA expression and nonplatinum chemotherapy otherwise, experienced superior response rates (51% vs. 39%, p = .02) [15].
Based on such reports suggesting clinical utility of ERCC1 testing as well as its recognition in current National Comprehensive Cancer Network guidelines [21], several commercial laboratories have offered ERCC1 testing to assist medical oncologists and their patients when deciding whether to administer platinum-based chemotherapy regimens. However, ERCC1 testing methodology and criteria for high and low results are nonstandardized and may vary considerably between laboratories. In addition, technical problems with ERCC1 assays have become apparent and prior results suggesting clinical utility have not been reproducible [22]. This study was performed to evaluate ERCC1 testing offered by three large commercial laboratories.
Methods
Participating Commercial Laboratories
Clarient, Inc. (Aliso, CA, http://www.clarientinc.com), Genzyme Corporation (Cambridge, MA, http://www.genzyme.com), and “Third Laboratory” (requesting anonymity after learning of study results) agreed to participate in the conduct of this study and confirmed the accuracy of this report. Each of these laboratories offered ERCC1 testing to facilitate NSCLC patient management, and all three assays had been used by physicians at Winthrop-University Hospital (Mineola, NY) prior to the initiation of this study. All three laboratories agreed to perform ERCC1 testing without charge when requested specifically for the purposes of this study. The design and conduct of this protocol were approved by the Winthrop-University Hospital Institutional Review Board as well as by each of the participating commercial laboratories.
Study Design
ERCC1 testing was performed on tumor specimens from three separate cohorts of NSCLC patients. First, we compared the prevalence of “high” versus “low” ERCC1 expression in NSCLC as reported by each independent laboratory. Second, we chose 18 patients treated at Winthrop-University Hospital whose tumors had already been tested for ERRC1 expression at Clarient and arranged for retesting of their same tumor specimens at Genzyme and Third Laboratory.
Finally, we retrospectively reviewed the charts of more than 300 NSCLC patients treated at Winthrop-University Hospital to identify 12 who were “platinum responders” and another 12 who were “platinum nonresponders” to platinum-based chemotherapy administered as first-line treatment for metastatic disease. Platinum responders were required to have achieved a partial response, whereas nonresponders were required to have had progressive disease while receiving systemic platinum-based chemotherapy using either World Health Organization (WHO) [23] or Response Evaluation Criteria in Solid Tumors (RECIST) [24] criteria. In addition, charts were reviewed to confirm the impression of clinical benefit in responders and worsening in nonresponders. Slides were cut from a single pretreatment tumor block for each of these 24 tumors, divided equally, and sent to each of the three commercial laboratories for their independent ERCC1 testing. Specimens were deidentified and coded so that no laboratory knew tumor response status or any other tumor- or patient-related characteristics before reporting its ERCC1 assay results.
ERCC1 Testing Methodologies
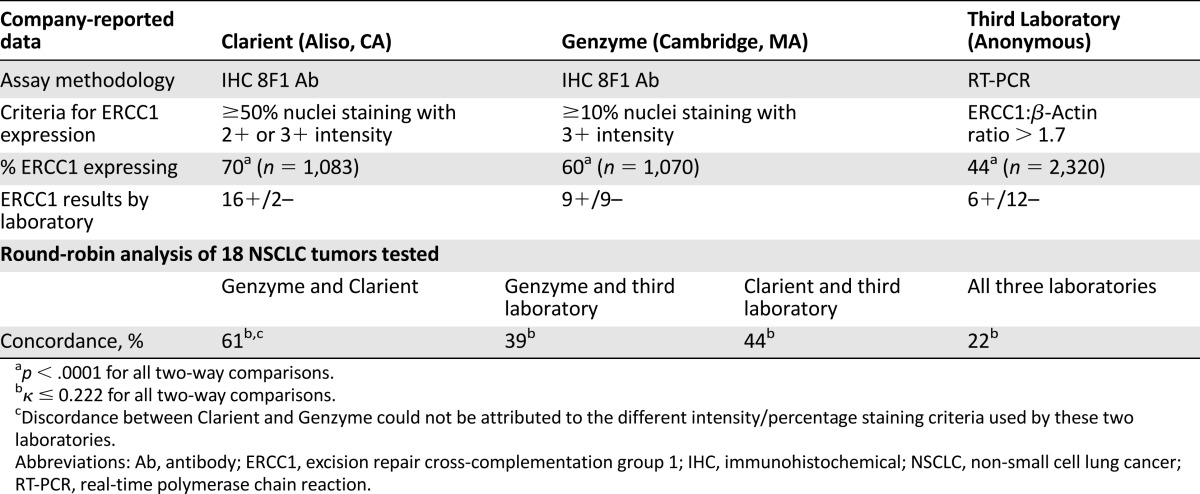
Each of the three laboratories performed ERCC1 testing according to the same protocol and standards as incorporated in its own commercially offered test. Clarient and Genzyme determined ERCC1 protein expression by immunohistochemical (IHC) analysis using the same antibody reagent, designated 8F1, and protocol as reported by Olaussen et al. [18, 25] The proportion (expressed as a percentage) and the intensity (0, 1+, 2+, or 3+) of IHC-positive staining tumor nuclei were determined in identical fashion by both Clarient and Genzyme. However, each laboratory had developed its own criteria for reporting a final test result as “positive” or “negative” for ERCC1 expression. For Genzyme, a positive result required 3+ staining in at least 10% of nuclei. A positive result for Clarient required 2+ or 3+ staining in at least 50% of examined tumor nuclei. Importantly, after recognizing a change in the performance of newer 8F1 antibody lots, Clarient chose to modify its commercial assay by retitering the 8F1 antibody from 1:200 to 1:8000. These modified assay conditions took effect commercially in November 2009 and were applied to only the third cohort of patients in this study.
Third Laboratory determined ERCC1 mRNA gene expression by quantitative real-time polymerase chain reaction (RT-PCR) assay according to a proprietary procedure [26a] as previously reported by Lord et al. [26b] ERCC1 RNA levels were reported as the ratio of ERCC1 gene transcripts to β-actin reference gene transcripts. Ratios above 1:7 were reported as “positive” for ERCC1 expression and lower ratios were reported as “negative.”
We do recognize that because of posttranscriptional and posttranslational regulatory mechanisms, the accurate measurement of ERCC1 mRNA and protein expression for ERCC1 need not be concordant. However, all of these ERCC1 assays ultimately report a clinical result intended to reflect platinum sensitivity or resistance. We chose to compare all three laboratory results on the basis of this clinical predictive outcome measure.
Statistical Analysis
For cohort 1, involving separate patient populations tested at each of the three participating laboratories, descriptive statistics were analyzed by proportion. A two-sample test of proportions was used to compare these independent ERCC1 expression rates. For cohort 2, Cochran’s Q test was used to determine concordance for ERCC1 results in identical patients tested at all three laboratories [27]. McNemer’s test of paired proportions was applied to determine discordance between laboratories. After applying Bonferroni correction for multiple comparisons, none of our results changed from being statistically significant to nonsignificant using p < .05 as a measure of statistical significance. κ statistics were calculated to evaluate interlaboratory concordance. For cohort 3, clinically determined chemotherapy response provided a “gold standard” against which each laboratory’s results were compared using McNemer’s test. Because we were rigorous in our definition of platinum-sensitive and -resistant tumor specimens, our cohort 3 sample size was limited and no a priori power analysis was performed. As such, this gold standard analysis should be considered exploratory. Sensitivity, specificity, positive predictive value, and negative predictive value were calculated using exact binomial proportions [28]. Calculations were performed using SAS 9.3 (SAS Institute, Cary, NC, http://www/sas/com) and Stata/SE 10.0 for Windows (StataCorp, College Station, TX, http://www.stata.com).
Results
Aggregate ERCC1 Expression at Three Different Laboratories
Considerable variation in the prevalence of ERCC1 expression was noted at Clarient (70%, n = 1,083), Genzyme (60%, n = 1,070), and Third Laboratory (44%, n = 2,320). These differences in aggregate ERCC1 expression rates, as shown in Table 1, were highly significant (p < .0001 for all two-way comparisons) and motivated us to compare ERCC1 expression results that these three laboratories found on identical tumor specimens.
Table 1.
Commercial laboratory testing of ERCC1 in non-small cell lung cancer
ERCC1 Expression in Identical Tumor Blocks at Three Different Laboratories
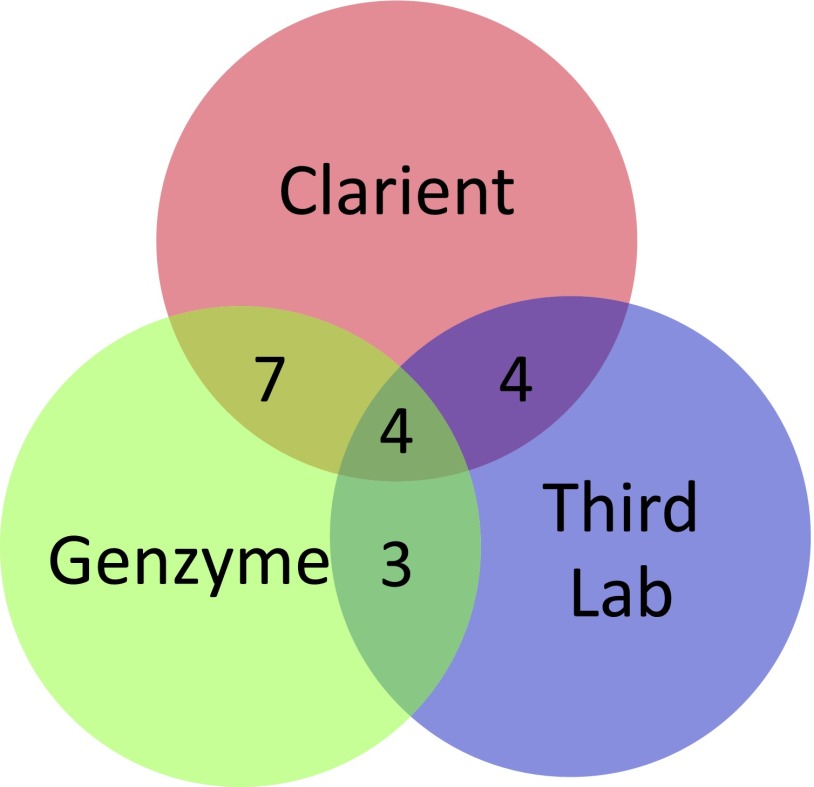
Tumors from 18 NSCLC patients were tested for ERCC1 expression at all three commercial laboratories. Results are delineated in Table 1 and interlaboratory concordance is depicted in Figure 1. As shown, all three laboratories reported the same “positive” or “negative” result in only 4 of these 18 specimens. Clarient and Genzyme, the two laboratories using identical IHC testing reagents and methodology, reported concordant results in 11 of the 18 tumors (61%) tested. All 7 of the discordant cases were reported as positive by Clarient and negative by Genzyme. The results reported by these two laboratories were shown to correlate poorly (κ: 0.222; 95% CI: 0.017–0.55) and the difference between ERCC1 expression reported by Clarient (89%) and Genzyme (50%) on identical tumor blocks was highly significant (p = .015). Discordance could not be attributed to the different intensity/percentage staining criteria used by these two laboratories in determining ERCC1 expression. This was determined by repeating comparisons after applying Clarient criteria to Genzyme specimens and alternatively after applying Genzyme criteria to Clarient specimens. Third Laboratory’s ERCC1 results were concordant with those of Genzyme in only 39% (κ: –0.222; 95% CI: −0.646–0.202) and with Clarient in only 44% (κ: 0.117; 95% CI: −0.053–0.289) of cases.
Figure 1.
Laboratory concordance for excision repair cross-complementation group 1 (ERCC1) expression results. Venn diagram with overlap indicating concordant ERCC1 results. As depicted, all three laboratories were concordant for “positive” or “negative” ERCC1 result in only 4 of 18 identical tumor blocks examined in round-robin analysis.
ERCC1 Expression in Selected Platinum Responders and Nonresponders Tested at Three Laboratories
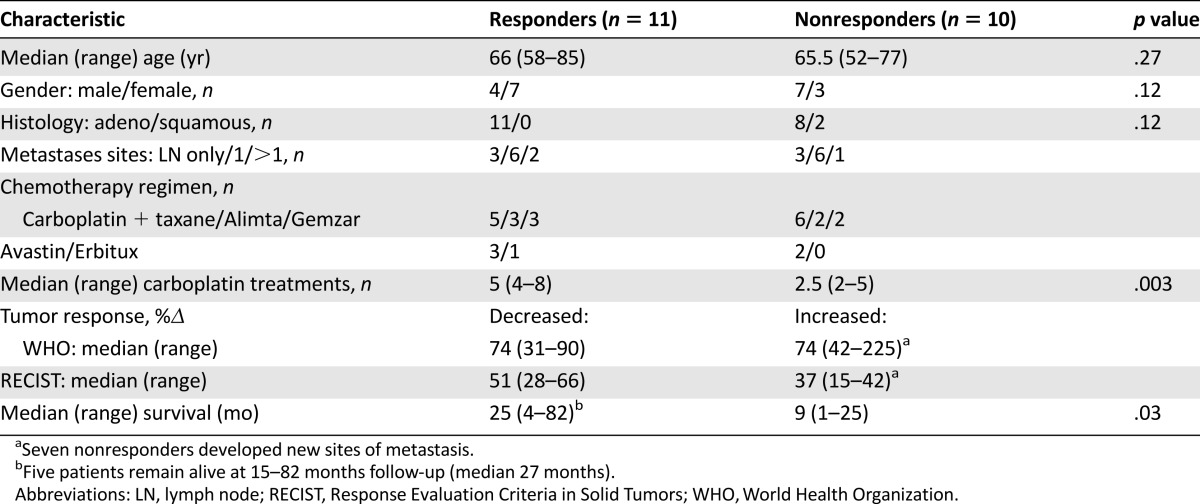
Specimens from 2 of the 12 selected platinum nonresponders and 1 of the 12 responders were deemed inadequate for ERCC1 determination by all three laboratories. Patient and tumor characteristics for each of the remaining 11 responders and 10 nonresponders are shown in Table 2. Responders and nonresponders were similar in age, gender, and the carboplatin-doublet chemotherapy regimens they received. Histology was adenocarcinoma in all patients, except for 2 nonresponders with squamous cell carcinoma. Per selection criteria for this study, all responders had partial response defined by WHO (n = 2), RECIST (n = 2), or both (n = 7) criteria, whereas all nonresponders had progression of disease defined by both WHO and RECIST criteria, except for 1 patient who had just 15% increase in maximum tumor diameter (RECIST criteria), but 42% increase in product of bidimensional target lesion measurements (WHO criteria) [24, 29]. Not surprisingly, responders received more cycles of carboplatin than nonresponders (median, 5 vs. 2.5 cycles, p = .003) and lived longer (median 25 vs. 9 months, p = .03). Only 3 nonresponders survived longer than 9 months, and all achieved major objective treatment response to second-line nonplatinum therapies.
Table 2.
Characteristics of platinum responders and nonresponders
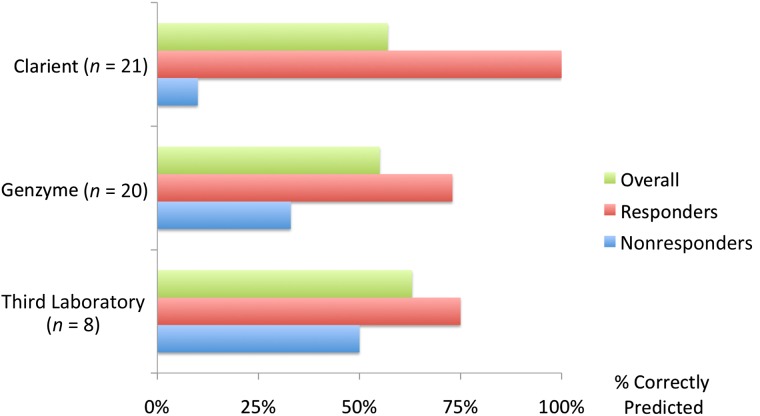
The ability of commercial laboratory ERCC1 assays to correctly identify our platinum-responsive and -nonresponsive patients is shown in Figure 2 and Table 3. Results were reported by Clarient for 21 tumors and by Genzyme for 20, whereas Third Laboratory was able to provide results for only 8, reporting “insufficient material” in the remaining 13 cases. This was somewhat unexpected because one of the perceived advantages of Third Laboratory’s RT-PCR assay was that it might provide an enhanced ability to derive results from smaller specimens not amenable to IHC testing.
Figure 2.
Accuracy of excision repair cross-complementation group 1 results in predefined platinum responders and nonresponders.
Table 3.
Sensitivity, specificity, and positive and negative predictive values for excision repair cross-complementation group 1 predicting platinum resistance at three commercial laboratories
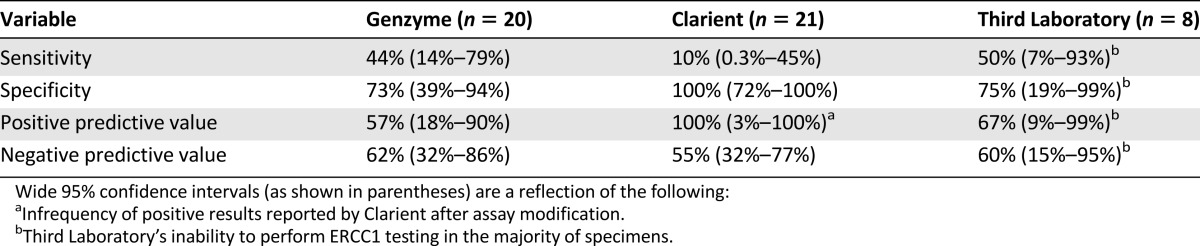
Initially, we were also surprised to find that Clarient, the laboratory that had previously been observed to report the highest rate of ERCC1 expression (Table 1), only reported one high ERCC1 expression result in this entire cohort of patients. Only after we queried Clarient about these unexpected results did we learn that its commercial assay had been deliberately modified to decrease the prevalence of high ERCC1 results, with that change having been implemented and applied between the second and third cohorts of this study (as above). Consequently, Clarient’s assay reported low ERCC1 results that correctly predicted response in all 11 platinum responders, but also incorrectly predicted platinum response in 9 of 10 nonresponders. Genzyme reported low ERCC1 results in 8 of 11 responders, but also in 5 of 9 nonresponders. Third Laboratory was unable to perform its RT-PCR-based mRNA assay on the majority of specimens, reporting low ERCC1 results in 3 of 4 responders and 2 of 4 nonresponders. Disappointingly, none of the three commercial assays reported a significantly higher frequency of low ERCC1 results in responders as compared with nonresponders in this selected cohort. The sensitivity, specificity, positive predictive value, and negative predictive value for each laboratory are shown in Table 3.
Discussion
Having entered an era in which cancer treatments are increasingly predicated on the results of tumor biomarker assays, the reliability of such assays is of the utmost importance. Our data demonstrate that ERCC1 testing, as performed in each of these three laboratories, is highly unreliable. Significant interlaboratory discrepancies in ERCC1 expression rates were confirmed by retesting specimens from the same tumor blocks at all three laboratories. Given that the test is reported as a simple dichotomous result (positive or negative), the degree of discordance that we observed between laboratories was both disappointing and alarming. Our results demonstrated that in 78% of cases, an ERCC1 result reported by one of these three laboratories would have been different if the test had been performed at one of the other two laboratories. In other words, if ERCC1 testing were used to decide whether to administer or withhold platinum-based chemotherapy, then that decision would most often be determined by which laboratory the test was sent to rather than any true difference in the biology of the tumor being tested. In the case of Clarient, the treatment decision would have likely been different if the test had been sent before or after their ERCC1 assay had been modified.
None of the ERCC1 assays that we studied could reliably distinguish between patients carefully selected as platinum responders and nonresponders. Because all patients were treated with platinum-doublet therapies, it is conceivable that responders who were determined to have high ERCC1 expression may have been truly resistant to the platinum therapy, but sensitive to the other coadministered cytotoxic agent. Nonresponder patients were, however, selected on the basis of being highly resistant to all administered agents in the platinum regimen, making their classification as “platinum nonresponders” more straightforward. Nevertheless, even in these refractory patients, none of the three commercially available assays correctly predicted refractory disease (high ERCC1) in more than half of tested specimens. In other words, none of the assays that we evaluated could predict platinum resistance on the basis of ERCC1 expression with a specificity of greater than 50%.
Recently, Friboulet et al. reported that none of the 16 currently available 8F1 antibody lots could reliably identify the ERCC1 isoform responsible for nucleotide excision repair and platinum resistance [22]. ERCC1 expression measured by these newer lots of 8F1 did not provide predictive utility in a newly studied cohort of patients receiving adjuvant cisplatin-based chemotherapy or in a reassessment of the original International Adjuvant Lung Trial cohort in which 8F1 had been previously validated. Importantly, when ERCC1 expression was measured with currently available 8F1 antibody lots, results were discordant with those previously observed with the original, and no longer available, 8F1 antibody. These authors suggest that the inadequacy of ERCC1 assays may reflect a change in the performance of currently available 8F1 antibody, but problems with ERCC1 measurement by 8F1 have been apparent since at least 2007 [30–32].
Prior reports have raised several other potential explanations as to why ERCC1 testing might not provide a reliable means of selecting patients for platinum or nonplatinum therapies. First, the very notion that measuring the expression of a single protein could adequately assess the complex process of DNA repair has been questioned [7, 32]. Second, low expression of ERCC1 has been shown to be associated with other genomic alterations, including those that affect sensitivity to nonplatinum chemotherapies [33, 34]. Third, others have previously noted that thresholds for ERCC1 “expression” had been arbitrarily assigned and inadequately validated [35]. Fourth, several prior studies had been unable to confirm the predictive utility of ERCC1 expression in platinum-treated lung cancer patients [31, 36]. Fifth, the measurement of ERCC1 protein by IHC and mRNA by RT-PCR has shown inconsistent correlation [37]. Sixth, the 8F1 antibody, most frequently used to measure ERCC1 protein levels, is unable to differentiate between normal and ERCC1-deficient cell lines, possibly due to excessive background cytoplasmic staining by 8F1 [30, 38, 39].
Based on such considerations, two consensus panels have appropriately recommended against the use of ERCC1 testing in routine clinical practice [2, 40]. Even the authors of studies previously reporting possible clinical utility of ERCC1 testing have responded to stated concerns by declaring that the test is still “not applicable for standard use in the everyday clinic” [41]. Yet, current National Comprehensive Cancer Network guidelines state that “Multiple translational investigations have provided evidence for the predictive use of ERCC-1 levels to assess the efficacy of platinum-based chemotherapies in NSCLC” [21] and the test remains commercially available.
Conclusion
This report provides direct evidence that currently marketed ERCC1 assays are unreliable. Initially perceived discrepancies between individual laboratory rates of ERCC1 expression suggested that some of these results were suspect. Discordance in laboratory round-robin analysis demonstrated that most were inconsistent. Sensitivity and specificity analysis among defined platinum responders and nonresponders confirmed that all were inadequate. We conclude that commercial laboratories should not offer ERCC1 testing until assays are better standardized and results are more thoroughly validated.
Two important lessons may be gleaned from the oncology community’s recent experience with ERCC1 testing. First, the process by which tumor biomarkers are validated and allowed to enter routine clinical practice is currently inadequate. Guideline committees, thought leaders, and clinicians should insist on more robust and consistent data before adopting tools that may alter patient care. Second, even after such tumor biomarker assays are incorporated into clinical practice, postmarketing scrutiny remains essential. This report suggests that such scrutiny may be achieved by simply demanding consistency of same-specimen results as well as by more ambitious efforts to develop gold standard specimens to ensure reliability of laboratory results.
Acknowledgments
This study was not supported by any external funding. The three commercial laboratories participating in this study performed ERCC1 assays at no charge when specimens were submitted specifically for the conduct of this study. Those three companies had no role in the analysis of data or in the writing of this manuscript. All three companies reviewed a final draft of this manuscript and confirmed that all technical aspects of their performance in this study have been accurately reported. We acknowledge and appreciate the cooperation of each of the three commercial laboratories participating in this study. Each laboratory participated with the knowledge that results might prove unfavorable to the marketing of its ERCC1 assays, but also with the understanding that such postmarketing surveillance might provide valuable insights for clinicians involved in the management of non-small cell lung cancer patients.
Footnotes
For Further Reading: Jared M. Weiss, Thomas E. Stinchcombe. Second-Line Therapy for Advanced NSCLC. The Oncologist 2013;18:947–953.
Implications for Practice: The landscape of first-line treatment of non-small cell lung cancer has generated challenges for clinical decisions in second-line therapy. For the patient treated with standard chemotherapy in the first line who has a treatable molecular change, this change should be targeted. More specifically, the patient with an epidermal growth factor receptor (EGFR) mutation should be treated with an EGFR tyrosine kinase inhibitor, and the patient with EML4/ALK rearrangement should be treated with crizotinib. However, these agents are increasingly being used in the first line, and most patients do not have these molecular changes. This leaves the clinician with many challenging questions regarding second-line therapy. How should the patient without treatable mutations be treated? Which clinical trials are most promising? How should the patient treated with a targeted agent in the first line be treated in the second line? This review addresses these issues, exploring the key existing data available to help guide informed clinical decisions.
Author Contributions
Conception/design: Jeffrey G. Schneider, Nosha Farhadfar, Abirami Sivapiragasam, Matthew Geller, Elena Selbs
Provision of study material or patients: Jeffrey G. Schneider, Nosha Farhadfar, Abirami Sivapiragasam, Matthew Geller, Elena Selbs
Collection and/or assembly of data: Jeffrey G. Schneider, Nosha Farhadfar, Abirami Sivapiragasam, Matthew Geller
Data analysis and interpretation: Jeffrey G. Schneider, Nosha Farhadfar, Abirami Sivapiragasam, Shahidul Islam
Manuscript writing: Jeffrey G. Schneider, Nosha Farhadfar, Abirami Sivapiragasam, Shahidul Islam, Elena Selbs
Final approval of manuscript: Jeffrey G. Schneider, Nosha Farhadfar, Abirami Sivapiragasam, Matthew Geller, Shahidul Islam, Elena Selbs
Disclosures
The authors indicated no financial relationships.
References
- 1.Schiller JH, Harrington D, Belani CP, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346:92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 2.Azzoli CG, Baker S, Jr, Temin S, et al. American Society of Clinical Oncology Clinical Practice Guideline update on chemotherapy for stage IV non-small-cell lung cancer. J Clin Oncol. 2009;27:6251–6266. doi: 10.1200/JCO.2009.23.5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arriagada R, Bergman B, Dunant A, et al. Cisplatin-based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer. N Engl J Med. 2004;350:351–360. doi: 10.1056/NEJMoa031644. [DOI] [PubMed] [Google Scholar]
- 4.Butts CA, Ding K, Seymour L, et al. Randomized phase III trial of vinorelbine plus cisplatin compared with observation in completely resected stage IB and II non-small-cell lung cancer: Updated survival analysis of JBR-10. J Clin Oncol. 2010;28:29–34. doi: 10.1200/JCO.2009.24.0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pignon JP, Tribodet H, Scagliotti GV, et al. Lung adjuvant cisplatin evaluation: A pooled analysis by the LACE Collaborative Group. J Clin Oncol. 2008;26:3552–3559. doi: 10.1200/JCO.2007.13.9030. [DOI] [PubMed] [Google Scholar]
- 6.Breathnach OS, Freidlin B, Conley B, et al. Twenty-two years of phase III trials for patients with advanced non-small-cell lung cancer: Sobering results. J Clin Oncol. 2001;19:1734–1742. doi: 10.1200/JCO.2001.19.6.1734. [DOI] [PubMed] [Google Scholar]
- 7.Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 8.Altaha R, Liang X, Yu JJ, et al. Excision repair cross complementing-group 1: Gene expression and platinum resistance. Int J Mol Med. 2004;14:959–970. [PubMed] [Google Scholar]
- 9.De Silva IU, McHugh PJ, Clingen PH, et al. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol Cell Biol. 2000;20:7980–7990. doi: 10.1128/mcb.20.21.7980-7990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niedernhofer LJ, Odijk H, Budzowska M, et al. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol Cell Biol. 2004;24:5776–5787. doi: 10.1128/MCB.24.13.5776-5787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reed E. ERCC1 measurements in clinical oncology. N Engl J Med. 2006;355:1054–1055. doi: 10.1056/NEJMe068162. [DOI] [PubMed] [Google Scholar]
- 12.Gazdar AF. DNA repair and survival in lung cancer—The two faces of Janus. N Engl J Med. 2007;356:771–773. doi: 10.1056/NEJMp068308. [DOI] [PubMed] [Google Scholar]
- 13.Soria JC. ERCC1-tailored chemotherapy in lung cancer: The first prospective randomized trial. J Clin Oncol. 2007;25:2648–2649. doi: 10.1200/JCO.2007.11.3167. [DOI] [PubMed] [Google Scholar]
- 14.Ceppi P, Volante M, Novello S, et al. ERCC1 and RRM1 gene expressions but not EGFR are predictive of shorter survival in advanced non-small-cell lung cancer treated with cisplatin and gemcitabine. Ann Oncol. 2006;17:1818–1825. doi: 10.1093/annonc/mdl300. [DOI] [PubMed] [Google Scholar]
- 15.Cobo M, Isla D, Massuti B, et al. Customizing cisplatin based on quantitative excision repair cross-complementing 1 mRNA expression: A phase III trial in non-small-cell lung cancer. J Clin Oncol. 2007;25:2747–2754. doi: 10.1200/JCO.2006.09.7915. [DOI] [PubMed] [Google Scholar]
- 16.Holm B, Mellemgaard A, Skov T, et al. Different impact of excision repair cross-complementation group 1 on survival in male and female patients with inoperable non-small-cell lung cancer treated with carboplatin and gemcitabine. J Clin Oncol. 2009;27:4254–4259. doi: 10.1200/JCO.2008.18.8631. [DOI] [PubMed] [Google Scholar]
- 17.Jiang J, Liang X, Zhou X, et al. ERCC1 expression as a prognostic and predictive factor in patients with non-small cell lung cancer: A meta-analysis. Mol Biol Rep. 2012;39:6933–6942. doi: 10.1007/s11033-012-1520-4. [DOI] [PubMed] [Google Scholar]
- 18.Olaussen KA, Dunant A, Fouret P, et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med. 2006;355:983–991. doi: 10.1056/NEJMoa060570. [DOI] [PubMed] [Google Scholar]
- 19.Reynolds C, Obasaju C, Schell MJ, et al. Randomized phase III trial of gemcitabine-based chemotherapy with in situ RRM1 and ERCC1 protein levels for response prediction in non-small-cell lung cancer. J Clin Oncol. 2009;27:5808–5815. doi: 10.1200/JCO.2009.21.9766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vilmar AC, Santoni-Rugiu E, Sørensen JB. ERCC1 and histopathology in advanced NSCLC patients randomized in a large multicenter phase III trial. Ann Oncol. 2010;21:1817–1824. doi: 10.1093/annonc/mdq053. [DOI] [PubMed] [Google Scholar]
- 21.Panel NCCN-NSCLC. NCCN clinical practice guidelines in oncology. Non-small cell lung cancer. Available at: http://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf. Accessed on August 11, 2013.
- 22.Friboulet L, Olaussen KA, Pignon JP, et al. ERCC1 isoform expression and DNA repair in non-small-cell lung cancer. N Engl J Med. 2013;368:1101–1110. doi: 10.1056/NEJMoa1214271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller AB, Hoogstraten B, Staquet M, et al. Reporting results of cancer treatment. Cancer. 1981;47:207–214. doi: 10.1002/1097-0142(19810101)47:1<207::aid-cncr2820470134>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 24.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 25.Olaussen KA, Fouret P, Kroemer G. ERCC1-specific immunostaining in non-small-cell lung cancer. N Engl J Med. 2007;357:1559–1561. doi: 10.1056/NEJMc072007. [DOI] [PubMed] [Google Scholar]
- 26a.Danenberg K, Danenberg PV, Swenson S. inventors; University of Southern California, assignee. Method for isolation of RNA from formalin-fixed paraffin-embedded tissue specimens. US patent 6 248 535. June 19, 2001. [Google Scholar]
- 26b.Lord RV, Brabender J, Gandara D, et al. Low ERCC1 expression correlates with prolonged survival after cisplatin plus gemcitabine chemotherapy in non-small cell lung cancer. Clin Cancer Res. 2002;8:2286–2291. [PubMed] [Google Scholar]
- 27.Siegel S, Castellan NJ, McGraw H. 2nd ed. New York, NY: McGraw-Hill; 1988. Nonparametric statistics for the Behavioral Sciences. [Google Scholar]
- 28.Fleiss JL. Statistical Methods for Rates and Proportions. 3rd ed. New York, NY: John Wiley & Sons; 2003. [Google Scholar]
- 29.Park JO, Lee SI, Song SY, et al. Measuring response in solid tumors: Comparison of RECIST and WHO response criteria. Jpn J Clin Oncol. 2003;33:533–537. doi: 10.1093/jjco/hyg093. [DOI] [PubMed] [Google Scholar]
- 30.Niedernhofer LJ, Bhagwat N, Wood RD. ERCC1 and non-small-cell lung cancer [letter] N Engl J Med. 2007;356:2538–2540; author reply 2540–2541. doi: 10.1056/NEJMc070742. [DOI] [PubMed] [Google Scholar]
- 31.Booton R, Ward T, Ashcroft L, et al. ERCC1 mRNA expression is not associated with response and survival after platinum-based chemotherapy regimens in advanced non-small cell lung cancer. J Thorac Oncol. 2007;2:902–906. doi: 10.1097/JTO.0b013e318155a637. [DOI] [PubMed] [Google Scholar]
- 32.Tan DS, Ng QS, Tan IB, et al. Truth about ERCC1 in lung cancer [letter] J Clin Oncol. 2010;28:e162. doi: 10.1200/JCO.2009.26.6270. author reply e164. [DOI] [PubMed] [Google Scholar]
- 33.Bepler G, Kusmartseva I, Sharma S, et al. RRM1 modulated in vitro and in vivo efficacy of gemcitabine and platinum in non-small-cell lung cancer. J Clin Oncol. 2006;24:4731–4737. doi: 10.1200/JCO.2006.06.1101. [DOI] [PubMed] [Google Scholar]
- 34.Zheng Z, Chen T, Li X, et al. DNA synthesis and repair genes RRM1 and ERCC1 in lung cancer. N Engl J Med. 2007;356:800–808. doi: 10.1056/NEJMoa065411. [DOI] [PubMed] [Google Scholar]
- 35.Tepeli E, Caner V, Büyükpınarbaşılı N, et al. Expression of ERCC1 and its clinicopathological correlations in non-small cell lung cancer. Mol Biol Rep. 2012;39:335–341. doi: 10.1007/s11033-011-0743-0. [DOI] [PubMed] [Google Scholar]
- 36.Tibaldi C, Giovannetti E, Vasile E, et al. Correlation of CDA, ERCC1, and XPD polymorphisms with response and survival in gemcitabine/cisplatin-treated advanced non-small cell lung cancer patients. Clin Cancer Res. 2008;14:1797–1803. doi: 10.1158/1078-0432.CCR-07-1364. [DOI] [PubMed] [Google Scholar]
- 37.Friboulet L, Barrios-Gonzales D, Commo F, et al. Molecular characteristics of ERCC1-negative versus ERCC1-positive tumors in resected NSCLC. Clin Cancer Res. 2011;17:5562–5572. doi: 10.1158/1078-0432.CCR-11-0790. [DOI] [PubMed] [Google Scholar]
- 38.Bhagwat NR, Roginskaya VY, Acquafondata MB, et al. Immunodetection of DNA repair endonuclease ERCC1-XPF in human tissue. Cancer Res. 2009;69:6831–6838. doi: 10.1158/0008-5472.CAN-09-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Panasci LC. Different impact of excision repair cross-complementing group 1 on survival [letter]. J Clin Oncol 2010;28:e163; author reply e164. [DOI] [PubMed]
- 40.Felip E, Gridelli C, Baas P, et al. Metastatic non-small-cell lung cancer: Consensus on pathology and molecular tests, first-line, second-line, and third-line therapy: First ESMO Consensus Conference in Lung Cancer; Lugano 2010. Ann Oncol. 2011;22:1507–1519. doi: 10.1093/annonc/mdr150. [DOI] [PubMed] [Google Scholar]
- 41.Holm B, Mellemgaard A, Skov T, et al. Reply to L.C. Panasci and D.S.-W. Tan et al. [letter] J Clin Oncol. 2010;28:e164. [Google Scholar]