Abstract
Background
The relationship between uncoupling protein (UCP) 1–3 polymorphisms and susceptibility to obesity has been investigated in several genetic studies. However, the impact of these polymorphisms on obesity is still under debate, with contradictory results being reported. Until this date, no meta-analysis evaluated the association of UCP polymorphisms with body mass index (BMI) variability. Thus, this paper describe a meta-analysis conducted to evaluate if the -3826A/G (UCP1); -866G/A, Ala55Val and Ins/Del (UCP2) and -55C/T (UCP3) polymorphisms are associated with BMI changes.
Methods
A literature search was run to identify all studies that investigated associations between UCP1-3 polymorphisms and BMI. Weighted mean differences (WMD) were calculated for different inheritance models.
Results
Fifty-six studies were eligible for inclusion in the meta-analysis. Meta-analysis results showed that UCP2 55Val/Val genotype was associated with increased BMI in Europeans [Random Effect Model (REM) WMD 0.81, 95% CI 0.20, 1.41]. Moreover, the UCP2 Ins allele and UCP3-55T/T genotype were associated with increased BMI in Asians [REM WMD 0.46, 95% CI 0.09, 0.83 and Fixed Effect Model (FEM) WMD 1.63, 95% CI 0.25, 3.01]. However, a decreased BMI mean was observed for the UCP2-866 A allele in Europeans under a dominant model of inheritance (REM WMD −0.18, 95% CI −0.35, −0.01). There was no significant association of the UCP1-3826A/G polymorphism with BMI mean differences.
Conclusions
The meta-analysis detected a significant association between the UCP2-866G/A, Ins/Del, Ala55Val and UCP3-55C/T polymorphisms and BMI mean differences.
Introduction
Obesity is the most common nutritional disease in industrialized countries, constituting a priority health problem. It is associated with the development of cardiovascular diseases, type 2 diabetes mellitus and various cancers, which leads to higher morbidity and mortality rates, reducing the quality and expectancy of life of sufferers [1]. Obesity is characterized by an excessive accumulation of body fat resulting from an imbalance between energy intake and expenditure [2]. This imbalance can be due to genetic or environmental risk factors, or more likely due to a combination of both, an intertwining of genetics and environment [3]. A general consensus is that about 65% of variation in obesity is genetic [4], and candidate genes include those that control energy expenditure and thermogenesis, such as those encoding mitochondrial uncoupling proteins (UCPs) [5].
Uncoupling proteins 1, 2 and 3 are members of an anion-carrier protein family located in the mitochondrial inner membrane [5]–[7]. These proteins share structural similarities, but have different tissue expressions [7]. The original UCP, UCP1, is mainly expressed in brown adipose tissue [8], [9]. Uncoupling protein 2 is distributed across a wide range of tissue and cell types, whereas UCP3 is mainly restricted to skeletal muscle [9]. Several studies have shown that UCPs decrease metabolic efficiency by uncoupling substrate oxidation in mitochondria from ATP synthesis by the mitochondrial respiratory chain. This is probably accomplished by promoting net translocation of protons from the intermembrane space, across the inner mitochondrial membrane, to the mitochondrial matrix, thereby dissipating the potential energy available for ATP synthesis, and consequently, decreasing ATP production. This uncoupling effect leads to homologue- and tissue-specific functions, such as thermogenesis and energy expenditure (UCP1), regulation of free-fatty acids (FFAs) metabolism (UCP2 and UCP3), reduction in ROS formation (UCP1-3), and regulation of ATP-dependent processes (UCP2) [5], [6], [9], [10].
The relationship between UCP1-3 loci and susceptibility to obesity has been investigated in several genetic studies and particular attention has been focused on the -3826A/G (rs1800592) polymorphism in the UCP1 gene, the -866G/A polymorphism (rs659366), the Ala55Val (C/T; rs660339) polymorphism and the Ins/Del polymorphism in the UCP2 gene, and the -55C/T (rs1800849) polymorphism in the UCP3 gene [5], [10], [11]. However, the impact of these polymorphisms on obesity susceptibility is still under debate, with contradictory results being reported. In this context, Qian et al. [12] evaluated the association of the UCP2-886G/A, UCP2 Ala55Val, and UCP3-55C/T polymorphisms with obesity in a meta-analysis of 22 articles. The meta-analysis revealed that the UCP2-866G/A polymorphism may be a risk factor for obesity in Europeans, but not in Asian subjects. In agreement with this data, Andersen et al. [13] published a meta-analysis showing an association between the UCP2-866G/A polymorphism and obesity in subjects of European descent. Until this date, no meta-analysis evaluated the association of UCP polymorphisms with body mass index (BMI) variability. Thus, to further investigate the potential role of UCP1-3 polymorphisms in influencing BMI changes, we performed a systematic review and meta-analysis of the literature on the subject.
Materials and Methods
Search Strategy, Eligibility Criteria and Data Extraction
This study was designed and described in accordance with current guidelines for execution of systematic reviews and meta-analyses [14], [15]. Both PubMed and Embase repositories were searched systemically to identify all available genetic studies which analyzed associations between obesity and the most-often studied polymorphisms of UCP1-3 genes: -3826A/G (rs1800592) polymorphism in the promoter region of the UCP1 gene, -866G/A (rs659366) polymorphism in the promoter region, the Ala55Val (C/T; rs660339) polymorphism in exon 4 and the Ins/Del polymorphism, which is an insertion/deletion of 45 bp in the 3′ untranslated region (3′UTR) of exon 8 of the UCP2 gene, and the -55C/T (rs1800849) polymorphism in the promoter region of the UCP3 gene. The following medical subject headings (MeSH) were searched: (‘‘Obesity’’ OR “Body mass index”) AND (‘‘mitochondrial uncoupling protein’’ OR ‘‘SLC25A27 protein, human’’ OR ‘‘mitochondrial uncoupling protein 2″ OR ‘‘mitochondrial uncoupling protein 3″) AND (‘‘mutation’’ OR ‘‘frameshift mutation’’ OR ‘‘germ-line mutation’’ OR ‘‘INDEL mutation’’ OR ‘‘mutation, missense’’ OR ‘‘point mutation’’ OR ‘‘codon, nonsense’’ OR ‘‘sequence deletion’’ OR ‘‘polymorphism, genetic’’ OR ‘‘polymorphism, single nucleotide’’ OR ‘‘polymorphism, restriction fragment length’’). The search was limited to human studies and English or Spanish language papers and was completed on January, 2014. All of the articles identified were also searched manually to identify any other relevant citations.
Two investigators (B.M.S and A.P.B.) independently reviewed the titles and abstracts of all articles selected in order to evaluate whether the studies were eligible for inclusion in the meta-analysis. Disagreements were resolved by discussion between them and when necessary a third reviewer (D.C.) was consulted. When abstracts did not provide sufficient information to fulfill the inclusion and exclusion criteria, the full text of the article was retrieved for evaluation. We included observational studies that compared BMI (mean ± SD) among different genotypes of one or more of the UCP1-3 polymorphisms in question. Studies were excluded from the analysis if they did not have sufficient data or if they did not employ validated genotyping methods. If data were duplicated and had been published more than once, the most complete study was chosen.
Data were independently extracted by two investigators (L.A.B. and T.S.A.) using a standardized abstraction form, and consensus was sought in all extracted items. When consensus could not be reached, differences in data extraction were resolved by referencing the original publication or by consulting a third reviewer (D.C.). The information extracted from each individual study was as follows: name of first author, publication year, ethnicity, number of subjects, age, gender, and BMI in different genotypes of UCP1-3 polymorphisms.
Statistical Analyses for Meta-analyses
Subjects’ genotype distributions were tested for conformity with Hardy-Weinberg Equilibrium (HWE) using a goodness-of-fitness χ2 test. Some articles only reported the mean BMI according to the different genotypes after stratification by sex or other characteristic such as presence of diabetes or obesity, so we had to calculate the pooled mean BMI to include this information in our meta-analysis. Heterogeneity among studies was tested using a χ2-based Cochran’s Q statistic and inconsistency was assessed with the I2 metric [16], [17]. Heterogeneity was considered statistically significant at P<0.10 for the Q statistic and I2>50% for the I2 metric statistics. Where significant heterogeneity was detected, the DerSimonian and Laird random effect model (REM) was used to calculate weighted mean differences (95% CI) in BMI for each individual study and for the pooled effect; where heterogeneity was not significant, the fixed effect model (FEM) was used for this calculation [16], [17]. Weighted mean differences (95% CI) in BMI were evaluated for additive, recessive and dominant models of inheritance [18], [19].
Meta-regression and sensitivity analyses were carried out to identify key studies with a substantial impact on inter-study heterogeneity. The factors investigated by meta-regression were age, gender and ethnicity. Sensitivity analyses were performed after stratifying the studies by ethnicity, given that the UCP polymorphisms show variable frequencies across ethnic groups. Risk of publication bias was assessed using funnel plot graphics, analyzed both visually and with the Begg and Egger test [20]. The significance of the intercept was determined by the t test, as proposed by Egger, with P<0.10 considered indicative of statistically significant publication bias. All statistical analyses were performed using Stata 11.0 software (StataCorp, College Station, TX, USA).
Results
Literature Search and Characteristics of Eligible Studies
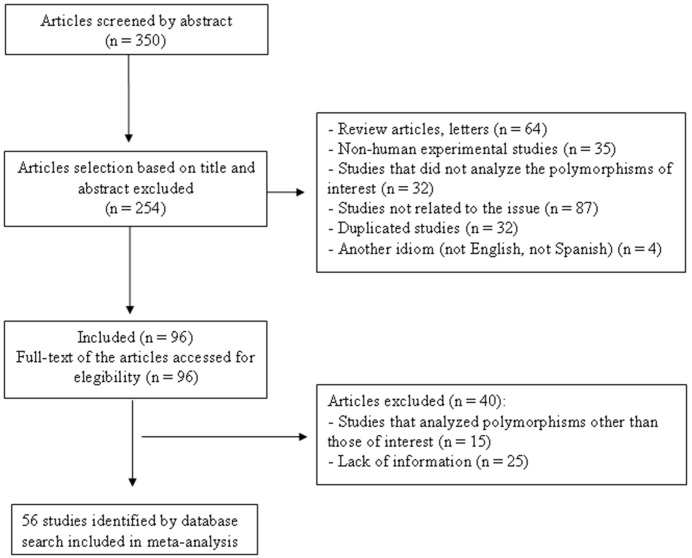
Figure 1 is a flow diagram illustrating the strategy used to identify and select studies for inclusion in the meta-analysis. A total of 350 potentially relevant citations were retrieved by searching the electronic databases and 254 of them were excluded during the review of titles and abstracts. Ninety-six articles therefore appeared to be eligible at this point and their full texts were evaluated. However, after careful analysis of the full texts, another 40 studies were excluded because of missing information, ineligible study designs or because they genotyped other UCP polymorphisms, but not the ones of interest here. A total of 56 articles fulfilled the eligibility criteria and were included in the meta-analyses [21]–[77]. The available characteristics of studies included in primary analyses are shown in Table S1. The median age of analyzed subjects was 46.0 (minimum 8.4–maximum 65.5 values) years, and the median BMI was 37.0 (20.3–50.2). Sixty-one percent of the subjects were men, 25.3% had diabetes, and 57.1% had obesity. Most of the studies were performed in European populations.
Figure 1. Flowchart illustrating the search strategy used to identify association studies of UCP1-3 polymorphisms with body mass index changes for inclusion in the meta-analysis.
Table S2 shows the mean BMI ± SD according to the different UCP1-3 genotypes in all studies included in the meta-analysis. Twenty studies analyzed the UCP1-3826A/G polymorphism (6191 subjects), 18 studies analyzed the UCP2-886G/A polymorphism (12072 subjects), 6 analyzed the UCP2 Ala55Val polymorphism (15060 subjects), 8 analyzed the UCP2 Ins/Del polymorphism (8490 subjects), and 8 investigated the UCP3-55C/T polymorphism (6663 subjects).
Quantitative Synthesis
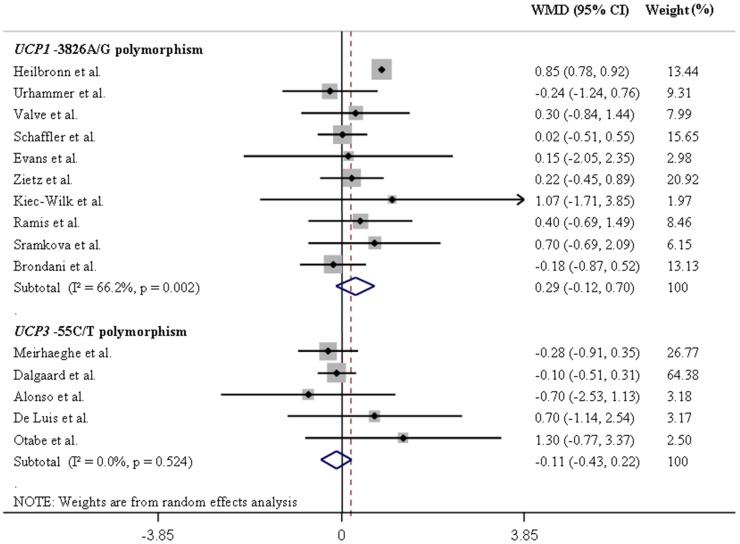
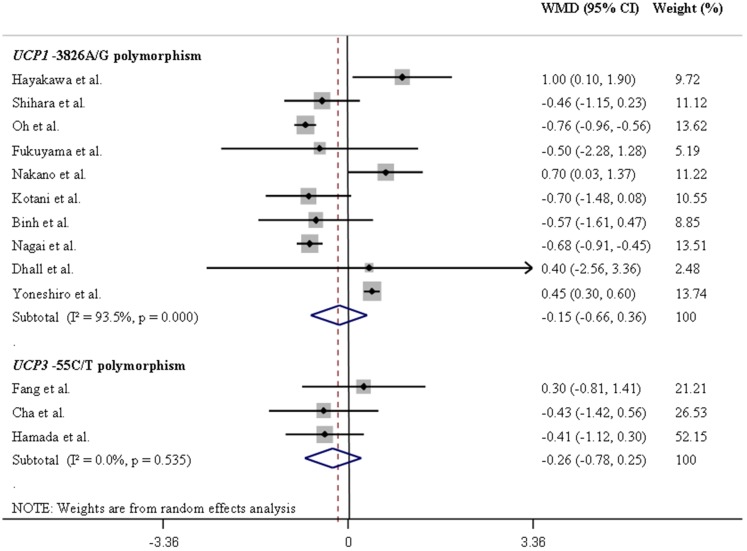
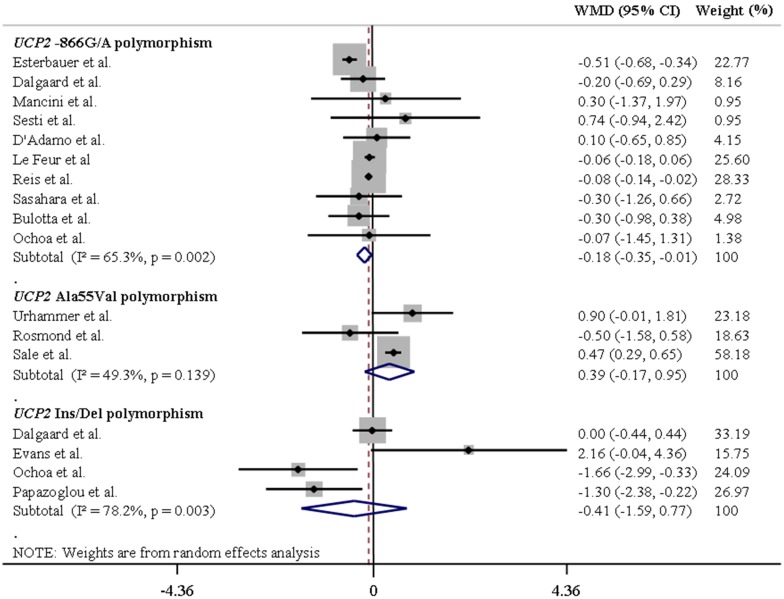
Table 1 summarizes the results of the pooled analyses for the associations between UCP polymorphisms and differences in mean BMI values. Figure 2 and Figure 3 illustrate pooled WMD in BMI values for the UCP1-3826A/G and UCP3-55C/T polymorphisms in Europeans and Asians, respectively, assuming a dominant inheritance model. Figure 4 and Figure 5 show pooled WMD in BMI for the UCP2-866G/A, Ala55Val and Ins/Del polymorphisms in Europeans and Asians, respectively, also under a dominant model.
Table 1. Pooled measures for associations between UCP1-3826A/G, UCP2-866G/A, UCP2 Ala55Val, UCP2 Ins/Del and UCP3-55C/T polymorphisms and body mass index mean differences.
| Inheritance model | n studies | n patients | I2 % | WMD (95% CI) | Sensitivity analysis | |
| I2 % | WMD (95% CI) | |||||
| UCP1- 3826 A/G | ||||||
| Additive | 17 | 3578 | 93.8 | 0.07 (−0.48, 0.63) | ||
| Asian | 9 | 888 | 95.0 | −0.11 (−0.78, 0.59) | ||
| European | 8 | 2690 | 76.7 | 0.34 (−0.61, 1.29) | 0.0 | −0.02 (−0.58, 0.53)a |
| Recessive | 17 | 5909 | 91.0 | 0.12 (−0.30, 0.54) | ||
| Asian | 9 | 1620 | 94.1 | 0.00 (−0.53, 0.54) | ||
| European | 8 | 4289 | 68.4 | 0.29 (−0.51, 1.09) | 0.0 | −0.02 (−0.56, 0.53)a |
| Dominant | 20 | 6191 | 93.3 | 0.05 (−0.33, 0.42) | ||
| Asian | 10 | 1713 | 93.5 | −0.15 (−0.66, 0.36) | ||
| European | 10 | 4478 | 59.1 | 0.29 (−0.12, 0.70) | 0.0 | 0.09 (−0.20, 0.39)a |
| UCP2- 866 G/A | ||||||
| Additive | 16 | 7125 | 99.4 | −0.07 (−0.52, 0.37) | ||
| Asian | 6 | 1440 | 72.9 | 0.37 (−0.47, 1.22) | ||
| European | 10 | 5685 | 98.6 | −0.30 (−1.02, 0.41) | ||
| Recessive | 17 | 12455 | 99.4 | −0.16 (−1.07, 0.76) | ||
| Asian | 6 | 2794 | 98.7 | −1.07 (−4.39, 2.25) | ||
| European | 11 | 9661 | 98.7 | 0.44 (−0.27, 1.15) | ||
| Dominant | 17 | 10471 | 85.3 | −0.07 (−0.26, 0.11) | ||
| Asian | 7 | 2894 | 71.0 | 0.14 (−0.50, 0.78) | ||
| European | 10 | 7577 | 65.3 | −0.18 (−0.35, −0.01) | 0.0 | −0.08 (−0.13, −0.02)b |
| UCP2 I/D | ||||||
| Additive | 5 | 5796 | 68.4 | 0.22 (−1.20, 1.64) | ||
| European | 3 | 5072 | 82.3 | −0.07 (−2.79, 2.66) | ||
| Recessive | 5 | 5796 | 53.8 | 0.19 (−0.94, 1.31) | 0.0 | 0.67 (−0.06, 1.29)c |
| European | 3 | 5072 | 76.2 | −0.05 (−2.31, 2.21) | 0.0 | 0.75 (−0.02, 1.52)c |
| Dominant | 8 | 8490 | 78.1 | 0.24 (−0,41, 0.89) | ||
| Asian | 3 | 3508 | 15.5 | 0.46 (0.09, 0.83) | ||
| European | 4 | 4893 | 78.2 | −0.41 (−1.59, 0.77) | ||
| UCP2 Ala55val | ||||||
| Additive | 5 | 7488 | 92.7 | 0.39 (−0.32, 1.10) | ||
| European | 3 | 894 | 70.2 | 0.74 (−0.24, 1.72) | ||
| Recessive | 6 | 17002 | 96.6 | 0.48 (0.01, 0.95) | ||
| European | 4 | 3974 | 97.0 | 0.81 (0.20, 1.41) | ||
| Dominant | 6 | 15060 | 72.4 | 0.27 (−0.06, 0.59) | ||
| Asian | 2 | 1276 | 55.1 | 0.46 (−0.52, 1.44) | ||
| European | 3 | 1728 | 49.3 | 0.39 (−0.17, 0.95) | ||
| UCP3- 55C/T | ||||||
| Additive | 6 | 3568 | 71.9 | 0.46 (−0.77, 1.68) | ||
| Asian | 3 | 434 | 81.7 | 0.32 (−2.07, 2.71) | 0.0 | 1.46 (0.02, 2.90)c |
| European | 3 | 3134 | 64.6 | 0.62 (−0.92, 2.16) | 0.0 | 0.26 (−0.43, 0.94)d |
| Recessive | 6 | 6131 | 73.1 | 0.54 (−0.67, 1.76) | ||
| Asian | 3 | 796 | 83.8 | 0.46 (−1.98, 2.90) | 0.0 | 1.63 (0.25, 3.01)c |
| European | 3 | 5335 | 61.8 | 0.62 (−0.83, 2.07) | 0.0 | 0.29 (−0.38, 0.96)d |
| Dominant | 8 | 6663 | 0.0 | −0.15 (−0.43, 0.12) | ||
| Asian | 3 | 796 | 0.0 | −0.26 (−0.78, 0.25) | ||
| European | 5 | 5867 | 0.0 | −0.11 (−0.43, 0.22) | ||
Figure 2. Forest plots showing individual and pooled weighted mean differences (95% CI) for the associations between UCP1-3826A/G and UCP3-55C/T polymorphisms and body mass index in Europeans, under a dominant inheritance model (A/A vs. G/G + A/G for the UCP1-3826A/G polymorphism, and C/C vs. T/T + C/T for the UCP3-55C/T polymorphism).
The area of the squares reflects the study-specific weight. The diamond shows the summary random-effect weighted mean differences estimated from the studies.
Figure 3. Forest plots showing individual and pooled weighted mean differences (95% CI) for the associations between UCP1-3826A/G and UCP3-55C/T polymorphisms and body mass index in Asians, under a dominant inheritance model (A/A vs. G/G + A/G for the UCP1-3826A/G polymorphism, and C/C vs. T/T + C/T for the UCP3-55C/T polymorphism).
The area of the squares reflects the study-specific weight. The diamond shows the summary random-effect weighted mean differences estimated from the studies.
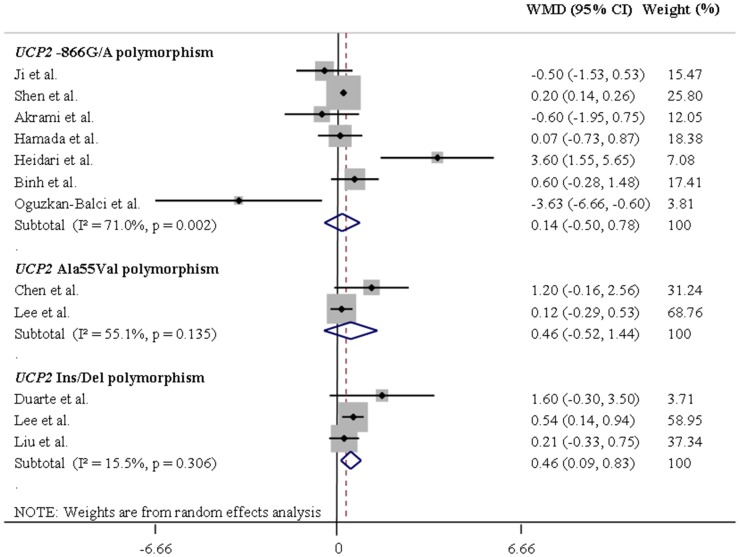
Figure 4. Forest plots showing individual and pooled weighted mean differences (95% CI) for the associations between UCP2-866G/A, Ala55Val and Ins/Del polymorphisms and body mass index in Europeans, under a dominant inheritance model (G/G vs. A/A + G/A for the UCP2-866G/A polymorphism, Ala/Ala vs. Val/Val + Ala/Val for the UCP2 Ala55Val polymorphism, and Del/Del vs. Ins/Ins+Ins/Del for the UCP2 Ins/Del polymorphism).
The area of the squares reflects the study-specific weight. The diamond shows the summary random-effect weighted mean differences estimated from the studies.
Figure 5. Forest plots showing individual and pooled weighted mean differences (95% CI) for the associations between UCP2-866G/A, Ala55Val and Ins/Del polymorphisms and body mass index in Asians, under a dominant inheritance model (G/G vs. A/A + G/A for the UCP2-866G/A polymorphism, Ala/Ala vs. Val/Val + Ala/Val for the UCP2 Ala55Val polymorphism, and Del/Del vs. Ins/Ins + Ins/Del for the UCP2 Ins/Del polymorphism).
The area of the squares reflects the study-specific weight. The diamond shows the summary random-effect weighted mean differences estimated from the studies.
UCP1-3826A/G and UCP3-55C/T polymorphisms were not associated with any change in BMI irrespectively of the inheritance model or after stratification by ethnicity ( Table 1 ). The A allele of the UCP2-866G/A polymorphism was significantly associated with decreased WMD in BMI in Europeans (REM WMD = −0.18, 95% CI −0.35 – −0.01, for the dominant model) but not in Asians. On the other hand, the Ins allele of the UCP2 Ins/Del polymorphism was associated with increased WMD in BMI in Asians (REM WMD = 0.46, 95% CI 0.09−0.83, dominant model) but not in Europeans. The UCP2 Ala55Val polymorphism was also associated with increased WMD in BMI in Europeans, under a recessive model (REM WMD = 0.81, 95% CI 0.20−1.41; Ala/Ala + Ala/Val vs. Val/Val).
As can be observed in Table 1 , there were significant heterogeneities (I2>50%) among studies in almost all comparisons of BMI according to the UCP1-3 genotypes under the different inheritance models. To investigate this finding in greater depth, sex, age and ethnicity were used as covariates in meta-regression analyses performed for the five UCP polymorphisms under different inheritance models. None of the covariates used in univariate and multivariate meta-regression analyses could individually explain the heterogeneities observed (data not shown).
Sensitivity analyses were carried out in order to estimate the influence of each individual study in the meta-analysis results. This was performed by repeating the meta-analyses omitting a different study each time. No individual study significantly influenced the inter-study heterogeneity for the UCP2 Ala55Val polymorphism (data not shown). In contrast, some studies seem to be responsible for the inter-study heterogeneity observed in the meta-analyses of UCP1-3826A/G, UCP2-866G/A and Ins/Del, and UCP3-55C/T polymorphisms ( Table 1 ). Exclusion of these studies from the respective meta-analysis abolished the heterogeneity; however, the pooled WMD in BMI only was significantly altered after the exclusion of one specific study [70] from the analyses of the UCP3-55C/T polymorphism in Asians (WMD = 1.46, 95% CI 0.02−2.90, for the additive model; and WMD = 1.63, 95% CI 0.25−3.01, for the recessive model; Table 1 ).
Importantly, funnel plots and Egger’s tests were performed to detect publication bias of the literature, as illustrated in Figures S1, S2 and S3. Symmetrical funnel plots were generated for each of the five UCP polymorphisms tested in the additive, dominant and recessive inheritance models. Egger’s tests further confirmed no publication bias for any of the UCP polymorphisms analyzed, which suggests that our data are statistically reliable.
Discussion
Uncoupling protein 1, UCP2 and UCP3 are candidate genes for obesity because they decrease mitochondrial membrane potential and mediate proton leak [5], [8], [9]. Mutations reducing the activity of these proteins could reduce energy expenditure by increasing coupling of oxidative phosphorylation, thereby contributing to BMI changes. These are the reasons why the roles played by UCP1-3826A/G, UCP2-866G/A, UCP2 Ala55Val, UCP2 Ins/Del and UCP3-55C/T polymorphisms in obesity risk and BMI changes have been extensively studied, although the results of these associations are still inconclusive (reviewed in [5], [6], [78]). Therefore, to further investigate if these UCP polymorphisms are associated with changes in BMI, we conducted a meta-analysis of 56 published articles from different populations.
Our results suggest that the UCP1-3826A/G polymorphism is not associated with BMI changes. The A allele of the UCP2-866G/A polymorphism was associated with a 0.18 reduction in BMI in Europeans. On the other hand, the Ins allele of the UCP2 Ins/Del polymorphism was associated with an increase of 0.46 units in BMI in Asians, while the Val/Val genotype of the UCP2 Ala55Val polymorphism was associated with an increase of 0.81 units in BMI in Europeans under a recessive model of inheritance. Moreover, the T/T genotype of the UCP3-55C/T polymorphism was associated with increased of 1.6 units in BMI in Asians but only after sensitivity analysis. These results show that associations of UCP2 and UCP3 polymorphisms with BMI changes are probably under an important influence of ethnicity. This may be explained by known differences in lifestyle and body weight distributions between Asian and European populations as well as by differences in the genotype frequencies of the analyzed polymorphisms between ethnicities. In this context, Luan et al. [79] found that effects of genetic polymorphisms on obesity and related-disorders could be changed by nutritional characteristic of the population. Thus, it might be possible that different diet patterns between European and Asian populations could modulate the effect of UCP polymorphisms in BMI changes.
It is worth noting that two previous meta-analyses investigated one or more of the UCP polymorphisms included in our meta-analysis, although only regarding their associations with susceptibility to obesity. Qian [12] et al. included 22 studies in the meta-analysis, and their results support an association between the A allele of the UCP2-866G/A polymorphism and protection for obesity in Europeans, which is in agreement with our data. However, they were not able to find any association of the UCP2 Ala55Val and UCP3-55C/T polymorphisms with obesity, which is in conflict with our data. One possible explanation for this discrepancy is that the outcome of Qian et al. [12] was presence of obesity while our outcome was BMI. It is reasonable to suppose that UCP polymorphisms may be associated with small changes in BMI; therefore, analyzing BMI changes would have a higher power to detect smaller associations with obesity. Furthermore, our meta-analysis included 56 studies, whereas the meta-analysis of Qian et al. [12] included only 22 studies. Andersen et al. [13] performed a meta-analysis of 8 studies that analyzed the association between the UCP2-886G/A polymorphism and obesity. In agreement with our data, they reported an association between the UCP2-866A allele and protection for obesity in Europeans. Moreover, they also observed an association between UCP2-866G/A polymorphism and insulin resistance among 5781 middle-aged Danes and 377 young health Danes, where subjects carrying the G allele consistently had lower insulin sensitivity [13].
The UCP2-866G/A polymorphism seems to be involved in putative binding sites for specific transcription factors and, in fact, several studies have shown that this promoter variant changes reporter gene activity [39], [77]. However, data in human tissues are conflicting, reporting either increased or decreased UCP2 mRNA levels being associated with the -866A allele (reviewed in [6]). In differentiated adipocytes, the A allele has a 22% more effective transcriptional activity [39]; therefore, the association of the UCP2-866A allele with decreased BMI in Europeans appears to be biologically plausible since an increased UCP2 gene expression in adipocytes would be associated with increased energy expenditure.
Our data indicate that the UCP2 Val55Val genotype is associated with the highest change in BMI (WMD = 0.81) in Europeans. However, this polymorphism causes a conservative amino acid change at position 55 of exon 4 and, until now, there had been no evidence that this alteration generates a functional change in the protein [6]. It is therefore possible that this polymorphism may not be a true disease-causing variant, but could simply be reflecting the effects of a functional polymorphism elsewhere in the UCP2/UCP3 loci. The biological significance of the UCP2 Ins/Del polymorphism is not well known. However, its location in the 3′UTR suggests that this polymorphism could alter the mRNA processing or the stability of the transcript. Any reduction in the UCP2 mRNA stability could compromise the ability to remove excess calories through thermogenesis, especially in a person with a propensity to obesity from other genetic or environmental influences [10]. This hypothesis is in agreement with our data showing that the UCP2 Ins/Del polymorphism is associated with increased BMI in Asians.
The UCP3-55C/T polymorphism is located 55 bp upstream of the most commonly used transcription initiation site of skeletal muscle [80], and thus appears to be functional. Here, this polymorphism was associated with a significant increase in BMI in Asians but only after sensitivity analysis. Thus, this data should be interpreted with caution since was based in only two studies. Further studies are still needed to elucidate the functional effects of the UCP3-55C/T polymorphism on UCP3 expression as well as to confirm its association with obesity. It is worth noting that a previous meta-analysis from our group showed an association of the UCP3-55C/T polymorphism with risk for type 2 diabetes mellitus in Asians (OR = 1.22, 95% CI 1.04–1.44, allele contrast model) [81], further indicating a role for this polymorphism in obesity-related features.
The results of the present meta-analysis should be interpreted within the context of a few limitations. First, meta-analysis is prone to publication bias, and although we have attempted to trace unpublished studies, we cannot be sure that small negative studies were overlooked. Moreover, we did not detect any significant publication bias in our analysis, showing that our data is statistically reliable. Second, because of the difficulty in getting the full texts of articles published in several languages, we only included studies published in English and Spanish. Third, inter-studies heterogeneity is common in meta-analyses for genetic association studies [82] and this can be a significant problem when interpreting their results. Our meta-analysis showed significant inter-study heterogeneity for most of the UCP polymorphisms analyzed. To investigate this problem in greater depth, meta-regression analyses were performed and showed that age, gender and ethnicity did not make significant contributions to inter-studies heterogeneity. The heterogeneity observed could be due to differences in sample selection, genotyping methods or gene-environment interactions and, without detailed information on the metabolic and clinical characteristics of the studies reviewed; we could not fully exclude the possibility that the heterogeneity observed might reduce our power to detect true associations. “Leave one out” sensitivity analyses showed that after excluding some studies, the heterogeneity for these analyses decreased significantly. However, despite the exclusion of these studies, only the pooled WMD obtained for the UCP3-55C/T polymorphism changed significantly: after the exclusion of Hamada et al. [70], the UCP3-55C/T polymorphism became associated with increase BMI in Asians. Fourth, obesity is a complex disorder that involves interactions of genes and environment. Many studies revealed different effects of the UCP1-3 polymorphisms depending on the physical activity and lifestyles [83]–[85]; however, we had insufficient data to take these confounder factors into account in our meta-analysis. Fifth, we also cannot rule out the possibility of type II error when analyzing associations of the UCP polymorphisms and BMI changes after stratifying by ethnicity. For the whole sample, we had at least 80% power (α = 0.05) to detect even modest changes in WMD for all analyzed polymorphisms, which is an evidence that our results are robust.
In conclusion, our results indicate that the UCP1-3826A/G polymorphism is not associated with significant changes in BMI. However, our results suggest that the UCP2-866A allele is associated with decreased BMI in Europeans, while the UCP2 Val55Val genotype is associated with increased BMI in Europeans. The UCP2 Ins allele and the UCP3-55T/T genotype seem to be associated with increased BMI in Asians. It is important to keep in mind that BMI is one phenotype of obesity; thus, we can not exclude the possibility that these polymorphisms could have different effects in other phenotypes, such as fat body mass index, waist circumference, and intra-abdominal adipose tissue. Since small sample sizes were obtained for some of the analyses performed in Asians, further additional larger studies that allow stratification for ethnicity and gene-gene and gene-environment interactions should also be conducted in order to elucidate the roles possibly played by UCP polymorphisms in obesity and BMI changes.
Supporting Information
Funnel plot for studies of the UCP1- 3826A/G polymorphism under a dominant model of inheritance.
(TIF)
Funnel plot for studies of the UCP2- 866G/A, Ala55Val and Ins/Del polymorphisms under a dominant model of inheritance.
(TIF)
Funnel plot for studies of the UCP3- 55C/T polymorphism under a dominant model of inheritance.
(TIF)
Characteristics of the studies included in the meta-analyses.
(XLS)
Body mass index means according to different genotypes of UCP1- 3826A/G, UCP2- 886G/A, UCP2 Ala55Val, UCP2 Ins/del and UCP3- 55C/T polymorphisms for the studies included in the meta-analysis.
(DOC)
PRISMA Checklist.
(DOC)
PRISMA Flow Diagram.
(DOC)
Funding Statement
This study was partially supported by grants from the Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS), the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), the Fundo de Incentivo à Pesquisa e Eventos (FIPE) at the Hospital de Clínicas de Porto Alegre and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Mitchell NS, Catenacci VA, Wyatt HR, Hill JO (2011) Obesity: overview of an epidemic. Psychiatr Clin North Am 34: 717–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moustafa JS, Froguel P (2014) From obesity genetics to the future of personalized obesity therapy. Nat Rev Endocrinol 10: 4. [DOI] [PubMed] [Google Scholar]
- 3. Drummond EM, Gibney ER (2013) Epigenetic regulation in obesity. Curr Opin Clin Nutr Metab Care 16: 392–397. [DOI] [PubMed] [Google Scholar]
- 4. Nan C, Guo B, Warner C, Fowler T, Barrett T, et al. (2012) Heritability of body mass index in pre-adolescence, young adulthood and late adulthood. Eur J Epidemiol 27: 247–253. [DOI] [PubMed] [Google Scholar]
- 5. Brondani LA, Assmann TS, Duarte GC, Gross JL, Canani LH, et al. (2012) The role of the uncoupling protein 1 (UCP1) on the development of obesity and type 2 diabetes mellitus. Arq Bras Endocrinol Metabol 56: 215–225. [DOI] [PubMed] [Google Scholar]
- 6. Souza BM, Assmann TS, Kliemann LM, Gross JL, Canani LH, et al. (2011) The role of uncoupling protein 2 (UCP2) on the development of type 2 diabetes mellitus and its chronic complications. Arq Bras Endocrinol Metabol 55: 239–248. [DOI] [PubMed] [Google Scholar]
- 7. Fisler JS, Warden CH (2006) Uncoupling proteins, dietary fat and the metabolic syndrome. Nutr Metab (Lond) 3: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Azzu V, Brand MD (2010) The on-off switches of the mitochondrial uncoupling proteins. Trends Biochem Sci 35: 298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ricquier D (2005) Respiration uncoupling and metabolism in the control of energy expenditure. Proc Nutr Soc 64: 47–52. [DOI] [PubMed] [Google Scholar]
- 10. Jia JJ, Zhang X, Ge CR, Jois M (2009) The polymorphisms of UCP2 and UCP3 genes associated with fat metabolism, obesity and diabetes. Obes Rev 10: 519–526. [DOI] [PubMed] [Google Scholar]
- 11. Jia JJ, Tian YB, Cao ZH, Tao LL, Zhang X, et al. (2010) The polymorphisms of UCP1 genes associated with fat metabolism, obesity and diabetes. Mol Biol Rep 37: 1513–1522. [DOI] [PubMed] [Google Scholar]
- 12. Qian L, Xu K, Xu X, Gu R, Liu X, et al. (2013) UCP2-866G/A, Ala55Val and UCP3-55C/T Polymorphisms in Association with Obesity Susceptibility - A Meta-Analysis Study. PLoS One 8: e58939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Andersen G, Dalgaard LT, Justesen JM, Anthonsen S, Nielsen T, et al. (2013) The frequent UCP2-866G>A polymorphism protects against insulin resistance and is associated with obesity: a study of obesity and related metabolic traits among 17 636 Danes. Int J Obes (Lond) 37: 175–181. [DOI] [PubMed] [Google Scholar]
- 14.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med 151: 264–269, W264. [DOI] [PubMed]
- 15. Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, et al. (2000) Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA 283: 2008–2012. [DOI] [PubMed] [Google Scholar]
- 16. Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ 327: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Higgins JP, Thompson SG (2002) Quantifying heterogeneity in a meta-analysis. Stat Med 21: 1539–1558. [DOI] [PubMed] [Google Scholar]
- 18. Minelli C, Thompson JR, Abrams KR, Thakkinstian A, Attia J (2005) The choice of a genetic model in the meta-analysis of molecular association studies. Int J Epidemiol 34: 1319–1328. [DOI] [PubMed] [Google Scholar]
- 19. Zintzaras E, Lau J (2008) Synthesis of genetic association studies for pertinent gene-disease associations requires appropriate methodological and statistical approaches. J Clin Epidemiol 61: 634–645. [DOI] [PubMed] [Google Scholar]
- 20. Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Urhammer SA, Fridberg M, Sørensen TI, Echwald SM, Andersen T, et al. (1997) Studies of genetic variability of the uncoupling protein 1 gene in Caucasian subjects with juvenile-onset obesity. J Clin Endocrinol Metab 82: 4069–4074. [DOI] [PubMed] [Google Scholar]
- 22. Valve R, Heikkinen S, Rissanen A, Laakso M, Uusitupa M (1998) Synergistic effect of polymorphisms in uncoupling protein 1 and beta3-adrenergic receptor genes on basal metabolic rate in obese Finns. Diabetologia 41: 357–361. [DOI] [PubMed] [Google Scholar]
- 23. Hayakawa T, Nagai Y, Taniguchi M, Yamashita H, Takamura T, et al. (1999) Phenotypic characterization of the beta3-adrenergic receptor mutation and the uncoupling protein 1 polymorphism in Japanese men. Metabolism 48: 636–640. [DOI] [PubMed] [Google Scholar]
- 24. Schäffler A, Palitzsch KD, Watzlawek E, Drobnik W, Schwer H, et al. (1999) Frequency and significance of the A–>G (-3826) polymorphism in the promoter of the gene for uncoupling protein-1 with regard to metabolic parameters and adipocyte transcription factor binding in a large population-based Caucasian cohort. Eur J Clin Invest 29: 770–779. [DOI] [PubMed] [Google Scholar]
- 25. Evans D, Minouchehr S, Hagemann G, Mann WA, Wendt D, et al. (2000) Frequency of and interaction between polymorphisms in the beta3-adrenergic receptor and in uncoupling proteins 1 and 2 and obesity in Germans. Int J Obes Relat Metab Disord 24: 1239–1245. [DOI] [PubMed] [Google Scholar]
- 26. Heilbronn LK, Kind KL, Pancewicz E, Morris AM, Noakes M, et al. (2000) Association of -3826 G variant in uncoupling protein-1 with increased BMI in overweight Australian women. Diabetologia 43: 242–244. [DOI] [PubMed] [Google Scholar]
- 27. Shihara N, Yasuda K, Moritani T, Ue H, Uno M, et al. (2001) Synergistic effect of polymorphisms of uncoupling protein 1 and beta3-adrenergic receptor genes on autonomic nervous system activity. Int J Obes Relat Metab Disord 25: 761–766. [DOI] [PubMed] [Google Scholar]
- 28. Zietz B, Watzlawek E, Palitzsch KD, Scholmerich J, Schaffler A (2001) GG-genotype in the promotor region of uncoupling-protein-1 gene is associated with lower level of dehydroepiandrosterone in type 2 diabetes. Exp Clin Endocrinol Diabetes 109: 102–106. [DOI] [PubMed] [Google Scholar]
- 29. Kieć-Wilk B, Wybrańska I, Malczewska-Malec M, Leszczyńska-Gołabek L, Partyka L, et al. (2002) Correlation of the -3826A>G polymorphism in the promoter of the uncoupling protein 1 gene with obesity and metabolic disorders in obese families from southern Poland. J Physiol Pharmacol 53: 477–490. [PubMed] [Google Scholar]
- 30. Ramis JM, González-Sánchez JL, Proenza AM, Martínez-Larrad MT, Fernández-Pérez C, et al. (2004) The Arg64 allele of the beta 3-adrenoceptor gene but not the -3826G allele of the uncoupling protein 1 gene is associated with increased leptin levels in the Spanish population. Metabolism 53: 1411–1416. [DOI] [PubMed] [Google Scholar]
- 31. Fukuyama K, Ohara T, Hirota Y, Maeda K, Kuno S, et al. (2006) Association of the -112A>C polymorphism of the uncoupling protein 1 gene with insulin resistance in Japanese individuals with type 2 diabetes. Biochem Biophys Res Commun 339: 1212–1216. [DOI] [PubMed] [Google Scholar]
- 32. Nakano T, Shinka T, Sei M, Sato Y, Umeno M, et al. (2006) A/G heterozygote of the A-3826G polymorphism in the UCP-1 gene has higher BMI than A/A and G/G homozygote in young Japanese males. J Med Invest 53: 218–222. [DOI] [PubMed] [Google Scholar]
- 33. Sramkova D, Krejbichova S, Vcelak J, Vankova M, Samalikova P, et al. (2007) The UCP1 gene polymorphism A-3826G in relation to DM2 and body composition in Czech population. Exp Clin Endocrinol Diabetes 115: 303–307. [DOI] [PubMed] [Google Scholar]
- 34. Kotani K, Sakane N, Saiga K, Adachi S, Shimohiro H, et al. (2008) Relationship between A-3826G polymorphism in the promoter of the uncoupling protein-1 gene and high-density lipoprotein cholesterol in Japanese individuals: a cross-sectional study. Arch Med Res 39: 142–146. [DOI] [PubMed] [Google Scholar]
- 35. Binh TQ, Nakahori Y, Hien VT, Khan NC, Lam NT, et al. (2011) Correlations between genetic variance and adiposity measures, and gene×gene interactions for obesity in postmenopausal Vietnamese women. J Genet 90: 1–9. [PubMed] [Google Scholar]
- 36. Nagai N, Sakane N, Tsuzaki K, Moritani T (2011) UCP1 genetic polymorphism (-3826 A/G) diminishes resting energy expenditure and thermoregulatory sympathetic nervous system activity in young females. Int J Obes (Lond) 35: 1050–1055. [DOI] [PubMed] [Google Scholar]
- 37. Dhall M, Chaturvedi MM, Rai U, Kapoor S (2012) Sex-dependent effects of the UCP1-3826 A/G polymorphism on obesity and blood pressure. Ethn Dis 22: 181–184. [PubMed] [Google Scholar]
- 38. Yoneshiro T, Aita S, Matsushita M, Kameya T, Nakada K, et al. (2011) Brown adipose tissue, whole-body energy expenditure, and thermogenesis in healthy adult men. Obesity (Silver Spring) 19: 13–16. [DOI] [PubMed] [Google Scholar]
- 39. Esterbauer H, Schneitler C, Oberkofler H, Ebenbichler C, Paulweber B, et al. (2001) A common polymorphism in the promoter of UCP2 is associated with decreased risk of obesity in middle-aged humans. Nat Genet 28: 178–183. [DOI] [PubMed] [Google Scholar]
- 40. Dalgaard LT, Andersen G, Larsen LH, Sørensen TI, Andersen T, et al. (2003) Mutational analysis of the UCP2 core promoter and relationships of variants with obesity. Obes Res 11: 1420–1427. [DOI] [PubMed] [Google Scholar]
- 41. Mancini FP, Sabatino L, Colantuoni V, Pasanisi F, Finelli C, et al. (2003) Variants of uncoupling protein-2 gene and obesity: interaction with peroxisome proliferator-activated receptorgamma2. Clin Endocrinol (Oxf) 59: 817–822. [DOI] [PubMed] [Google Scholar]
- 42. Sesti G, Cardellini M, Marini MA, Frontoni S, D’Adamo M, et al. (2003) A common polymorphism in the promoter of UCP2 contributes to the variation in insulin secretion in glucose-tolerant subjects. Diabetes 52: 1280–1283. [DOI] [PubMed] [Google Scholar]
- 43. D’Adamo M, Perego L, Cardellini M, Marini MA, Frontoni S, et al. (2004) The -866A/A genotype in the promoter of the human uncoupling protein 2 gene is associated with insulin resistance and increased risk of type 2 diabetes. Diabetes 53: 1905–1910. [DOI] [PubMed] [Google Scholar]
- 44. Ji Q, Ikegami H, Fujisawa T, Kawabata Y, Ono M, et al. (2004) A common polymorphism of uncoupling protein 2 gene is associated with hypertension. J Hypertens 22: 97–102. [DOI] [PubMed] [Google Scholar]
- 45. Le Fur S, Le Stunff C, Dos Santos C, Bougnères P (2004) The common -866 G/A polymorphism in the promoter of uncoupling protein 2 is associated with increased carbohydrate and decreased lipid oxidation in juvenile obesity. Diabetes 53: 235–239. [DOI] [PubMed] [Google Scholar]
- 46. Reis AF, Dubois-Laforgue D, Bellanne-Chantelot C, Timsit J, Velho G (2004) A polymorphism in the promoter of UCP2 gene modulates lipid levels in patients with type 2 diabetes. Mol Genet Metab 82: 339–344. [DOI] [PubMed] [Google Scholar]
- 47. Bulotta A, Ludovico O, Coco A, Di Paola R, Quattrone A, et al. (2005) The common -866G/A polymorphism in the promoter region of the UCP-2 gene is associated with reduced risk of type 2 diabetes in Caucasians from Italy. J Clin Endocrinol Metab 90: 1176–1180. [DOI] [PubMed] [Google Scholar]
- 48. Shen H, Qi L, Tai ES, Chew SK, Tan CE, et al. (2006) Uncoupling protein 2 promoter polymorphism -866G/A, central adiposity, and metabolic syndrome in Asians. Obesity (Silver Spring) 14: 656–661. [DOI] [PubMed] [Google Scholar]
- 49. Akrami SM, Heidari J, Heshmat R, Amiri P, Fakhrzadeh H, et al. (2007) The common -866G/A polymorphism of the UCP2 gene in healthy Iranians compared with world populations. Hum Biol 79: 103–110. [DOI] [PubMed] [Google Scholar]
- 50. Ochoa MC, Santos JL, Azcona C, Moreno-Aliaga MJ, Martínez-González MA, et al. (2007) Association between obesity and insulin resistance with UCP2-UCP3 gene variants in Spanish children and adolescents. Mol Genet Metab 92: 351–358. [DOI] [PubMed] [Google Scholar]
- 51. Hamada T, Kotani K, Fujiwara S, Sano Y, Domichi M, et al. (2008) The UCP2-866 A/A genotype is associated with low density lipoprotein particle sizes in the general population. Med Sci Monit 14: CR107–111. [PubMed] [Google Scholar]
- 52. Heidari J, Akrami SM, Heshmat R, Amiri P, Fakhrzadeh H, et al. (2010) Association study of the -866G/A UCP2 gene promoter polymorphism with type 2 diabetes and obesity in a Tehran population: a case control study. Arch Iran Med 13: 384–390. [PubMed] [Google Scholar]
- 53. Lapice E, Pinelli M, Pisu E, Monticelli A, Gambino R, et al. (2010) Uncoupling protein 2 G(-866) A polymorphism: a new gene polymorphism associated with C-reactive protein in type 2 diabetic patients. Cardiovasc Diabetol 9: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Martinez-Hervas S, Mansego ML, de Marco G, Martinez F, Alonso MP, et al. (2012) Polymorphisms of the UCP2 gene are associated with body fat distribution and risk of abdominal obesity in Spanish population. Eur J Clin Invest 42: 171–178. [DOI] [PubMed] [Google Scholar]
- 55. Oguzkan-Balci S, Col-Araz N, Nacak M, Araz M, Sabanci H, et al. (2013) Mitochondrial uncoupling protein 2 (UCP2) gene polymorphisms are associated with childhood obesity and related metabolic disorders. J Pediatr Endocrinol Metab 26: 277–283. [DOI] [PubMed] [Google Scholar]
- 56. Rosmond R, Bouchard C, Björntorp P (2002) Lack of association between the uncoupling protein-2 Ala55Val gene polymorphism and phenotypic features of the Metabolic Syndrome. Biochim Biophys Acta 1588: 103–105. [DOI] [PubMed] [Google Scholar]
- 57. Sale MM, Hsu FC, Palmer ND, Gordon CJ, Keene KL, et al. (2007) The uncoupling protein 1 gene, UCP1, is expressed in mammalian islet cells and associated with acute insulin response to glucose in African American families from the IRAS Family Study. BMC Endocr Disord 7: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chen HH, Lee WJ, Wang W, Huang MT, Lee YC, et al. (2007) Ala55Val polymorphism on UCP2 gene predicts greater weight loss in morbidly obese patients undergoing gastric banding. Obes Surg 17: 926–933. [DOI] [PubMed] [Google Scholar]
- 59. Lee YH, Kim W, Yu BC, Park BL, Kim LH, et al. (2008) Association of the ins/del polymorphisms of uncoupling protein 2 (UCP2) with BMI in a Korean population. Biochem Biophys Res Commun 371: 767–771. [DOI] [PubMed] [Google Scholar]
- 60. Bielinski SJ, Pankow JS, Boerwinkle E, Bray MS, Kao WH, et al. (2008) Lack of association between uncoupling protein-2 Ala55Val polymorphism and incident diabetes in the atherosclerosis risk in communities study. Acta Diabetol 45: 179–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dalgaard LT, Sørensen TI, Andersen T, Hansen T, Pedersen O (1999) An untranslated insertion variant in the uncoupling protein 2 gene is not related to body mass index and changes in body weight during a 26-year follow-up in Danish Caucasian men. Diabetologia 42: 1413–1416. [DOI] [PubMed] [Google Scholar]
- 62. Duarte NL, Colagiuri S, Palu T, Wang XL, Wilcken DE (2003) A 45-bp insertion/deletion polymorphism of uncoupling protein 2 in relation to obesity in Tongans. Obes Res 11: 512–517. [DOI] [PubMed] [Google Scholar]
- 63. Papazoglou D, Papathanasiou P, Papanas N, Papatheodorou K, Chatziangeli E, et al. (2012) Uncoupling protein-2 45-base pair insertion/deletion polymorphism: is there an association with severe obesity and weight loss in morbidly obese subjects? Metab Syndr Relat Disord 10: 307–311. [DOI] [PubMed] [Google Scholar]
- 64. Meirhaeghe A, Amouyel P, Helbecque N, Cottel D, Otabe S, et al. (2000) An uncoupling protein 3 gene polymorphism associated with a lower risk of developing Type II diabetes and with atherogenic lipid profile in a French cohort. Diabetologia 43: 1424–1428. [DOI] [PubMed] [Google Scholar]
- 65. Otabe S, Clement K, Dina C, Pelloux V, Guy-Grand B, et al. (2000) A genetic variation in the 5′ flanking region of the UCP3 gene is associated with body mass index in humans in interaction with physical activity. Diabetologia 43: 245–249. [DOI] [PubMed] [Google Scholar]
- 66. Dalgaard LT, Sørensen TI, Drivsholm T, Borch-Johnsen K, Andersen T, et al. (2001) A prevalent polymorphism in the promoter of the UCP3 gene and its relationship to body mass index and long term body weight change in the Danish population. J Clin Endocrinol Metab 86: 1398–1402. [DOI] [PubMed] [Google Scholar]
- 67. Alonso A, Martí A, Corbalán MS, Martínez-González MA, Forga L, et al. (2005) Association of UCP3 gene -55C>T polymorphism and obesity in a Spanish population. Ann Nutr Metab 49: 183–188. [DOI] [PubMed] [Google Scholar]
- 68. Fang QC, Jia WP, Yang M, Bao YQ, Chen L, et al. (2005) Effect of polymorphism of uncoupling protein 3 gene -55 (C>T) on the resting energy expenditure, total body fat and regional body fat in Chinese. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 22: 485–488. [PubMed] [Google Scholar]
- 69. de Luis DA, Aller R, Izaola O, González Sagrado M, Conde R, et al. (2007) Lack of association of −55CT polymorphism of UCP3 gene with fat distribution in obese patients. Ann Nutr Metab 51: 374–378. [DOI] [PubMed] [Google Scholar]
- 70. Hamada T, Kotani K, Fujiwara S, Sano Y, Domichi M, et al. (2008) The common -55 C/T polymorphism in the promoter region of the uncoupling protein 3 gene reduces prevalence of obesity and elevates serum high-density lipoprotein cholesterol levels in the general Japanese population. Metabolism 57: 410–415. [DOI] [PubMed] [Google Scholar]
- 71. Urhammer SA, Dalgaard LT, Sørensen TI, Møller AM, Andersen T, et al. (1997) Mutational analysis of the coding region of the uncoupling protein 2 gene in obese NIDDM patients: impact of a common amino acid polymorphism on juvenile and maturity onset forms of obesity and insulin resistance. Diabetologia 40: 1227–1230. [DOI] [PubMed] [Google Scholar]
- 72. Yanovski JA, Diament AL, Sovik KN, Nguyen TT, Li H, et al. (2000) Associations between uncoupling protein 2, body composition, and resting energy expenditure in lean and obese African American, white, and Asian children. Am J Clin Nutr 71: 1405–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cha MH, Shin HD, Kim KS, Lee BH, Yoon Y (2006) The effects of uncoupling protein 3 haplotypes on obesity phenotypes and very low-energy diet-induced changes among overweight Korean female subjects. Metabolism 55: 578–586. [DOI] [PubMed] [Google Scholar]
- 74. Oh HH, Kim KS, Choi SM, Yang HS, Yoon Y (2004) The effects of uncoupling protein-1 genotype on lipoprotein cholesterol level in Korean obese subjects. Metabolism 53: 1054–1059. [DOI] [PubMed] [Google Scholar]
- 75. Liu X, Zhang B, Shen Y, Li J, Zhao N, et al. (2012) A 45-bp insertion/deletion polymorphism in uncoupling protein 2 is not associated with obesity in a Chinese population. Biochem Genet 50: 784–796. [DOI] [PubMed] [Google Scholar]
- 76. Brondani LA, Duarte GC, Canani LH, Crispim D (2014) The Presence of At Least Three Alleles of the ADRB3 Trp64Arg (C/T) and UCP1-3826A/G Polymorphisms Is Associated with Protection to Overweight/Obesity and with Higher High-Density Lipoprotein Cholesterol Levels in Caucasian-Brazilian Patients with Type 2 Diabetes. Metab Syndr Relat Disord 12: 16–24. [DOI] [PubMed] [Google Scholar]
- 77. Sasahara M, Nishi M, Kawashima H, Ueda K, Sakagashira S, et al. (2004) Uncoupling protein 2 promoter polymorphism -866G/A affects its expression in beta-cells and modulates clinical profiles of Japanese type 2 diabetic patients. Diabetes 53: 482–485. [DOI] [PubMed] [Google Scholar]
- 78. Jia JJ, Zhang X, Ge CR, Jois M (2009) The polymorphisms of UCP2 and UCP3 genes associated with fat metabolism, obesity and diabetes. Obes Rev 10: 519–526. [DOI] [PubMed] [Google Scholar]
- 79. Luan JA, Wong MY, Day NE, Wareham NJ (2001) Sample size determination for studies of gene-environment interaction. Int J Epidemiol 30: 1035–1040. [DOI] [PubMed] [Google Scholar]
- 80. Esterbauer H, Oberkofler H, Krempler F, Strosberg AD, Patsch W (2000) The uncoupling protein-3 gene is transcribed from tissue-specific promoters in humans but not in rodents. J Biol Chem 275: 36394–36399. [DOI] [PubMed] [Google Scholar]
- 81. de Souza BM, Assmann TS, Kliemann LM, Marcon AS, Gross JL, et al. (2012) The presence of the -866A/55Val/Ins haplotype in the uncoupling protein 2 (UCP2) gene is associated with decreased UCP2 gene expression in human retina. Exp Eye Res 94: 49–55. [DOI] [PubMed] [Google Scholar]
- 82. Munafò MR, Flint J (2004) Meta-analysis of genetic association studies. Trends Genet 20: 439–444. [DOI] [PubMed] [Google Scholar]
- 83. Berentzen T, Dalgaard LT, Petersen L, Pedersen O, Sorensen TI (2005) Interactions between physical activity and variants of the genes encoding uncoupling proteins −2 and −3 in relation to body weight changes during a 10-y follow-up. Int J Obes (Lond) 29: 93–99. [DOI] [PubMed] [Google Scholar]
- 84. Jun HS, Kim IK, Lee HJ, Kang JH, Kim JR, et al. (2009) Effects of UCP2 and UCP3 variants on the manifestation of overweight in Korean children. Obesity (Silver Spring) 17: 355–362. [DOI] [PubMed] [Google Scholar]
- 85. Rosmond R (2003) Association studies of genetic polymorphisms in central obesity: a critical review. Int J Obes Relat Metab Disord 27: 1141–1151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Funnel plot for studies of the UCP1- 3826A/G polymorphism under a dominant model of inheritance.
(TIF)
Funnel plot for studies of the UCP2- 866G/A, Ala55Val and Ins/Del polymorphisms under a dominant model of inheritance.
(TIF)
Funnel plot for studies of the UCP3- 55C/T polymorphism under a dominant model of inheritance.
(TIF)
Characteristics of the studies included in the meta-analyses.
(XLS)
Body mass index means according to different genotypes of UCP1- 3826A/G, UCP2- 886G/A, UCP2 Ala55Val, UCP2 Ins/del and UCP3- 55C/T polymorphisms for the studies included in the meta-analysis.
(DOC)
PRISMA Checklist.
(DOC)
PRISMA Flow Diagram.
(DOC)