Abstract
The architectural integrity of the endoplasmic reticulum (ER) network depends on the function of membrane-associated dynamin-like GTPases that include metazoan atlastins, plant RHD3 and yeast Sey1p. The evidence that these proteins are sufficient to drive membrane fusion of reconstituted proteoliposomes, and that loss-of-function mutations lead to conspicuous ER shape defects indicates that atlastins, RHD3 and Sey1p promote ER membrane fusion. However, complementation experiments in reciprocal loss-of-function backgrounds have also suggested that RHD3 and Sey1p may be not functionally equivalent, supporting that ER fusion mechanisms may be not entirely conserved in eukaryotes. In this Letter, we provide a brief overview of the field as well as evidence that may explain the functional differences of the plant and yeast ER-shaping dynamin-like GTPases.
Keywords: ER, Root Hair Defective 3 (RHD3), dynamin-like GTPases, Arabidopsis
The endoplasmic reticulum (ER) is an essential organelle necessary for the synthesis, modification and quality control of all the secretory proteins, which generally account for one third of the eukaryotic proteome. The ER structure is based on a dynamic web-like network of interconnected tubules and cisternae, which are continuously remodeled by the incessant interconversion of ER tubules and cisternae, as well as by tubule formation, sliding, and fusion.1,2 Homotypic fusion of ER membranes is mediated by a family of structurally-conserved dynamin-like GTPases known as atlastins in metazoans, RHD3 proteins in plants and Sey1p in yeast.3-6 These are integral membrane proteins with a large cytosolic GTPase domain at the N-terminus, followed by a linker region, connecting two transmembrane (TM) domains and a cytosolic tail at the C-terminus. Genetic ablation of these proteins or overexpression of dominant negative forms of the GTPase domain leads to conspicuous defects in the ER organization with the appearance of long, unbranched ER tubules in cultured mammalian cells and in plant cells7-9 but ER fragmentation in Drosophila melanogaster.10 Whether the formation of the long tubules is linked to a loss of tubule fusion or branching is yet to be tested in vivo, however fusogenic activity of ATLs, RHD3 and Sey1p has been demonstrated in vitro with reconstituted proteoliposomes.8,10-12 Intriguingly, genetic complementation analyses have shown that the ER phenotype in yeast due to concomitant ablation of Sey1p and tubule-shaping proteins, Rtn1p or Yop1p, can be restored by expression of either Sey1p, human ATL, or RHD3,8,13 supporting that these proteins are functional orthologs. Interestingly however, while a fluorescent protein fusion to Sey1p localizes to the ER in plants, it does not seem to be able to rescue the defective growth phenotype associated with the Arabidopsis rhd3 loss-of-function mutation.7 It has been suggested that when Sey1p or RHD3 are expressed at non-endogenous levels in a heterologous system, they may affect the structure of the ER possibly due to unbalanced or uncontrolled activity of the proteins.13 Similarly and consistently with this hypothesis, RHD3 overexpression leads to partial collapse of the ER structure in plant cells.14 An overabundance of the GTPases due to overexpression may lead to excessive membrane fusion, but it is also possible that other factors may have a critical role. For example, unique protein determinants may account for the observed inability of Sey1p to complement loss of RHD3. In this view, RHD3 could functionally complement the Sey1p yeast mutant because RHD3 would be sufficient to satisfy the requirements for ER homotypic fusion in yeast cells; however, Sey1p might not have attributes that are present in RHD3 to facilitate ER membrane fusion in plant cells. To examine this possibility, in this work we have compared the C-terminal (CT) regions of RHD3 and Sey1p, which to date have not been studied in detail, and found that these domains exert different effects on ER integrity in plants. These observations along with the identification of a unique functional domain in the CT extension of RHD3 support that the differences in the function of the ER-shaping dynamin-like GTPases in vivo may be dictated by the presence of distinct protein attributes.
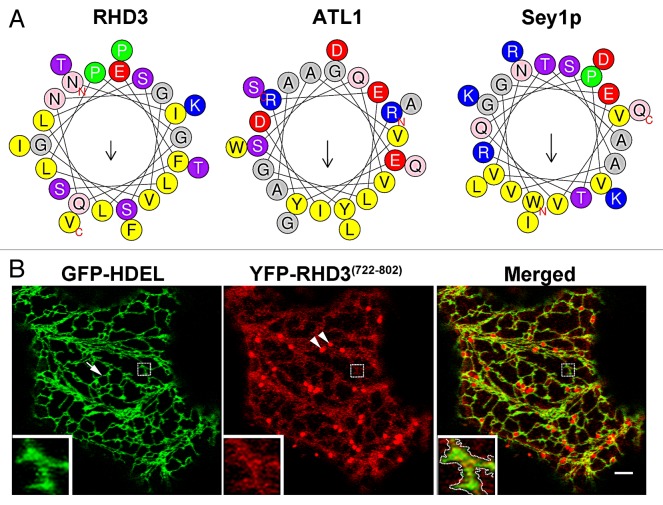
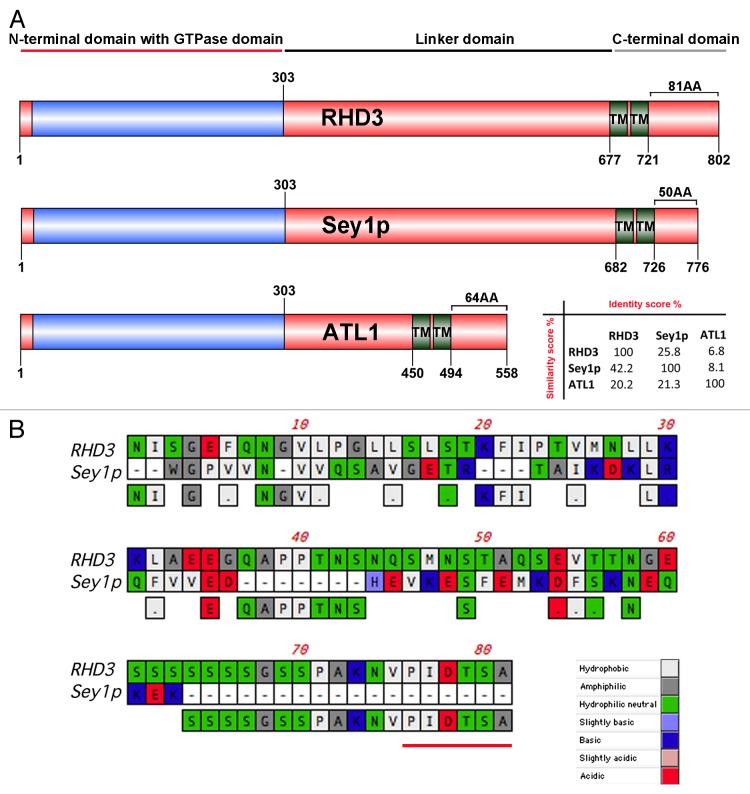
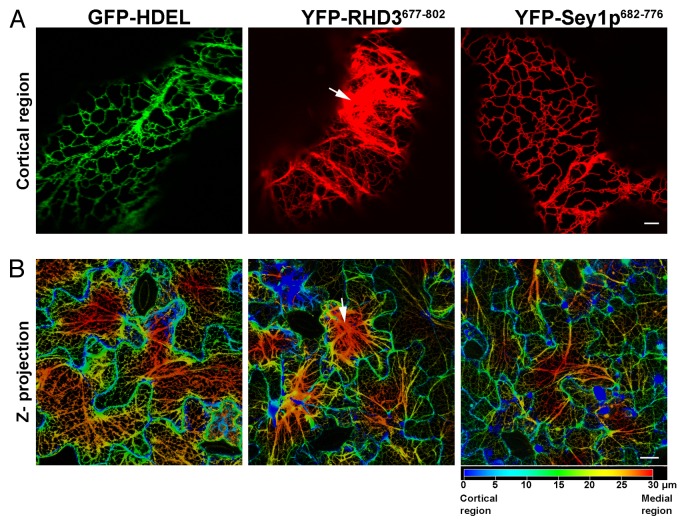
To date most of the characterization of ER-shaping dynamin-like GTPases has been focused on the N-terminal region. For example, crystal structures of the N-terminal region of atlastins have been produced,12,15,16 and the kinetics of the GTPase activity of the recombinant N-terminal region have been studied for atlastins, RHD3 and Sey1p.8,10,13 The current view supports that a GTP hydrolysis-induced conformational change of the N-terminal cytosolic domain is necessary for membrane fusion. Nonetheless, it has also been demonstrated that the region encompassing the two TM domains and the CT extension is required for efficient membrane fusion by ATL. Specifically, the TM domains are important for protein dimerization while the CT extension, which exhibits amphipathic properties, interacts directly with the lipid bilayer.17 Such an interaction is likely to destabilize the bilayer and facilitate membrane fusion.17 To acquire insights on the characteristics of this protein region in RHD3 and Sey1p for comparison with ATL1, we analyzed the first 25 amino acid stretch in CT extension after the second TM domain of these proteins. We found that similar to the equivalent region in ATL1, those of RHD3 and Sey1p show amphipathic characteristics (Fig. 1A). To test whether the entire CT extension of RHD3 (aa 722–802; Fig. 2A) could associate with membranes as it would be predicted if it contained an amphipathic anchor, we expressed it as a N-terminal fluorescent protein fusion in tobacco leaf epidermal cells for confocal imaging, following established procedures.18,19 Indeed subcellular localization analyses of YFP fusion to the RHD3 CT-extension (RHD3722–802) established that this region can be found in association with the ER and the Golgi as well as in the cytosol (Fig. 1B). Based on these observations, we speculate that the CT extension can associate with membranes but that saturation of available membrane binding sites leads to a partial distribution of the chimera in the cytosol. Although these findings denote similarities among the C-terminal region of the ER shaping proteins, the size of the CT extension varies noticeably when comparing those of ATL1, Sey1p and RHD3 with the CT extension of RHD3 being the longest (81 aa) and that of Sey1p being the shortest (50 aa) (Fig. 2A). The degree of conservation of amino acid sequence of the RHD3 CT extension with Sey1p and ATL1 is also rather low (RHD3/Sey1p 6.2% identity, 17.3% similarity; RHD3/ATL1 9.9% identity, 29.6% similarity) (Fig. 2B). These observations raise the question whether the properties of the CT extension are equivalent among the ER-shaping GTPases. To explore this in more detail, we tested whether fluorescent protein fusions to the C-terminal region of RHD3 and Sey1p exhibited a different behavior in plant cells when driven by the same promoter (CaMV 35S). As shown in Figure 3, YFP fusions to the regions encompassing the two TM domains and the entire CT extension of either RHD3 (YFP-RHD3677–802) or Sey1p (YFP-Sey1p682–776) localized to the ER. When we analyzed cells with similar levels of expression, which was established by quantitative analyses of pixel intensity in ER tubules with normal morphology, we found that YFP-RHD3677–802 lead to disruption of the cortical ER network; in contrast, YFP-Sey1p682–776 did not show any significant alteration of the ER organization. These data support that that the C-terminal region of RHD3 can exert a dominant negative role in ER integrity and that this protein domain has distinct properties compared with the equivalent region of Sey1p. The TM domain of ATL has been shown to be necessary for ATL molecules to form oligomers, a requisite for its function in vivo.17 RHD3 is known to oligomerize.7 Therefore, expression of the C-terminal region that encompasses the TM and CT extension of RHD3 most likely exerts a dominant negative role by interfering with the ability of the endogenous RHD3 to form functional oligomers. In this view, expression of the equivalent protein domain of Sey1p would not affect the ER structure in plants if Sey1p were not able to oligomerize with a plant ortholog. This hypothesis may explain the observed differences in the effect of the expression of the RHD3 and Sey1p on the ER structure (Fig. 3), and suggests that RHD3 and Sey1p may have different functional requirements in vivo. Indeed, a close inspection of the RHD3 CT extension reveals the presence of a short peptide signature, which is annotated as a PDZ-binding peptide by the eukaryotic linear motif resource20 and is absent in Sey1p (Fig. 2B). PDZ domains are considered protein-protein interaction modules, which interact with short amino acid motifs in target proteins.21 It is possible that the predicted PDZ binding peptide at the carboxyl-terminus of RHD3 may facilitate the interaction of RHD3 with other proteins that could modulate its function in vivo. Therefore, expression of YFP-RHD3722–802 fusion could exert a dominant negative effect by compromising protein-protein interactions of the endogenous RHD3. On the other hand, if homo- or hetero- interactions of RHD3 have a critical role in the function of this protein, the absence of PDZ binding peptide at the carboxyl-terminus of Sey1p may at least partially explain the inability of this protein to complement the loss of rhd3.7 While this hypothesis awaits experimental validation, the results provided here open new perspectives on the mechanisms underlying functional equivalence and dissimilarities among ER-shaping proteins and suggest that the requirements for ER membrane fusion may be not entirely conserved across kingdoms.
Figure 1. The C-terminal extension of RHD3 can associate with the ER. (A) Helical wheel projections of the first 25 amino acids in the C-terminal region of RHD3 (aa 722–747), ATL1 (aa 495–520), and Sey1p (aa 727–752) drawn by HeliQuest.22 The diagram shows a differential distribution of hydrophobic or hydrophilic amino acids that is similar for the three proteins. (B) Subcellular distribution of YFP-RHD3722–802 in Nicotiana tabacum epidermal cells. The protein can associate with the ER (labeled by the ER lumen marker, GFP-HDEL, arrow), as shown in the merged image. Note that in addition to an obvious discrete signal at the ER (dashed line inside the inset enclosing the ER), a more diffuse signal is present in the cytosol and on punctate structures. The latter are Golgi, as established by colocalization assays with a Golgi marker (not shown). Inset = magnified area of main image. Bar = 5 μm.
Figure 2. The overall structure of RHD3 and Sey1p is largely conserved although variations occur in the size of the CT extension. (A) Schematic representation of protein domains and motifs found in RHD3 (NCBI accession number: P93042), ATL1 (NCBI accession number: NP_056999) and Sey1p (NCBI accession number: NP_014808). Numbers indicate the amino acids. TM: transmembrane. Protein identity (%) vs. similarity (%) is indicated in the matrix. (B) Sequences of RHD3 (aa 722–802) and Sey1p (aa 727–776) were aligned using CLUSTALW and the GONNET matrix. Predicted PDZ binding motif in RHD3 is underlined in red.
Figure 3. RHD3677–802 but not Sey1p682–776 alters ER structure. Confocal microscopy images of the ER in cells expressing either the ER marker GFP-HDEL, YFP-RHD3677–802 or YFP-Sey1p682–776, viewed as single plane for single cells (A) or in a 30 μm z-depth code (B, 0 μm-upper level = blue color; 30 μm-lower depth = red color). In A, the images show that differently from cells expressing GFP-HDEL or YFP-Sey1p682–776, expression of YFP-RHD3677–802 causes aberrant organization of the ER structure. Arrows in A and B show defects in the ER structures that are visible only in cells expressing YFP-RHD3677–802. Bars = 5 μm (A); 20 μm (B).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This study was supported by NSF (MCB 0948584 and MCB1243792).
Footnotes
Previously published online: www.landesbioscience.com/journals/cellularlogistics/article/28217
References
- 1.Griffing LR. Networking in the endoplasmic reticulum. Biochem Soc Trans. 2010;38:747–53. doi: 10.1042/BST0380747. [DOI] [PubMed] [Google Scholar]
- 2.Sparkes I, Runions J, Hawes C, Griffing L. Movement and remodeling of the endoplasmic reticulum in nondividing cells of tobacco leaves. Plant Cell. 2009;21:3937–49. doi: 10.1105/tpc.109.072249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J, Doyle C, Qi X, Zheng H. The endoplasmic reticulum: a social network in plant cells. J Integr Plant Biol. 2012;54:840–50. doi: 10.1111/j.1744-7909.2012.01176.x. [DOI] [PubMed] [Google Scholar]
- 4.McNew JA, Sondermann H, Lee T, Stern M, Brandizzi F. GTP-dependent membrane fusion. Annu Rev Cell Dev Biol. 2013;29:529–50. doi: 10.1146/annurev-cellbio-101512-122328. [DOI] [PubMed] [Google Scholar]
- 5.Chen S, Novick P, Ferro-Novick S. ER structure and function. Curr Opin Cell Biol. 2013;25:428–33. doi: 10.1016/j.ceb.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee H, Sparkes I, Gattolin S, Dzimitrowicz N, Roberts LM, Hawes C, Frigerio L. An Arabidopsis reticulon and the atlastin homologue RHD3-like2 act together in shaping the tubular endoplasmic reticulum. New Phytol. 2013;197:481–9. doi: 10.1111/nph.12038. [DOI] [PubMed] [Google Scholar]
- 7.Chen J, Stefano G, Brandizzi F, Zheng H. Arabidopsis RHD3 mediates the generation of the tubular ER network and is required for Golgi distribution and motility in plant cells. J Cell Sci. 2011;124:2241–52. doi: 10.1242/jcs.084624. [DOI] [PubMed] [Google Scholar]
- 8.Hu J, Shibata Y, Zhu PP, Voss C, Rismanchi N, Prinz WA, Rapoport TA, Blackstone C. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell. 2009;138:549–61. doi: 10.1016/j.cell.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stefano G, Renna L, Moss T, McNew JA, Brandizzi F. In Arabidopsis, the spatial and dynamic organization of the endoplasmic reticulum and Golgi apparatus is influenced by the integrity of the C-terminal domain of RHD3, a non-essential GTPase. Plant J. 2012;69:957–66. doi: 10.1111/j.1365-313X.2011.04846.x. [DOI] [PubMed] [Google Scholar]
- 10.Orso G, Pendin D, Liu S, Tosetto J, Moss TJ, Faust JE, Micaroni M, Egorova A, Martinuzzi A, McNew JA, et al. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature. 2009;460:978–83. doi: 10.1038/nature08280. [DOI] [PubMed] [Google Scholar]
- 11.Zhang M, Wu F, Shi J, Zhu Y, Zhu Z, Gong Q, Hu J. ROOT HAIR DEFECTIVE3 family of dynamin-like GTPases mediates homotypic endoplasmic reticulum fusion and is essential for Arabidopsis development. Plant Physiol. 2013;163:713–20. doi: 10.1104/pp.113.224501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Byrnes LJ, Sondermann H. Structural basis for the nucleotide-dependent dimerization of the large G protein atlastin-1/SPG3A. Proc Natl Acad Sci U S A. 2011;108:2216–21. doi: 10.1073/pnas.1012792108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang M, Wu F, Shi J, Zhu Y, Zhu Z, Gong Q, Hu J. ROOT HAIR DEFECTIVE3 family of dynamin-like GTPases mediates homotypic endoplasmic reticulum fusion and is essential for Arabidopsis development. Plant Physiol. 2013;163:713–20. doi: 10.1104/pp.113.224501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng H, Chen J. Emerging aspects of ER organization in root hair tip growth: lessons from RHD3 and Atlastin. Plant Signal Behav. 2011;6:1710–3. doi: 10.4161/psb.6.11.17477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Byrnes LJ, Singh A, Szeto K, Benvin NM, O’Donnell JP, Zipfel WR, Sondermann H. Structural basis for conformational switching and GTP loading of the large G protein atlastin. EMBO J. 2013;32:369–84. doi: 10.1038/emboj.2012.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bian X, Klemm RW, Liu TY, Zhang M, Sun S, Sui X, Liu X, Rapoport TA, Hu J. Structures of the atlastin GTPase provide insight into homotypic fusion of endoplasmic reticulum membranes. Proc Natl Acad Sci U S A. 2011;108:3976–81. doi: 10.1073/pnas.1101643108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu TY, Bian X, Sun S, Hu X, Klemm RW, Prinz WA, Rapoport TA, Hu J. Lipid interaction of the C terminus and association of the transmembrane segments facilitate atlastin-mediated homotypic endoplasmic reticulum fusion. Proc Natl Acad Sci U S A. 2012;109:E2146–54. doi: 10.1073/pnas.1208385109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brandizzi F, Snapp EL, Roberts AG, Lippincott-Schwartz J, Hawes C. Membrane protein transport between the endoplasmic reticulum and the Golgi in tobacco leaves is energy dependent but cytoskeleton independent: evidence from selective photobleaching. Plant Cell. 2002;14:1293–309. doi: 10.1105/tpc.001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stefano G, Osterrieder A, Hawes C, Brandizzi F. Endomembrane and Golgi traffic in plant cells. Methods Cell Biol. 2013;118:69–83. doi: 10.1016/B978-0-12-417164-0.00005-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dinkel H, Van Roey K, Michael S, Davey NE, Weatheritt RJ, Born D, et al. The eukaryotic linear motif resource ELM: 10 years and counting. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee HJ, Zheng JJ. PDZ domains and their binding partners: structure, specificity, and modification. Cell Commun Signal. 2010;8:8. doi: 10.1186/1478-811X-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gautier R, Douguet D, Antonny B, Drin G. HELIQUEST: a web server to screen sequences with specific α-helical properties. Bioinformatics. 2008;24:2101–2. doi: 10.1093/bioinformatics/btn392. [DOI] [PubMed] [Google Scholar]