This article, which is based on a lecture delivered as part of the 2013 Bob Pinedo Cancer Care Prize, reviews the impact of molecular biomarkers on the management of colorectal cancer, emphasizing changes that have occurred in recent years, and focuses on potential mechanisms of patient stratification and opportunities for novel therapeutic development based on enhanced biological understanding of colorectal cancer.
Abstract
In recent years, a number of protein and genomic-based biomarkers have begun to refine the prognostic information available for colorectal cancer (CRC) and predict defined patient groups that are likely to benefit from systemic treatment or targeted therapies. Of these, KRAS represents the first biomarker integrated into clinical practice for CRC. Microarray-based gene expression profiling has been used to identify prognostic signatures and, to a lesser extent, predictive signatures in CRC. Despite these advances, a number of major challenges remain. This article, which is based on a lecture delivered as part of the 2013 Bob Pinedo Cancer Care Prize, reviews the impact of molecular biomarkers on the management of CRC, emphasizing changes that have occurred in recent years, and focuses on potential mechanisms of patient stratification and opportunities for novel therapeutic development based on enhanced biological understanding of colorectal cancer.
Abstract
摘要
近年来,多种蛋白质类和基因组类生物标志物已开始为结直肠癌 (CRC) 提供更高质量的预后信息,并用于预测哪些患者群体更有可能受益于全身治疗或靶向治疗。其中,KRAS 是首个被纳入临床的 CRC 生物标志物。基于微阵列芯片的基因表达分析工具已被用以鉴定 CRC 的预后标志物,次之用于鉴定预测性标志物。尽管有了这些进展,目前仍存在若干重大挑战。本文基于一堂在 2013 年 Bob Pinedo 癌症治疗奖颁奖时发表的演讲,总结回顾了分子生物标志物对 CRC 治疗的影响,强调了近年来发生的一些变化,同时根据对结直肠癌生物机制的更深一步了解,重点分析了患者分层的潜在机制和新疗法的研发机遇。The Oncologist 2014;19:568–573

Patrick G. Johnston
Introduction
Our improved understanding of cancer biology in colorectal cancer, coupled with the implementation of a number of new protein- and genomic-based technologies, has demonstrated that colorectal cancer (CRC) should be viewed as a heterogeneous disease. Consequently, there is an increasing need to implement molecularly guided therapeutic strategies including combinations of targeted therapies and chemotherapy in CRC [1].
The addition of the novel cytotoxic agents oxaliplatin and irinotecan to standard 5-fluorouracil (5-FU) regimens along with the use of inhibitors of the vascular endothelial growth factor (VEGF) and the epidermal growth factor receptor (EGFR) pathways have enhanced overall survival to more than 20 months [2–5]. Although the majority of patients with CRC will still receive standard treatment with 5-FU and irinotecan (the FOLFIRI regimen) or 5-FU and oxaliplatin (the FOLFOX regimen), close to 50% will have no benefit from these treatments and will develop toxic side effects.
The recent improvements in anticancer treatments and patient outcome in CRC have been followed by a series of biomarker studies attempting to refine prognosis and predict patients who are likely to derive the most benefit from treatment. In CRC, only KRAS has recently entered routine clinical practice as a predictive marker for response to EGFR monoclonal antibody (mAb) therapies. Anti-EGFR-targeted mAbs represent the paradigm of personalized medicine in CRC and are used in combination with standard chemotherapy in wild-type KRAS CRC patients, improving overall survival to 23 months [6, 7]. EGFR-targeted therapies, however, have failed to show significant differences in overall survival, especially when administered as second- or third-line therapy, and a significant number of the wild-type KRAS patients do not benefit from EGFR-targeted treatment [8, 9].
VEGF-targeted therapies have also been shown to increase survival when added to first- and second-line standard chemotherapy; however, we urgently need markers that identify those patients who will have maximal benefit from this treatment [5, 10, 11].
Clinical and Molecular Risk Factors
In CRC we still rely primarily on histological analysis of resected tumor tissues for diagnosis and staging. The most widely used prognostic factors to assess recurrence risk and overall survival for patients are T stage (extent of invasion) and N stage (number of lymph node metastases). Those patients with stage III colon cancer are offered postoperative adjuvant chemotherapy; however, wide variations are seen in the outcomes for patients with stage III disease. Among patients with stage II colon cancer, additional clinical and pathological findings are considered, including number of lymph nodes sampled, evidence of obstruction and/or perforation, histological grade, and lymphovascular and perineural invasion [12, 13].
The search for prognostic factors for patients with colorectal carcinoma has included biomarkers such as microsatellite instability, loss of heterozygosity, p53, proliferation markers such as Ki-67, and key chemotherapeutic target enzymes such as thymidylate synthase (TS) and angiogenic factors such as VEGF [14–17].
Mutations in p53 have been associated with decreased sensitivity to several classes of chemotherapy, including DNA-damaging agents such as irinotecan and oxaliplatin [14, 15]. However, p53 immunohistochemistry analysis does not correlate well with direct sequencing results and, consequently, is rarely used. Moreover, the association of p53 overexpression with poor clinical outcome has not been shown consistently in clinical trials.
Several studies have reported that patients with cancer who overexpress TS have a lower response rate to treatment with 5-FU [16, 17]. A number of studies had shown that overexpression of TS predicts a poorer response and survival to fluoropyrimidines; however, other studies have not been able to verify these findings.
The tumor necrosis factor-related apoptosis-inducing ligand, or TRAIL, death receptors DR4 and DR5 have also been an area of interest and have been shown to be important for assessing response to fluoropyrimidines in xenograft models [18]. High expression of DR4 has also been identified as a negative prognostic factor for patients receiving adjuvant therapy, with a relative risk of recurrence of 2.2 for patients who were high expressers [19]. Another recent study from our group has suggested that high levels of cellular FLICE-inhibitory protein and TRAIL may be independent adverse prognostic markers in stage II and stage III CRC and might identify patients most at risk for relapse [20]. Hector et al. recently demonstrated the importance of the apoptosome-dependent caspase activation pathway (procaspase 3 and APAF-1 proteins) for predicting both prognosis and response to adjuvant 5-FU treatment in stage II and stage III CRC [21]. Although these studies have been interesting, none have proven reliable in larger validation studies, and thus their clinical utility remains limited.
Mismatch Repair Status
One indicator that has proven to be reliable for prognosis is DNA mismatch repair status (MMR). Approximately 15% of patients with sporadic CRC have tumors that are characterized by deficient DNA mismatch repair due to mutations or silencing of genes such as MLH1, MSH2, MSH6, and PMS2, leading to microsatellite instability (MSI) [22]. These tumors are pathologically distinct from CRC tumors arising from the chromosomal instability pathway and also harbor significant differences in clinical behavior. A number of important retrospective studies in patients with stage II or stage III CRC have shown that patients with MSI-high (MSI-H) tumors have a more favorable prognosis than patients with microsatellite-stable (MSS) or MSI-low (MSI-L) tumors [23–27]. Sinicrope et al. found that patients with MSI-H cancers had improved time to recurrence and overall survival with surgery alone compared with MSI-L or MSS patients [24]. Recently, Roth et al. looked at the importance of KRAS and BRAF mutations as prognostic markers, alongside MSI, SMAD4, and 18qLOH, in patients treated as part of the adjuvant PETACC3 chemotherapy trial [25]. MSI-H status and SMAD4 focal loss of expression were identified as independent prognostic factors with better relapse-free survival (RFS) and overall survival for MSI-H status and poor RFS and overall survival for SMAD4 loss. They also found that the impact of MSI status was stronger in stage II compared with stage III CRC patients. Notably, the subgroup of stage III tumors with MSI-H status that had retained SMAD4 expression had outcomes similar to those patients with stage II disease. These studies highlight the importance of assessing molecular markers alongside TNM staging because this may provide a more accurate prediction of prognosis than by using TNM staging alone.
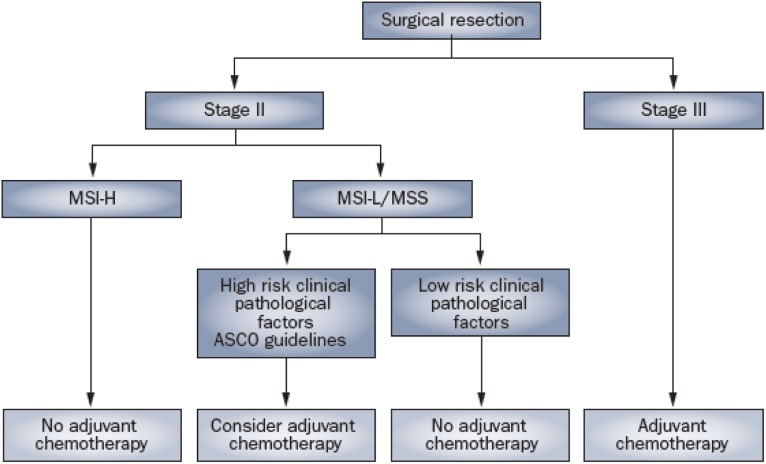
Most studies that have examined the role of MSI in predicting response to chemotherapy have noted a lack of benefit from adjuvant 5-FU treatment in patients with MSI-H CRC [26, 27]. Several have shown that in patients with stage II or stage III CRC, 5-FU-based adjuvant chemotherapy significantly reduces the recurrence risk in MSS and MSI-L patients but not in MSI-H patients. It is recommended that patients with stage II colon cancer with MSI-H tumors should not receive 5-FU as adjuvant treatment, given their already favorable prognosis and lack of benefit from 5-FU. The MMR phenotype is now considered to be a robust prognostic biomarker in the adjuvant disease setting and is of use in clinical decision making, particularly for patients with stage II disease (Fig. 1).
It is recommended that patients with stage II colon cancer with MSI-H tumors should not receive 5-FU as adjuvant treatment, given their already favorable prognosis and lack of benefit from 5-FU. The MMR phenotype is now considered to be a robust prognostic biomarker in the adjuvant disease setting and is of use in clinical decision making, particularly for patients with stage II disease.
Figure 1.
Algorithm for utilizing microsatellite instability alongside clinical pathological factors in stage II and III colorectal cancer. First published in Nat Rev Clinic Oncol 2011;8:222–232, doi:10.1038/nrclinonc.2011.15. Reproduced with permission of Nature Publishing Group, a division of Macmillan Publishers Limited.
Abbreviations: ASCO, American Society of Clinical Oncology; MSI-H, microsatellite instability high; MSI-L, microsatellite instability low; MSS, microsatellite stable.
Gene Expression Profiling
With the advent of high-throughput technologies, assessment of the entire genome has become possible. High-throughput genotyping has enabled analysis of genes such as KRAS and BRAF and PI3 kinase in individual tumors. Moreover, the use of gene expression profiling to discover and develop robust genetic signatures that can be used as reliable prognostic and predictive biomarkers has become widespread. Over the last several years, a number of gene expression signatures have been developed for use in stage II and III colon cancer [28].
Oncotype DX (Genomic Health Inc., Redwood City, CA, http://www.oncotypedx.com) was developed as a 12-gene colon cancer recurrence score (RS) using reverse transcription-polymerase chain reaction expression analysis of 761 candidate genes on formalin-fixed paraffin-embedded (FFPE) tissues from 1,851 patients enrolled in four National Surgical Adjuvant Breast and Bowel Project adjuvant colon cancer trials and Cleveland Clinic observational studies [29]. The final gene list was composed of seven genes associated with recurrence and a distinct set of six genes associated with clinical benefit from FU/LV (i.e., 5-FU with leucovorin) chemotherapy. The RS identified low-, intermediate-, and high-recurrence risk groups within the study and was subsequently validated in 1,436 stage II patients from the Quick and Simple and Reliable, or QUASAR, study. Pathological factors were also assessed, as was MMR status, by immunohistochemistry for MMR proteins. The 3-year risk of recurrence of the low, intermediate, and high RS groups were 12%, 18%, and 22%, respectively, in the surgery-only arm and 9%, 13%, and 16%, respectively, in the chemotherapy arm [29]. More recently, the Oncotype DX test has been clinically validated as a prognostic signature for patients with stage II colon cancer in a subsequent large clinical study (NSABP C-07) [30]. Although the test is not routinely used, it may identify patients at increased risk for recurrence who might be considered for adjuvant therapy.
ColDx (Almac Group Ltd, Craigavon, U.K., http://www.almacgroup.com) was developed as a 634-gene signature from 215 stage II patients using FFPE tissue and analyzed using the Almac Dx colorectal cancer disease-specific array [31, 32]. Multivariate analysis included tumor stage, grade, location, patient age and gender, mucinous type, lymphovascular invasion, and number of lymph nodes examined, but microsatellite instability status was not included. The study included a training set from 142 low-risk and 73 high-risk patients, and an independent validation set of 144 patients enriched for recurrence included 85 low-risk and 59 high-risk patients. The hazard ratios (HRs) for 5-year relapse-free survival and cancer-related death were 2.53 and 2.21, respectively, and the test outperformed conventional clinicopathological risk factors. The ColDx has recently been validated in 393 patients enrolled into the CALGB 9581 trial of edrecolomab versus observation in patients with stage II CRC [33]. This study confirmed that ColDx predicted stage II CRC patients at low and high risk for relapse (HR: 2.0) independent of other clinocopathological criteria and MSI status. Further studies will be required to define the role of the 634-gene signature as a prognostic and predictive marker of treatment benefit in CRC.
Salazar et al. identified an 18-gene signature (ColoPrint; Agendia Inc., Irvine, CA, http://www.agendia.com) using fresh frozen tissue [34]. The training set consisted of CRC samples from 100 patients with stage II disease, and the validation cohort consisted of CRC samples collected prospectively from 114 independent patients. In the patients with stage II disease, two thirds were classified as low risk and one third were classified as high risk, with 5-year RFS rates of 91% and 74%, respectively. The ColoPrint assay significantly improved the prognostic accuracy of traditional pathological factors and microsatellite instability and identified stage II CRC patients at low risk for recurrence [34]. A large phase II clinical trial to validate the ColoPrint test in stage II CRC (PARSC study, NCT00903565) is currently under way [35].
ColoGuide Ex is a 13-gene signature derived from 207 stage I–IV colorectal cancer samples from two independent Norwegian clinical series using Affymetrix exon-based microarrays (Affymetrix, Santa Clara, CA, http://www.affymetrix.com). Validation was performed in patients with stage II colorectal cancer in a second Norwegian series and in 108 stage II colorectal cancer samples from other populations (U.S. and Australia). An optimal 13-gene expression classifier for prediction of relapse specific to patients with stage II disease was developed with an HR for recurrence of 6.5 (p = .001) in the second validation set [36]. The same Norwegian group assessed global gene expression profiles from 387 stage II and III colorectal cancer tissue samples from three independent patient series. A seven-gene prognostic classifier (ColoGuidePro) stratified patients in a test series that was then externally validated in 215 stage II and III CRC patients (HR: 3.7) [37].
The use of genome expression profiling to develop genomic stratification markers is a highly complex and challenging technical area that requires use of the correct technology platforms, robust developmental studies, and in-depth analytical validation including assessment of reproducibility, accuracy, and transferability, alongside clinical validation in prospective randomized clinical trials. One major challenge has been the need for sufficient high-quality RNA for successful transcriptional profiling and the need for fresh frozen tissue. This is often not feasible within the context of large clinical trials, for which tissue specimens are FFPE for subsequent histological examination. Although RNA fragmentation has been a major problem following RNA extraction from FFPE tissues, a number of microarray and reverse transcription-polymerase chain reaction platforms have been developed that are suitable for use in this context [29, 31]. The further application of these platforms in this way holds great promise for developing clinically robust signatures that may enable patient selection for adjuvant treatment.
Biomarkers of Response to Epidermal Growth Factor-Targeted Therapy
KRAS mutational testing has recently entered routine clinical practice as a predictive marker for response to EGFR-based therapies. KRAS mutations are found in 45% of patients with CRC and occur mainly in exon 2 (codon 12 and 13) and less frequently in exon 3 (codon 61) and exon 4 (codon 146) [38, 39].
A number of large phase II and III randomized studies have now confirmed the negative predictive value of KRAS mutations for response to the EGFR-targeted therapies cetuximab and panitumumab. The clinical study of panitumumab and best supportive care versus best supportive care alone in patients with chemorefractory CRC demonstrated response rates of 17% and 0% for the KRAS wild-type and KRAS mutant groups, respectively [6]. On this basis, panitumumab was approved by the European Medicines Agency as third-line treatment for patients with KRAS wild-type tumors. This finding was also replicated for cetuximab by retrospective analysis of data in the National Cancer Institute of Canada Clinical Trials Group study of best supportive care plus cetuximab versus best supportive care, and this study also found that the benefit of cetuximab was confined to patients with KRAS wild-type tumors [40].
Other randomized trials have substantiated the negative predictive value of KRAS mutations for response to EGFR treatment, and it is widely accepted as the major determinant as to whether a patient should receive EGFR-targeted therapy [8, 41]. Retrospective analysis of data from the OPUS and CRYSTAL studies has shown that addition of cetuximab to first-line FOLFOX and FOLFIRI chemotherapy has no benefit in patients with KRAS mutant tumors [8, 42].
The high prevalence of KRAS mutations in CRC and its strong negative predictive value for EGFR mAb therapies have led to the rapid acceptance of KRAS as a valuable biomarker. The European Medicines Agency, the U.S. Food and Drug Administration, and the American Society of Clinical Oncology now recommend that all patients with metastatic CRC who are candidates for anti-EGFR mAb therapy should be tested for KRAS mutations, and if a KRAS mutation in codon 12 or 13 is detected, then patients should not receive anti-EGFR antibody therapy.
The KRAS biomarker story is important because it represents the first biomarker integrated into clinical practice in CRC. It has also become clear that a significant number (30%) of patients with KRAS wild-type tumors will not benefit from treatment, highlighting the need to further refine the selection of KRAS wild-type patients for anti-EGFR-mAbs.
The BRAF V600E mutation occurs in approximately 8%–15% of patients with CRC and is mutually exclusive with mutations in KRAS [42–45]. Tumors with the BRAF V600E mutation tend to be mucinous, are high grade, are located in the proximal colon, and have an MSI-H phenotype. A number of studies suggest that KRAS and BRAF mutant CRC tumors induce different gene expression profiles, suggesting that these tumors may have a distinct underlying biology. Recent data from the OPUS and CRYSTAL studies have shown that in chemotherapy-naïve patients with wild-type KRAS and mutant BRAF tumors, the addition of cetuximab to FOLFIRI treatment improves overall survival (14 months vs. 10 months), progression-free survival (7 months vs. 3.7 months), and response rate (22% vs. 13%) compared with FOLFIRI alone. These differences were not statistically significant because of the limited number of patients in this group [42].
The use of KRAS testing represents an important paradigm shift in personalized medicine in CRC. It has allowed us to more accurately determine who should receive anti-EGFR-targeted monoclonal antibodies, either alone or in combination with chemotherapy, and has changed our approach to drug development and clinical trials in this disease.
The use of KRAS testing represents an important paradigm shift in personalized medicine in CRC. It has allowed us to more accurately determine who should receive anti-EGFR-targeted monoclonal antibodies, either alone or in combination with chemotherapy, and has changed our approach to drug development and clinical trials in this disease.
Development of Novel Therapeutics in CRC
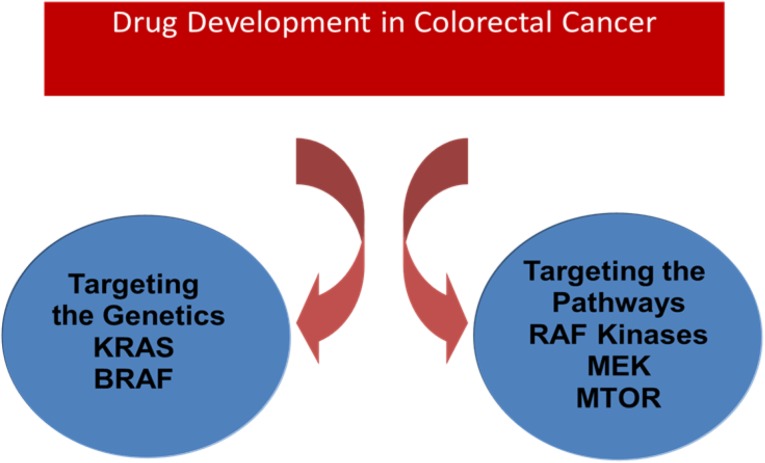
The ability to identify distinct molecular subtypes of colorectal cancer and to define their underlying biology has begun to unravel critical mutations in genes such as KRAS, BRAF, and PTEN and in PI3 kinase. Alterations in specific genes or groups of genes related to specific molecular subtypes encode proteins that, in themselves, may become targets for new therapeutics, particularly those encoding regulators of key pathways such as kinases. Although many mutated cancer genes may not be tractable targets for new drug discovery, screening for kinases and regulators of key signaling pathways that demonstrate synthetic lethality with mutated cancer genes such as KRAS are increasingly a major focus. The identification of key regulatory kinases and evaluation of targeting of KRAS downstream, such as inhibitors of RAF kinase, MEK, or MTOR, are currently under clinical evaluation [46]. These approaches demonstrate how understanding this biology may uncover potential therapeutic strategies that can be applied to these large subtypes of colorectal tumors (Fig. 2).
Figure 2.
Mutated colorectal cancer genes as the basis for discovery of new therapeutic targets for drug discovery.
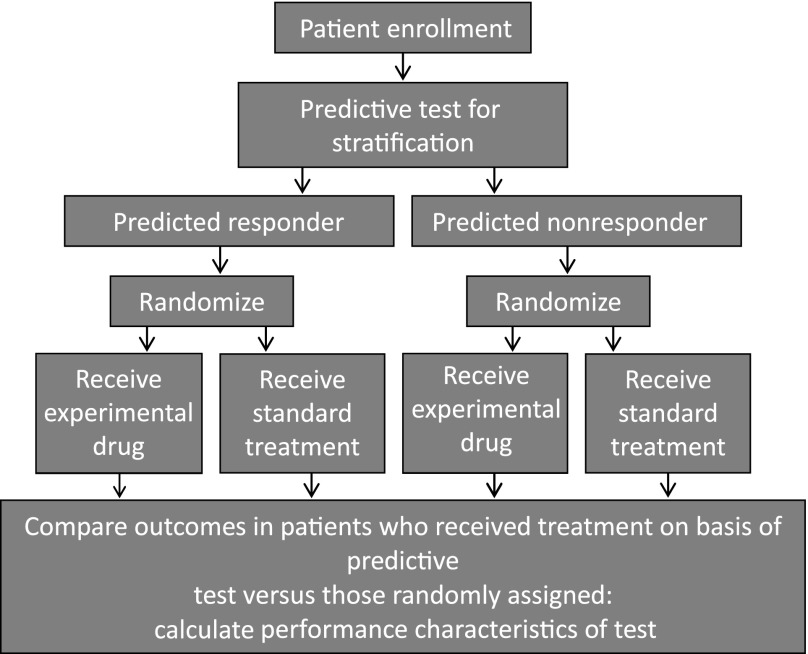
Initiatives such as the International Cancer Genome Consortium represent large-scale cancer genomic studies that will help further refine the definition of these distinct molecular subgroups in CRC and unravel additional therapeutic targets that may be important [47]. These developments will require clinical trials with enough statistical power and clearly defined endpoints in molecularly stratified patient populations, as outlined in Figure 3. Novel adaptive clinical trial designs that incorporate markers in prospective randomized phase II or III studies will enable clinical validation of these markers and may facilitate their implementation into routine medical practice.
Figure 3.
Process for comparing outcomes in patients who received treatment on the basis of stratification versus those randomly assigned.
Conclusion
In recent years our enhanced understanding of the molecular anomalies that drive the development and progression of CRC has resulted in a number of novel, targeted agents becoming part of standard treatment. Chemotherapy and targeted treatments have markedly reduced the risk of recurrence and mortality for CRC patients and have increased 5-year survival rates. MSI is now considered to be a robust prognostic biomarker in the adjuvant setting, and KRAS has been taken up as part of routine clinical practice as a predictive marker for response to EGFR-targeted therapies. The introduction of high-throughput technologies has enabled us to molecularly classify individual tumor types that traditionally have the same clinicopathological features and uncover molecular signatures that may be useful as prognostic and predictive biomarkers.
These technologies have already had an effect on drug development and clinical practice within colorectal cancer, as outlined previously. The pace of the research into cancer genomics will increase around coordinated global initiatives that generate full genome sequences for tens of thousands of genes that will uncover a new molecular taxonomy for colorectal cancer. In order to exploit the full clinical potential of this, we will have to move away from routine clinical trial approaches and begin to focus on designing trials based on molecularly stratified populations in this disease.
Acknowledgment
This article, which was delivered as part of the Bob Pinedo Cancer Care Prize Lecture (the Pinedo Lecture), is dedicated to the memory of Sir Allen McClay.
Footnotes
Editor’s Note: View Dr. Johnston's 2013 Bob Pinedo Cancer Care Prize Lecture, “Translational Cancer Medicine: A Journey from Concept to Reality,” online at http://sto-online.org.
Disclosures
Patrick G. Johnston: Chugai Pharmaceuticals, Sanofi-Aventis, Pfizer (C/A, H); Almac Diagnostics (E); Almac Diagnostics, Fusion Antibodies (OI).
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
References
- 1.Van Schaeybroeck S, Allen WL, Turkington RC, et al. Implementing prognostic and predictive biomarkers in CRC clinical trials. Nat Rev Clin Oncol. 2011;15:222–232. doi: 10.1038/nrclinonc.2011.15. [DOI] [PubMed] [Google Scholar]
- 2.de Gramont A, Figer A, Seymour M, et al. Leucovorin and fluorouracil with or without oxaliplatin as first-line treatment in advanced colorectal cancer. J Clin Oncol. 2000;18:2938–2947. doi: 10.1200/JCO.2000.18.16.2938. [DOI] [PubMed] [Google Scholar]
- 3.Douillard JY, Cunningham D, Roth AD, et al. Irinotecan combined with fluorouracil compared with fluorouracil alone as first-line treatment for metastatic colorectal cancer: A multicentre randomised trial. Lancet. 2000;355:1041–1047. doi: 10.1016/s0140-6736(00)02034-1. [DOI] [PubMed] [Google Scholar]
- 4.Van Cutsem E, Köhne CH, Hitre E, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408–1417. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 5.Allegra CJ, Yothers G, O’Connell MJ, et al. Phase III trial assessing bevacizumab in stages II and III carcinoma of the colon: Results of NSABP protocol C-08. J Clin Oncol. 2011;29:11–16. doi: 10.1200/JCO.2010.30.0855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 7.Lièvre A, Bachet JB, Boige V, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol. 2008;26:374–379. doi: 10.1200/JCO.2007.12.5906. [DOI] [PubMed] [Google Scholar]
- 8.Van Cutsem E, Köhne CH, Láng I, et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: Updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2011;29:2011–2019. doi: 10.1200/JCO.2010.33.5091. [DOI] [PubMed] [Google Scholar]
- 9.De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: A retrospective consortium analysis. Lancet Oncol. 2010;11:753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 10.Grothey A, Sugrue MM, Purdie DM, et al. Bevacizumab beyond first progression is associated with prolonged overall survival in metastatic colorectal cancer: Results from a large observational cohort study (BRiTE) J Clin Oncol. 2008;26:5326–5334. doi: 10.1200/JCO.2008.16.3212. [DOI] [PubMed] [Google Scholar]
- 11.Giantonio BJ, Catalano PJ, Meropol NJ, et al. Bevacizumab in combination with oxaliplatin, fluorouracil, and leucovorin (FOLFOX4) for previously treated metastatic colorectal cancer: Results from the Eastern Cooperative Oncology Group Study E3200. J Clin Oncol. 2007;25:1539–1544. doi: 10.1200/JCO.2006.09.6305. [DOI] [PubMed] [Google Scholar]
- 12.Gill S, Loprinzi C, Kennecke H, et al. Prognostic web-based models for stage II and III colon cancer: A population and clinical trials-based validation of numeracy and adjuvant! online. Cancer. 2011;117:4155–4165. doi: 10.1002/cncr.26003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engstrom PF, Arnoletti JP, Benson AB, III, et al. NCCN clinical practice guidelines in oncology: Colon cancer. J Natl Compr Canc Netw. 2009;7:778–831. doi: 10.6004/jnccn.2009.0056. [DOI] [PubMed] [Google Scholar]
- 14.Allegra CJ, Paik S, Colangelo LH, et al. Prognostic value of thymidylate synthase, Ki-67, and p53 in patients with Dukes’ B and C colon cancer: A National Cancer Institute-National Surgical Adjuvant Breast and Bowel Project collaborative study. J Clin Oncol. 2003;21:241–250. doi: 10.1200/JCO.2003.05.044. [DOI] [PubMed] [Google Scholar]
- 15.Westra JL, Schaapveld M, Hollema H, et al. Determination of TP53 mutation is more relevant than microsatellite instability status for the prediction of disease-free survival in adjuvant-treated stage III colon cancer patients. J Clin Oncol. 2005;23:5635–5643. doi: 10.1200/JCO.2005.04.096. [DOI] [PubMed] [Google Scholar]
- 16.Lenz HJ, Danenberg KD, Leichman CG, et al. p53 and thymidylate synthase expression in untreated stage II colon cancer: Associations with recurrence, survival, and site. Clin Cancer Res. 1998;4:1227–1234. [PubMed] [Google Scholar]
- 17.Johnston PG, Lenz HJ, Leichman CG, et al. Thymidylate synthase gene and protein expression correlate and are associated with response to 5-fluorouracil in human colorectal and gastric tumors. Cancer Res. 1995;55:1407–1412. [PubMed] [Google Scholar]
- 18.Wang S, El-Deiry WS. Inducible silencing of KILLER/DR5 in vivo promotes bioluminescent colon tumor xenograft growth and confers resistance to chemotherapeutic agent 5-fluorouracil. Cancer Res. 2004;64:6666–6672. doi: 10.1158/0008-5472.CAN-04-1734. [DOI] [PubMed] [Google Scholar]
- 19.van Geelen CM, Westra JL, de Vries EG, et al. Prognostic significance of tumor necrosis factor-related apoptosis-inducing ligand and its receptors in adjuvantly treated stage III colon cancer patients. J Clin Oncol. 2006;24:4998–5004. doi: 10.1200/JCO.2006.06.8809. [DOI] [PubMed] [Google Scholar]
- 20.McLornan DP, Barrett HL, Cummins R, et al. Prognostic significance of TRAIL signaling molecules in stage II and III colorectal cancer. Clin Cancer Res. 2010;16:3442–3451. doi: 10.1158/1078-0432.CCR-10-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hector S, Conlon S, Schmid J, et al. Apoptosome-dependent caspase activation proteins as prognostic markers in stage II and III colorectal cancer. Br J Cancer. 2012;106:1499–1505. doi: 10.1038/bjc.2012.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sinicrope FA, Sargent DJ. Molecular pathways: Microsatellite instability in colorectal cancer: Prognostic, predictive, and therapeutic implications. Clin Cancer Res. 2012;18:1506–1512. doi: 10.1158/1078-0432.CCR-11-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watanabe T, Wu TT, Catalano PJ, et al. Molecular predictors of survival after adjuvant chemotherapy for colon cancer. N Engl J Med. 2001;344:1196–1206. doi: 10.1056/NEJM200104193441603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sinicrope FA, Foster NR, Thibodeau SN, et al. DNA mismatch repair status and colon cancer recurrence and survival in clinical trials of 5-fluorouracil-based adjuvant therapy. J Natl Cancer Inst. 2011;103:863–875. doi: 10.1093/jnci/djr153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roth AD, Tejpar S, Yan P, et al. Stage-specific prognostic value of molecular markers in colon cancer: Results of the translational study on the PETACC 3-EORTC 40993-SAKK 60–00 trial. J Clin Oncol. 2009;27(suppl):4002a. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 26.Sargent DJ, Marsoni S, Monges G, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28:3219–3226. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sinicrope FA. DNA mismatch repair and adjuvant chemotherapy in sporadic colon cancer. Nat Rev Clin Oncol. 2010;7:174–177. doi: 10.1038/nrclinonc.2009.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelley RK, Venook AP. Prognostic and predictive markers in stage II colon cancer: Is there a role for gene expression profiling? Clin Colorectal Cancer. 2011;10:73–80. doi: 10.1016/j.clcc.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gray RG, Quirke P, Handley K, et al. Validation study of a quantitative multigene reverse transcriptase-polymerase chain reaction assay for assessment of recurrence risk in patients with stage II colon cancer. J Clin Oncol. 2011;29:4611–4619. doi: 10.1200/JCO.2010.32.8732. [DOI] [PubMed] [Google Scholar]
- 30.Yothers G, O’Connell MJ, Lee M, et al. Validation of the 12-gene Colon Cancer Recurrence Score in NSABP C-07 as a predictor of recurrence in patients with stage II and III colon cancer treated with fluorouracil and leucovorin (FU/LV) and FU/LV plus oxaliplatin. J Clin Oncol. 2013;31:4512–4519. doi: 10.1200/JCO.2012.47.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kennedy RD, Bylesjo M, Kerr P, et al. Development and independent validation of a prognostic assay for stage II colon cancer using formalin-fixed paraffin-embedded tissue. J Clin Oncol. 2011;29:4620–4626. doi: 10.1200/JCO.2011.35.4498. [DOI] [PubMed] [Google Scholar]
- 32.Allen WL, Jithesh PV, Oliver GR, et al. The colorectal cancer disease-specific transcriptome may facilitate the discovery of more biologically and clinically relevant information. BMC Cancer. 2010;10:687. doi: 10.1186/1471-2407-10-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niedzwiecki D, Frankel W, Venook AP, et al. Association between ColDx assay result and recurrence-free interval in stage II colon cancer patients on CALGB (Alliance) 9581. J Clin Oncol. 2014;32(suppl 3):455a. doi: 10.1200/JCO.2015.65.4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salazar R, Roepman P, Capella G, et al. Gene expression signature to improve prognosis prediction of stage II and III colorectal cancer. J Clin Oncol. 2011;29:17–24. doi: 10.1200/JCO.2010.30.1077. [DOI] [PubMed] [Google Scholar]
- 35.Salazar R, Capdevila J, Rosenberg R, et al. Comparison of ColoPrint risk classification with clinical risk in the prospective PARSC trial. J Clin Oncol. 2014;32(suppl 3):465a. [Google Scholar]
- 36.Agesen TH, Sveen A, Merok MA, et al. ColoGuideEx: A robust gene classifier specific for stage II colorectal cancer prognosis. Gut. 2012;61:1560–1567. doi: 10.1136/gutjnl-2011-301179. [DOI] [PubMed] [Google Scholar]
- 37.Sveen A, Ågesen TH, Nesbakken A, et al. ColoGuidePro: A prognostic 7-gene expression signature for stage III colorectal cancer patients. Clin Cancer Res. 2012;18:6001–6010. doi: 10.1158/1078-0432.CCR-11-3302. [DOI] [PubMed] [Google Scholar]
- 38.Siena S, Sartore-Bianchi A, Di Nicolantonio F, et al. Biomarkers predicting clinical outcome of epidermal growth factor receptor-targeted therapy in metastatic colorectal cancer. J Natl Cancer Inst. 2009;101:1308–1324. doi: 10.1093/jnci/djp280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andreyev HJ, Norman AR, Cunningham D, et al. Kirsten ras mutations in patients with colorectal cancer: The multicenter “RASCAL” study. J Natl Cancer Inst. 1998;90:675–684. doi: 10.1093/jnci/90.9.675. [DOI] [PubMed] [Google Scholar]
- 40.Au HJ, Karapetis CS, O’Callaghan CJ, et al. Health-related quality of life in patients with advanced colorectal cancer treated with cetuximab: Overall and KRAS-specific results of the NCIC CTG and AGITG CO.17 trial. J Clin Oncol. 2009;27:1822–1828. doi: 10.1200/JCO.2008.19.6048. [DOI] [PubMed] [Google Scholar]
- 41.Allegra CJ, Jessup JM, Somerfield MR, et al. American Society of Clinical Oncology provisional clinical opinion: Testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091–2096. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- 42.Bokemeyer C, Van Cutsem E, Rougier P, et al. Addition of cetuximab to chemotherapy as first-line treatment for KRAS wild-type metastatic colorectal cancer: Pooled analysis of the CRYSTAL and OPUS randomised clinical trials. Eur J Cancer. 2012;48:1466–1475. doi: 10.1016/j.ejca.2012.02.057. [DOI] [PubMed] [Google Scholar]
- 43.Loupakis F, Ruzzo A, Cremolini C, et al. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer. 2009;101:715–721. doi: 10.1038/sj.bjc.6605177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Di Nicolantonio F, Martini M, Molinari F, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 45.Laurent-Puig P, Cayre A, Manceau G, et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol. 2009;27:5924–5930. doi: 10.1200/JCO.2008.21.6796. [DOI] [PubMed] [Google Scholar]
- 46.Scholl C, Fröhling S, Dunn IF, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137:821–834. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 47.Hudson TJ, Anderson W, Artez A, et al. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]