Abstract
The mammalian gastrointestinal tract, the site of digestion and nutrient absorption, harbors trillions of beneficial commensal microbes from all three domains of life. Commensal bacteria, in particular, are key participants in the digestion of food, and are responsible for the extraction and synthesis of nutrients and other metabolites that are essential for the maintenance of mammalian health. Many of these nutrients and metabolites derived from commensal bacteria have been implicated in the development, homeostasis and function of the immune system, suggesting that commensal bacteria may influence host immunity via nutrient- and metabolite-dependent mechanisms. Here we review the current knowledge of how commensal bacteria regulate the production and bioavailability of immunomodulatory, diet-dependent nutrients and metabolites and discuss how these commensal bacteria-derived products may regulate the development and function of the mammalian immune system.
The mammalian gastrointestinal tract harbors trillions of beneficial commensal bacteria, a population composed of at least 1,000–5,000 species1,2. Across the animal kingdom, species as diverse as flies, fish, mice and humans have co-opted the faculties of beneficial microbes to support normal development and homeostasis3-14. Recent studies have highlighted that alterations in the composition of commensal bacterial populations are linked to multiple metabolic and inflammatory diseases in humans including but not limited to inflammatory bowel disease (IBD), obesity, type 2 diabetes, atherosclerosis, allergy and colon cancer9-15. These associations provoke fundamental questions regarding the cellular and molecular pathways through which commensal bacteria regulate mammalian gene expression and influence resistance or susceptibility to a broad range of clinically important diseases.
Recent studies have identified a critical role for commensal bacteria and their products in regulating the development, homeostasis and function of innate and adaptive immune cells15-24. Several recent comprehensive reviews have described how commensal bacteria are recognized by the innate immune system and how individual species or consortia of commensal bacterial species can influence distinct modules of the innate and adaptive immune response15-24. However, an emerging area that has received relatively little attention is how metabolites and nutrients derived from commensal bacteria regulate the host immune system.
Commensal bacteria are key regulators of digestion, a process that begins in the mouth and continues as ingested food and its digestive intermediates transit more than 20 feet (6 meters) to the end of the adult human gastrointestinal tract. Along the way, the digestive slurry is mixed with commensal bacteria, which are important for the extraction, synthesis and absorption of many nutrients and metabolites (Box 1), including bile acids, lipids, amino acids, vitamins and short-chain fatty acids (SCFAs; Fig. 1a). These nutrients and metabolites derived from commensal bacteria are directly linked to diet and digestion and are therefore considered to be diet-dependent microbial products (Box 1)14. In addition to producing diet-dependent metabolites and nutrients, commensal bacteria can produce many different diet-independent microbial products, examples of which are lipopolysaccharide and peptidoglycan (Box 1)11-14,23,24.
Box 1. Definitions.
Metabolites; intermediates and products of metabolism. This is a broad term that usually refers to small molecules.
Nutrients; a subset of metabolites that must be acquired from the environment to support cellular processes. In mammals, the diet and commensal bacteria provide most nutrients7-14.
Commensal bacteria; beneficial bacteria that reside on host barrier surfaces such as the intestine. Although the term ‘pcommensal’ implies that bacteria benefit and the host is unaffected, commensal bacteria and the host have a mutualistic relationship in which both derive benefit from the interaction15.
Dysbiosis a state in which commensal bacteria populations are altered or imbalanced15. Dysbiosis is associated with several human diseases45,58-68.
Diet-dependent microbial products: factors derived from commensal bacteria (for example, nutrients and metabolites) that are directly linked to diet or digestion14. Examples of diet-dependent microbial products include but are not limited to bile acids, short-chain fatty acids, vitamins, amino acids and fatty acids. The case of bile acids illustrates this concept. Commensal bacteria participate in the synthesis of bile acids, a family of metabolites that are stored in the gall bladder and secreted into the intestinal lumen in response to diet-dependent signals32-45. Bile salts then facilitate the digestion and absorption of lipids32. Therefore, bile acids derived from commensal bacteria are directly linked to both diet and digestion and can be considered to be diet-dependent microbial products.
Diet-independent microbial products: factors derived from commensal bacteria that are not directly linked to diet or digestion14. Examples include but are not limited to lipopolysaccharide and peptidoglycan.
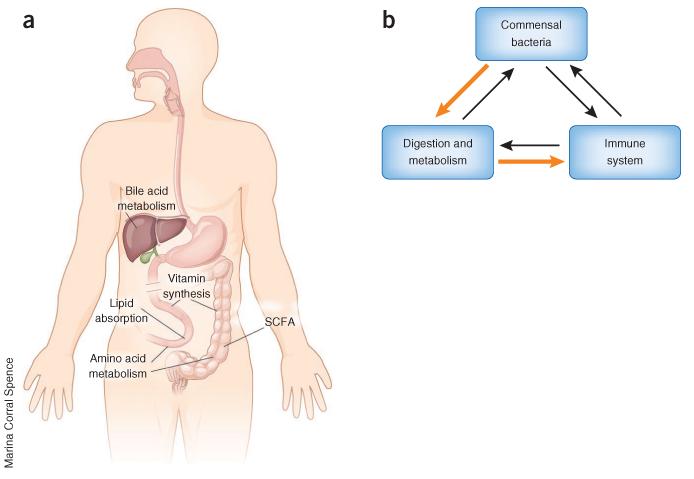
Figure 1.
Commensal bacteria at the interface of host metabolism and immunity. (a) Commensal bacteria regulate digestion by mediating bile acid synthesis, lipid absorption, amino acid metabolism, vitamin synthesis and SCFA production. (b) Commensal bacteria participate in digestion and regulate host metabolic homeostasis, and commensal bacteria–derived nutrients regulate the immune system. This commensal bacteria–metabolite–immune system axis (orange arrows) exists in a complex network of interactions among commensal bacteria, host digestion and the immune system (orange and black arrows).
Nutrients and metabolites derived from commensal bacteria may regulate immune cells via indirect and direct mechanisms. Commensal bacteria-mediated alterations in the availability or use of energy substrates (for example, by modulating oxidation of glucose and fatty acids) may indirectly influence the development, homeostasis and function of immune cells5,11-14,25-31, although direct experimental analyses of these interactions are limited at present. In addition, direct effects of metabolites can be mediated via metabolite-specific receptors that can activate signal-transduction pathways and transcriptional programs that control differentiation, proliferation, migration and effector function of immune cells. The roles of diet-independent microbial products in the regulation of immunity have been recently discussed elsewhere15-24; therefore in this Review, we will discuss how commensal bacteria-derived diet-dependent nutrients and metabolites regulate the host immune system (Fig. 1b). We describe how commensal bacteria can regulate the synthesis and/or availability of bile acids, SCFAs, vitamins, amino acids and fatty acids. In the context of each of these diet-dependent nutrients and metabolites, we discuss current knowledge of how these factors regulate the immune response in the context of health and disease.
Regulation of bile acids by commensal bacteria
Bile acids are a family of cholesterol-derived amphipathic molecules that solubilize dietary fat in the small intestine to support the digestion and absorption of fat and of fat-soluble vitamins. In addition to their roles in regulating digestion, bile acids act as signaling molecules that regulate metabolic homeostasis32,33 and, as we will discuss below, immune cell homeostasis and function. Commensal bacteria participate in the synthesis of bile acids in two main ways (Fig. 2a). First, commensal bacteria convert primary bile acids (for example, cholic and chenodeoxycholic acids), which are synthesized de novo by the liver, into secondary bile acids (for example, deoxycholic, lithocholic and muricholic acids) through dehydration reactions34. Both primary and secondary bile acids are subsequently reabsorbed in the ileum and returned via portal circulation to the liver, where they are conjugated to glycine (in humans) or taurine (in mice)34,35. These covalent modifications increase the efficiency of reabsorption of bile acids in subsequent rounds of digestion34. Second, commensal bacteria deconjugate bile acids to re-derive the unconjugated forms36,37. The reabsorption of unconjugated bile acids is inefficient; therefore, deconjugation of bile acids mediated by commensal bacteria promotes the excretion and elimination of bile acids32.
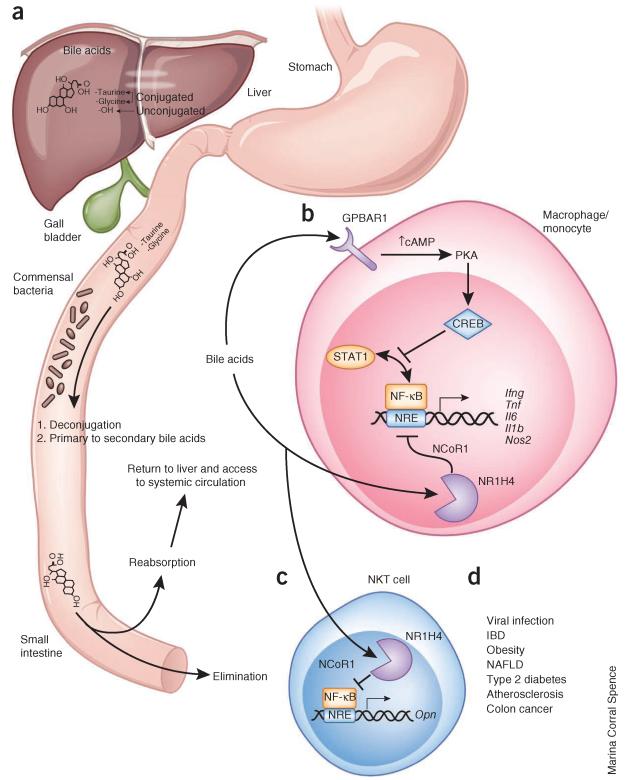
Figure 2.
Regulation of bile acid metabolism by commensal bacteria and effects of bile acids on immune cells. (a) Roles of commensal bacteria in synthesis of bile acids. Bile acids are synthesized in the liver from cholesterol-derived precursor molecules and are emptied into the small intestine. There, commensal bacteria deconjugate bile acids and convert primary into secondary bile acids. Bile acids are reabsorbed in the ileum and transported to the liver to complete enterohepatic circulation. Some bile acids gain access to systemic circulation. Molecular structure of bile acid is representative of some but not all bile acids. (b) Bile acid signaling in macrophages and monocytes via GPBAR1 and NR1H4. Both pathways lead to inhibition of NF-κB, albeit by different mechanisms. GPBAR1 signaling involves cAMP-PKA-mediated inhibition of STAT1 and NF-κB, and NR1H4 signaling leads to NR1H4-NCoR1–mediated repression of NF-κB–responsive elements (NRE). Bile acid stimulation of both pathways leads to less NF-κB–dependent gene expression. (c) Bile acid signaling in NKT cells. This cell type expresses NR1H4, and bile acid engagement of this nuclear receptor represses osteopontin expression. This molecule has many functions, including promoting neutrophil activation and chemotaxis as well as NKT cell activation in an autocrine or paracrine manner. (d) Pathogens and diseases associated with immune responses regulated by bile acids. NAFLD, non-alcoholic fatty liver disease.
Highlighting the influence of commensal bacteria on the synthesis of bile acids, mice and rats reared in the absence of commensal bacteria or treated with oral broad-spectrum antibiotics have fewer secondary bile acids and more conjugated bile acids in the fecal matter, ileum lamina propria, liver, kidney, heart and plasma compared to conventionally reared counterparts38-45. Consistent with these observations, some human patients with IBD have intestinal dysbiosis (Box 1) that is associated with lower concentrations of secondary bile acids in the feces and periphery as well as more conjugated bile acids in the feces compared to healthy subjects45. Such data suggest that commensal bacteria appear to be intimately involved in generating an array of bile acids, and at least a subset of these bile acid metabolites derived from commensal bacteria gain access to enterohepatic and systemic circulation (Fig. 2a).
Bile acid regulation of immune cells
The role of commensal bacteria in the production of bile acids raises the question of whether bile acids derived from commensal bacteria are associated with regulation of the host immune system. Some bile acids can regulate the function of immune cells via the G protein–coupled bile acid receptor 1 (GPBAR1; also known as TGR5 and membrane-type receptor for bile acids, M-BAR) and the nuclear receptor subfamily 1, group H, member 4 (NR1H4; also known as farnesoid X receptor, FXR), both of which are highly expressed in monocytes and macrophages as well as other immune cell types32,33. In macrophages and monocytes, bile acid signaling via these receptors is linked to a common anti-inflammatory response involving the inhibition of NF-κB activity and repression of NF-κB–dependent transcription (Fig. 2b)45-49. Treatment of macrophages with the highly selective GPBAR1 agonist INT-777 (ref. 50) is associated with higher cellular concentrations of cAMP and activation of protein kinase A (PKA) and of cAMP-responsive element-binding protein (CREB)49. The cAMP-PKA-CREB pathway lowers STAT1 phosphorylation and NF-κB transcriptional activity51-53. Macrophages treated with INT-777 exhibit lower NF-κB binding to the nitric oxide synthase 2 (Nos2) promoter and attenuated induction of the proinflammatory cytokines tumor necrosis factor (TNF), interleukin 6 (IL-6) and IL-1β in response to lipopolysaccharide48. These effects of INT-777 were not observed in Gpbar1−/− macrophages, indicating that GPBAR1 was required for the anti-inflammatory effects of INT-777. Consistent with these findings, loss-of-function studies indicate that Gpbar1−/− mice exhibited exaggerated colitis in response to treatment of dextran sodium sulfate (DSS) and trinitrobenzene sulfonate (TNBS), two mouse models of intestinal damage and inflammation54.
Similarly, treatment of intestinal lamina propria CD11b+ cells with the NR1H4-selective agonist INT-747 was associated with a decrease in the expression of NF-κB–dependent genes Tnf, Il6, Il1b, Ifng and Nos2 in response to treatment with lipopolysaccharide46. Treatment of wild-type mice exposed to DSS or TNBS with INT-747 was also associated with lower expression of IL-1β, IL-6 and monocyte chemotractant protein 1 (MCP-1) as well as lower colonic inflammation55. However, unlike bile acid–GPBAR1 pathway–dependent inhibition of NF-κB activity, NR1H4-mediated repression of NF-κB–dependent transcription appears to be regulated through stabilization of the nuclear receptor corepressor 1 (NCoR1) on NF-κB–responsive elements, thereby blocking access to NF-κB46. Although future studies are required, the anti-inflammatory properties of the bile acid–NR1H4 pathway are likely not specific to monocytes and macrophages, as NR1H4 is expressed in other immune cell types, including liver natural killer T (NKT) cells (Fig. 2c). For example, in liver NKT cells NR1H4 regulates the expression of osteopontin56,57, a pleiotropic protein that regulates many cell types, an example of which is neutrophils. Compared to wild-type mice, NR1H4-deficient mice have more liver NKT cell–derived osteopontin and increased liver neutrophils in concanavalin A–induced hepatitis56. Moreover, natural secondary bile acids, which can bind both GPBAR1 and NR1H4 (ref. 33), also appear to have anti-inflammatory effects in nonimmune cells, as treatment of intestinal epithelial cell lines such as Caco-2 cells with deoxycholic acid or lithocholic acid abrogates IL-1β–induced expression of IL-8 (ref. 45). Although GPBAR1 and NR1H4 are not fully redundant, as evidenced by the spontaneous intestinal inflammation observed at steady state in Nr1h4−/− mice with functional GPBAR1 but not Gpbar1−/− mice with functional NR1H4 (refs. 46,54), the above-mentioned studies indicate that bile-acid signaling pathways have a common anti-inflammatory function involving inhibition of NF-κB.
Bile acids in health and disease
The role of commensal bacteria in the production of bile acids and the anti-inflammatory effects of bile acids in some cell types raise questions about whether bile acids derived from commensal bacteria might influence the functions of the immune system in diseases associated with dysbiosis. Such diseases include IBD45,58,59, obesity6,60-63, non-alcoholic fatty liver disease64, type 2 diabetes62,63,65, atherosclerosis66,67, colon cancer68, viral infection69,70 and others (Fig. 2d). Here we will briefly discuss IBD and viral infection.
As reviewed above, bile acids appear to regulate the function of at least some immune cell types through GPBAR1 and NR1H4, both of which lead to the inhibition of NF-κB–dependent expression of proinflammatory genes. Mice lacking GPBAR1 or NR1H4 exhibit exaggerated colitis in models of intestinal inflammation, and stimulating these receptors appears to be associated with protection against colon tissue damage46,54,55,71. Patients with IBD, at least two diseases (Crohn’s disease and ulcerative colitis) with diverse phenotypes, are reported to have alterations in commensal bacteria as well as diminished concentrations of bile acids45,58,59 and, in the case of Crohn’s disease, diminished NR1H4 activity in the ileum72. These correlative studies provoke speculation about whether loss of bile acids derived from commensal bacteria might contribute to the development of at least some subsets of IBD and whether the use of analogs of bile acids might be a novel therapeutic strategy to treat this complex set of diseases. However, the conjugated bile acid taurocholic acid supports the growth of pathogenic bacteria that promote colitis in IL-10–deficient mice73, indicating that a better understanding of the commensal bacteria–bile acid–immune system axis is necessary to fully understand the role of bile acids in IBD. In this context, additional research will be necessary to investigate the possible roles of bile acid metabolites in clinically stratified IBD patients.
The bile acid–mediated decrease in NF-κB activity in macrophages and monocytes may also be associated with the impaired antiviral immunity observed in germ-free mice or mice with experimentally altered composition of commensal bacteria69,70. Macrophages from germ-free and antibiotic-treated mice have lower NF-κB–dependent gene expression and interferon responses in association with diminished CD8+ T cell and NKT cell function, diminished capacity to limit viral replication and greater susceptibility to viral infection69,70. These studies suggest that commensal microbiota provide instructive tonic signals that support the proper functioning of innate immune cells and the coordination of adaptive immune responses69,70. It is also possible, however, that germ-free and antibiotic-treated mice have impaired macrophage and NKT function because of elevated bile acid concentrations, which would be expected to inhibit NF-κB–dependent gene expression, such as the interferon response programs in macrophages and monocytes. Consistent with this idea are studies demonstrating that cells treated with bile acids have an impaired capacity to limit replication of hepatitis C virus and of porcine enteric calicivirus74,75. In addition, treatment of hepatocytes with the NR1H4 antagonist Z-guggulsterone improves the ability of hepatocytes treated with bile acids to limit replication of hepatitis C virus75. This bile acid–mediated effect is associated with activation of the PKA pathway and less STAT1 phosphorylation as well as impaired interferon responses74. Collectively, these studies suggest that an increase in bile acid signaling via GPBAR1 and NR1H4 may be associated with lower antiviral immunity in germ-free and antibiotic-treated mice. More studies will be required to investigate further whether bile acids regulate the functions of innate and adaptive immune cells in mice with experimentally altered composition of commensal bacteria.
Commensal bacteria–derived short-chain fatty acids
SCFAs are 1–6 carbons in length and are produced in the colon by bacterial fermentation of plant-derived nondigestible polysaccharides, such as cellulose (Fig. 3a)76-79. The most abundant SCFAs are butyrate (C4), propionate (C3) and acetate (C2) in the intestinal lumen80,81. Compared to the case in conventionally reared mice, the contents of colons of germ-free mice include more of the nondigestible plant oligosaccharide raffinose40, a bacterial fermentation substrate82, and markedly diminished concentrations of SCFAs83-85, indicating that commensal microbiota are essential for synthesis of SCFAs.
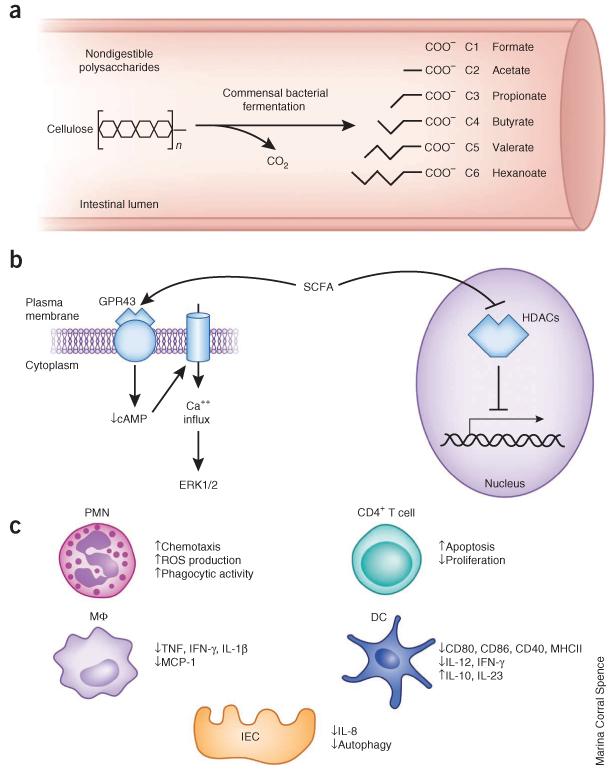
Figure 3.
Synthesis of SCFAs by commensal bacteria, and regulation of immunity by SCFAs. (a) Commensal bacteria ferment nondigestible polysaccharides ingested in the diet (for example, cellulose) to produce SCFAs. Various SCFAs and their molecular structures are shown. C1–C6 refers to the number of carbons. Isoforms of these SCFAs are not shown. (b) Known molecular pathways through which SCFA regulate populations of immune cells. SCFAs bind the G protein–coupled receptor GPR43, which leads to a decrease in cAMP levels, calcium influx and ERK1/2 activation. SCFAs also inhibit histone deacetylases (HDACs), which are transcriptional repressor proteins. (c) SCFA regulation of neutrophils, macrophages/monocytes (MΦ), dendritic cells (DCs), CD4+ T cells and intestinal epithelial cells (IEC). ROS, reactive oxygen species; MHCII, major histocompatibility class II.
SCFA regulation of immune cells
Several roles have been described for SCFAs, including the regulation of host metabolism86-88 and function of the immune system85,89,90. SCFAs might regulate the immune system in multiple ways, for example, through the activation GPR43 (free fatty acid receptor 2, FFA2)91-93, the inhibition of histone deacetylases84,94-97 or the regulation of autophagy84,98 (Fig. 3b). GPR43 is a G protein–coupled receptor that recognizes endogenous SCFA ligands, including acetate, propionate and butyrate91-93,99, and is highly expressed in neutrophils, macrophages and monocytes92,100. A similar receptor, GPR41 (FFA3), also recognizes SCFA ligands with a different rank-order of ligand potency91-93,99, but GPR41 expression is low or undetectable in neutrophils and monocytes92,100, and at present there is limited evidence to suggest a direct role for GPR41 in the regulation of the immune system.
SCFAs propionate and acetate, derived from commensal bacteria, promote neutrophil chemotaxis85,89,90,92. The SCFA-induced chemotactic response in neutrophils is associated with an increase in intracellular calcium levels, a decrease in cAMP concentration and activation of ERK1/2 (ref. 92). Unlike bone marrow–derived neutrophils (BMDNs) from wild-type mice, Gpr43−/− BMDNs do not exhibit calcium influx in response to acetate treatment and do not undergo acetate-induced chemotaxis85. Furthermore, production of reactive oxygen species and the phagocytic activity of wild-type BMDNs but not of Gpr43−/− BMDNs is increased with acetate treatment85, indicating that SCFAs promote not only neutrophil chemotaxis but also neutrophil function. Consistent with the role of SCFAs in promoting neutrophil chemotaxis, Gpr43−/− mice treated with DSS to induce colitis had lower neutrophil content in the colon compared to wild-type mice treated with DSS89. However, other studies report fewer colonic neutrophils in wild-type mice treated with DSS and acetate85 or DSS and butyrate101 compared to mice treated with DSS alone. It is not known why treatment with SCFAs and genetic deletion of the SCFA receptor GPR43 led to similar anti-inflammatory phenotypes in DSS-induced colitis, and further research will be required to investigate this issue.
Other cell types have also emerged as targets of SCFAs, including monocytes, dendritic cells, T cells and intestinal epithelial cells (Fig. 3c). Human peripheral blood mononuclear cells (PBMCs) express GPR43, and treatment of this cell type with SCFAs diminished expression of monocyte chemotactic protein-1 and production of TNF, IFN-γ and IL-10 in response to treatment with lipopolysac-charide100. In dendritic cells, treatment with butyrate is associated with decreased expression of the pro-inflammatory cytokines IL-12 and IFN-γ, and increased expression of IL-10 and IL-23 (refs. 102,103). There is also some evidence to suggest that butyrate may regulate the ability of dendritic cells to present antigen and to prime T cells. For example, butyrate treatment of dendritic cells is associated with downregulation of antigen-presentation machinery, including CD40, CD80, CD86 and major histocompatibility complex class II (refs. 102,103). In a dendritic cell and T cell co-culture system, however, butyrate-treated dendritic cells elicit increased T cell–derived production of IL-17 and IL-10 (ref. 102). In addition to regulating dendritic cell–T cell interactions, SCFAs might also directly regulate responses of T cells, as SCFAs may inhibit proliferation and promote apoptosis in CD4+ T cells104-107. Furthermore, SCFAs appear to regulate production of proinflammatory cytokines in nonimmune cells, as Caco-2 cells treated with butyrate and IL-1β produce less IL-8 than do Caco-2 cells treated with IL-1β alone, indicating that a colonic epithelial cell line is also responsive to butyrate108. In primary colonic epithelial cells, SCFA deficiency establishes an energy-deprived state and leads to activation of autophagy84, a process that is linked to many aspects of innate and adaptive immunity98 and the maintenance of intestinal barrier integrity109. The molecular mechanisms by which SCFAs regulate all of these cell types are not well understood, although treating cells with butyrate has been linked to the inhibition of histone deacetylases84,86,97,105,110 and alterations in histone acetylation that depend on the metabolic state of the cell95. Future studies will be required to define the full spectrum of cellular targets and molecular mechanisms through which SCFAs regulate development and function of immune cells.
SCFAs in health and disease
As discussed above, commensal bacteria are essential for the production of SCFAs, which appear to have anti-inflammatory properties in multiple types of immune cells. This raises fundamental questions about whether SCFAs might contribute to responses of immune cells in diseases associated with alterations in populations of commensal bacteria. Germ-free mice, which are deficient in SCFAs in the gastrointestinal tract83-85, exhibit an exaggerated response of the immune system to DSS-induced colitis in some studies85. Consistent with these findings, treatment of germ-free mice with the SCFA acetate is sufficient to ameliorate the intestinal inflammation in response to treatment with DSS85. This effect is not observed in Gpr43−/− mice, indicating that GPR43 is required for the anti-inflammatory effects of SCFA in this model of intestinal inflammation in germ-free mice85. In contrast, another study showed that conventionally reared Gpr43−/− mice exhibit less intestinal inflammation in response to treatment with DSS than wild-type mice but develop systemic bacterial dissemination and die of complications resulting from sepsis89, suggesting that SCFAs might regulate intestinal barrier function, a topic that warrants further investigation. Notably, treatment of IBD patients with butyrate enemas diminishes intestinal inflammation111, and in vitro treatment of IBD lesional biopsies with butyrate is associated with lower production of pro-inflammatory cytokines112. Collectively, these studies suggest that SCFAs might influence responses of immune cells in at least some forms of IBD. Despite these advances, future work will be required to define how SCFAs regulate responses of the immune system in the context of IBD.
Commensal bacteria–derived vitamins and immunity
Vitamins are organic nutrients that are necessary for normal cellular function. For humans, there are 13 essential vitamins: the fat-soluble vitamins A, D, E and K and the water-soluble vitamins B1, B2, B3, B5, B6, B7, B9, B12 and C. Some commensal bacterial species have the capacity to synthesize essential vitamins, especially of the B and K groups, and this has been proposed to be an important source of these vitamins113. The influence of vitamins on development and function of immune cells has been discussed in detail elsewhere114-122, and we will therefore briefly discuss this here only in the context of a recently discovered link between commensal bacteria–derived vitamin biosynthetic intermediates and immune cells that directly recognize these intermediates. In particular, the monomorphic major histocompatibility complex class I–related protein (also known as MR1) presents vitamin from the riboflavin (vitamin B2) biosynthetic pathway to mucosa-associated invariant T cells122, a population of T cells that produces IL-17 and IFN-γ and that is activated in response to microbe-derived products of the riboflavin biosynthetic pathway122-126. These data suggest that commensal bacteria–derived metabolites in vitamin biosynthetic pathways, not just the vitamin end product, may be previously unappreciated groups of molecules that regulate immune cells or that immune cells use to sense commensal bacteria. Whether immune cells recognize intermediates of other vitamin biosynthetic pathways has yet to be determined. In addition, it is currently unknown whether or how vitamin biosynthetic intermediates influence development, homeostasis or function of immune cells—topics that should be the subject of future inquiries.
Commensal bacteria–derived amino acids and immunity
There are 20 essential amino acids that must be acquired through the diet and absorbed via amino acid transport proteins in the intestine. Several studies suggest that commensal bacteria are important for the extraction, synthesis or absorption of some amino acids, including alanine, aspartate, glutamate, glycine and tryptophan, although many of the reported differences in these amino acids appear to be tissue-specific40,44,127-132. Amino acids serve as building blocks for protein synthesis, substrates that feed directly into metabolic pathways (for example, glutamine into the citric acid cycle) and cell-to-cell signaling molecules (for example, the neurotransmitter glycine). Some amino acids, such as arginine, leucine, glutamine and tryptophan, are also associated with regulation of the immune system119,130,133-140. It is not yet known whether amino acids that are regulated by commensal bacteria influence the development, homeostasis or function of immune cells, but emerging research suggests that this might be the case. For example, tryptophan concentrations in the lumen of the colon are lower in germ-free mice compared to conventionally reared mice, suggesting that production or absorption of this amino acid is altered in the absence of commensal bacteria131. Tryptophan influences proliferation of T cells by regulating passage through the G1 phase of the cell cycle137 and regulates immune responses in models of skin allo-graft rejection, tumor growth and autoimmune encephalomyelitis138 as well as in TNBS-induced colitis119. Additional research is required to investigate the emerging link between amino acids that are regulated by commensal bacteria and the immune system.
Alterations in the composition of commensal bacteria could be an important etiologic factor in protein energy malnutrition (PEM)127,128, a set of diseases characterized by immunodeficiency. For example, children with various forms of PEM have greater rates of infection141-143, higher infection-associated mortality143,144 and diminished vaccine efficacy145,146. Although insufficient dietary protein intake is considered to be a causal factor in the development of PEM, recent data suggest that commensal bacteria also have a role in PEM127,128, perhaps because of their ability to synthesize, extract or regulate the absorption of amino acids. Supporting this hypothesis, it has been demonstrated that twins discordant for kwashiorkor, a PEM disease, have distinct gut microbiomes127. In addition, colonization of germ-free mice with fecal matter from the kwashiorkor-affected twin leads to rapid weight loss as well as impaired weight gain when given ready-to-use therapeutic food, a clinical intervention used in areas of widespread undernutrition127. Moreover, treatment of kwashiorkor patients with ready-to-use therapeutic food plus an oral antibiotic cocktail resulted in increased weight gain and decreased mortality compared to kwashiorkor patients treated with ready-to-use therapeutic food that did not receive antibiotics128. This suggests that commensal bacteria contribute to the etiology of this PEM disease, a notion supported by studies of germ-free mice that suggest that commensal bacteria regulate bioavailability and/or uptake of amino acids40,44,129,131,132. The mechanisms through which commensal bacteria regulate host amino acid metabolism in various compartments are unclear, and understanding how commensal bacteria-dependent amino acid abnormalities affect the immune system will require additional functional analyses.
Commensal bacteria, fatty acids and immunity
Fatty acids are a large family of nutrients that serve as energy substrates and that regulate or support the function of many immune cells. Germ-free fish, mice and rats exhibit altered lipid metabolism3,5,40,41,44,147, and lipid homeostasis abnormalities in germ-free mice can be partially rescued by colonization with human fecal contents39. Furthermore, mice reared in a germ-free environment have lower fat mass compared to their conventionally reared counterparts5, a phenotype that is associated with a diminished capacity for harvesting energy from ingested food61 and a broadly altered lipid profile5,40,41,44,147. Commensal bacterial regulation of lipid homeostasis has also been reported in other species, as zebrafish raised in an axenic environment exhibit impaired lipid absorption and altered lipid profiles3. It therefore seems that commensal bacteria are necessary for the maintenance of lipid homeostasis.
This association raises the question of whether commensal bacteria regulate the immune system via modulation of systemic lipid homeostasis. Such a phenomenon could be mediated in at least two ways: the direct sensing of lipid moieties or the establishment of a metabolic program that favors some immune functions or the development of some cell types over others. The first mechanism is supported by data showing that invariant natural killer T (iNKT) cells, which recognize and respond to glycolipid antigens presented on the major histocompatibility complex class I–related molecule CD1d148 are altered in mice lacking commensal bacteria149,150. Germ-free mice and a wild-type strain with less Sphingomonas, a glycolipid-producing commensal bacteria taxa, have fewer iNKT cells compared to wild-type mice harboring more Sphingomonas150. Others have shown that there are more iNKT cells in the small intestine of germ-free mice, a tissue-specific effect associated with signals derived from commensal bacteria early in mouse development that regulate the ontogeny of iNKT cells150.
Commensal bacteria might also regulate immune cell function through the fatty acidy sensor proteins in the peroxisome proliferator-activated receptor (PPAR) family of nuclear receptors that recognize endogenous and exogenous lipid moieties and initiate transcriptional changes linked to metabolic reprogramming and immune function151-155. Commensal bacteria promote PPAR-γ–dependent export of NF-κB from the nucleus and decrease expression of the NF-κB–dependent gene Il8 in a PPAR-γ–dependent manner156. Moreover, treatment of human intestinal epithelial cells from new-borns with Enterococcus fecalis is associated with the activation of PPAR-γ1 and increased production of IL-10 (ref. 157). Therefore, commensal bacteria appear capable of producing or stimulating the production of PPAR-activating molecules. However, additional research is needed to investigate the molecular mechanisms by which metabolites derived from commensal bacteria regulate PPAR family members and to explore how commensal bacteria regulate the immune system via PPARs.
An alternative mechanism by which alterations in lipid homeostasis can alter immune system function is by establishing a metabolic state that favors some immune processes over others. For example, T cells rely on fatty-acid oxidation at steady state and proceed through multiple metabolic checkpoints as they become activated, expand, contract and differentiate into effector or memory T cells25,26,158. T helper cells, including TH1, TH2 and TH17 cells, tend to rely on glucose metabolism, whereas regulatory T cells and memory CD8+ T cells exhibit increased fatty acid oxidation27,28,159. Blocking glycolysis inhibits development of TH17 cells and promotes generation of T regulatory cells28. Treating mice with the anti-diabetes drug metformin or the immunosuppressant drug rapamycin, two pleiotropic molecules that promote oxidation of fatty acids via distinct pathways, is associated with increased CD8+ memory T cell formation and function159. Oxidation of fatty acids also regulates hematopoietic stem cell maintenance and immune cell hematopoiesis160. Thus, the metabolic characteristics of immune cells may dictate their development and function, and alterations in lipid homeostasis that are dependent on commensal bacteria may represent an unappreciated mechanism through which commensal bacteria influence the immune system. Additional functional studies will be required to understand how lipids derived from commensal bacteria and alterations in host lipid homeostasis mediated by commensal bacteria regulate immune responses.
Conclusions and perspectives
In this Review we discussed how beneficial commensal bacteria in the gastrointestinal tract regulate the production of immunomodulatory, diet-dependent nutrients and metabolites. Commensal bacteria are required for the production of bile acids and SCFAs, both of which have anti-inflammatory properties in multiple immune cell populations. In addition, commensal bacteria are important sources of vitamins and amino acids and regulate systemic lipid homeostasis, and alterations in the levels of these metabolites can influence immune function. Although important advances have been made in our under-standing of the commensal bacteria–metabolite–immune system axis, fundamental gaps in knowledge remain regarding how nutrients and other metabolites derived from commensal bacteria regulate host metabolism and immunity. Future challenges include interrogation of the molecular mechanisms through which nutrients and metabolites derived from commensal bacteria regulate immune responses and linking the commensal bacteria–metabolite–immune system axis to human diseases that are associated with alterations in populations of intestinal commensal bacteria. Elaborating our understanding of the commensal bacteria–metabolite–immune system axis in the context of health and disease may provide useful insights for the development of improved preventive and therapeutic agents of multiple infectious, inflammatory and metabolic disorders.
ACKNOWLEDGMENTS
We thank all members of the Artis laboratory for discussions and critical reading of the manuscript. Supported by US National Institutes of Health grants (AI061570, AI074878, AI083480, AI087990, AI095466, AI095608, AI097333 and AI102942 to D.A.), the Burroughs Wellcome Fund Investigator in Pathogenesis of Infectious Disease Award (D.A.) and the Crohn’s and Colitis Foundation of America (D.A.). J.R.B. is supported by National Institutes of Health grant T32-AI060516.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Semova I, et al. Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe. 2012;12:277–288. doi: 10.1016/j.chom.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shin SC, et al. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science. 2011;334:670–674. doi: 10.1126/science.1212782. [DOI] [PubMed] [Google Scholar]
- 5.Bäckhed F, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes linked to obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 7.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 8.Hooper LV, Midtvedt T, Gordon JI. Host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 9.Flint HJ, Scott KP, Louis P, Duncan SH. The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 2012;9:577–589. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- 10.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Musso G, Gambino R, Cassader M. Interactions between gut microbiota and host metabolism predisposing to obesity. Annu. Rev. Med. 2011;62:361–380. doi: 10.1146/annurev-med-012510-175505. [DOI] [PubMed] [Google Scholar]
- 12.Nicholson JK, et al. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 13.Holmes E, Li JV, Athanasiou T, Ashrafian H, Nicholson JK. Understanding the role of gut microbiome-host metabolic signal disruption in health and disease. Trends Microbiol. 2011;19:349–359. doi: 10.1016/j.tim.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 14.Tremaroli V, Bächked F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 15.Hill DA, Artis D. Intestinal bacteria and the regulation of immune cell homeostasis. Annu. Rev. Immunol. 2010;28:623–667. doi: 10.1146/annurev-immunol-030409-101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Littman DR, Pamer EG. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe. 2011;10:311–323. doi: 10.1016/j.chom.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chinen T, Rudensky AY. The effects of commensal microbiota on immune cell subsets and inflammatory responses. Immunol. Rev. 2012;245:45–55. doi: 10.1111/j.1600-065X.2011.01083.x. [DOI] [PubMed] [Google Scholar]
- 19.Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu. Rev. Immunol. 2012;30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Molloy MJ, Bouladoux N, Belkaid Y. Intestinal microbiota: shaping local and systemic immune responses. Semin. Immunol. 2012;24:58–66. doi: 10.1016/j.smim.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abt MC, Artis D. The dynamic influence of commensal bacteria on the immune response to pathogens. Curr. Opin. Microbiol. 2013;16:4–9. doi: 10.1016/j.mib.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamada N, Seo S, Chen GY, Núñez G. Role of gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 24.Abraham C, Medzhitov R. Interactions between the host innate immune system and microbes in inflammatory bowel disease. Gastroenerology. 2011;140:1729–1737. doi: 10.1053/j.gastro.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang R, Green DR. Metabolic checkpoints in activated T cells. Nat. Immunol. 2012;13:907–915. doi: 10.1038/ni.2386. [DOI] [PubMed] [Google Scholar]
- 26.Pearce EL, Pearce EJ. Metabolic pathways in immune cell activation and quiescence. Immunity. 2013;38:633–643. doi: 10.1016/j.immuni.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michalek RD, et al. Cutting Edge: Distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J. Immunol. 2011;186:3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi LZ, et al. HIF1α-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of Th17 and Treg cells. J. Exp. Med. 2011;208:1367–1376. doi: 10.1084/jem.20110278. References 27 and 28 demonstrate that distinct metabolic programs critically regulate differentiation of T cell subsets.
- 29.Haschemi A, et al. The sedoheptulose kinase CARKL directs macrophage polarization through control of glucose metabolism. Cell Metab. 2012;15:813–826. doi: 10.1016/j.cmet.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donohoe DR, Wali A, Brylawski BP, Bultman SJ. Microbial regulation of glucose metabolism and cell-cycle progression in mammalian coloncytes. PLoS ONE. 2012;7:e46589. doi: 10.1371/journal.pone.0046589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Odegaard JI, Chawla A. The immune system as a sensor of the metabolic state. Immunity. 2013;38:644–654. doi: 10.1016/j.immuni.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signaling for metabolic diseases. Nat. Rev. Drug Discov. 2008;7:678–693. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- 33.Fiorucci S, Mencarelli A, Palladino G, Cipriani S. Bile-acid-activated receptors: targeting TGR5 and farnesoid-X-receptor in lipid and glucose disorders. Trends Pharmacol. Sci. 2009;30:570–580. doi: 10.1016/j.tips.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 34.Ridlon JM, Kang DL, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 35.Trauner M, Boyer JL. Bile salt transporters: molecular characterization, function, and regulation. Physiol. Rev. 2003;83:633–671. doi: 10.1152/physrev.00027.2002. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka H, Hashiba H, Kok J, Mierau I. Bile salt hydrolase of Bifidobacterium longum-biochemical and genetic characterization. Appl. Environ. Microbiol. 2000;66:2502–2512. doi: 10.1128/aem.66.6.2502-2512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl. Acad. Sci. USA. 2008;105:13580–13585. doi: 10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sayin SI, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. This article comprehensively characterizes bile acid metabolism in multiple mouse tissues and provides insight into how beneficial commensal bacteria in the intestine regulate metabolism of bile acids.
- 39.Martin FP, et al. A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Mol. Syst. Biol. 2007;3:112. doi: 10.1038/msb4100153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Claus SP, et al. Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Mol. Syst. Biol. 2008;4:219. doi: 10.1038/msb.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin FP, et al. Panorganismal gut microbiome-host metabolic crosstalk. J. Proteome Res. 2009;8:2090–2105. doi: 10.1021/pr801068x. [DOI] [PubMed] [Google Scholar]
- 42.Martin FP, et al. Dietary modulation of gut functional ecology studied by fecal metabonomics. J. Proteome Res. 2010;9:5284–5295. doi: 10.1021/pr100554m. [DOI] [PubMed] [Google Scholar]
- 43.Swann JR, et al. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc. Natl. Acad. Sci. USA. 2011;108:4523–4530. doi: 10.1073/pnas.1006734107. This article comprehensively characterizes amounts of bile acid metabolites in multiple tissues of germ-free mice versus conventionally reared mice.
- 44.Claus SP, et al. Colonization-induced host-gut microbial metabolic interaction. MBio. 2011;2:e00271–10. doi: 10.1128/mBio.00271-10. References 39-42 and 44 compare metabolite levels in multiple compartments of conventionally reared mice versus germ-free mice using metabolomic approaches.
- 45.Duboc H, et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut. 2013;62:531–539. doi: 10.1136/gutjnl-2012-302578. [DOI] [PubMed] [Google Scholar]
- 46.Vavassori P, Mencarelli A, Renga B, Distrutti E, Fiorucci S. The bile acid receptor FXR is a modulator of intestinal innate immunity. J. Immunol. 2009;183:6251–6261. doi: 10.4049/jimmunol.0803978. This article demonstrates that FXR regulates intestinal inflammation in a model of IBD and provides mechanistic insight into how bile acid-FXR signaling inhibits activity of NF-κB.
- 47.Wang YD, Chen WD, Yu D, Forman BM, Huang W. The G-protein-coupled bile acid receptor, Gpbar1 (TGR5), negatively regulated hepatic inflammatory response through antagonizing nuclear factor κ light-chain enhancer of activated B cells (NF-κB) in mice. Hepatology. 2011;54:1421–1432. doi: 10.1002/hep.24525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pols TW, et al. TGR5 activation inhibits atherosclerosis by reducing macrophage inflammation and lipid loading. Cell Metab. 2011;14:747–757. doi: 10.1016/j.cmet.2011.11.006. This article demonstrates that the bile acid receptor TGR5 attenuates atherosclerosis by decreasing macrophage-associated inflammation.
- 49.Maruyama T, et al. Identification of membrane-type receptor for bile acids (M-BAR) Biochem. Biophys. Res. Commun. 2002;298:714–719. doi: 10.1016/s0006-291x(02)02550-0. [DOI] [PubMed] [Google Scholar]
- 50.Pellicciari R, et al. Discovery of 6alpha-ethyl-23(S)-methylcholic acid (S-EMCA, INT-777) as a potent and selective agonist for the TGR5 receptor, a novel target for diabesity. J. Med. Chem. 2009;52:7958–7961. doi: 10.1021/jm901390p. [DOI] [PubMed] [Google Scholar]
- 51.David M, Petricoin E, III, Larner AC. Activation of protein kinase A inhibits interferon induction of the Jak/Stat pathway in U266 cells. J. Biol. Chem. 1996;271:4585–4588. doi: 10.1074/jbc.271.9.4585. [DOI] [PubMed] [Google Scholar]
- 52.Lee EH, Rikihisa Y. Protein kinase A-mediated inhibition of gamma interferon-induced tyrosine phosphorylation of Janus kinases and latent cytoplasmic transcription factors in human monocytes by Ehrlichia chaffeensis. Infect. Immun. 1998;66:2514–2520. doi: 10.1128/iai.66.6.2514-2520.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wen AY, Sakamoto KM, Miller LS. The role of the transcription factor CREB in immune function. J. Immunol. 2010;185:6413–6419. doi: 10.4049/jimmunol.1001829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cipriani S, et al. The bile acid receptor GPBAR-1 (TGR5) modulates integrity of intestinal barrier and immune response to experimental colitis. PLoS ONE. 2011;6:e25637. doi: 10.1371/journal.pone.0025637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gadaleta RM, et al. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut. 2011;60:463–472. doi: 10.1136/gut.2010.212159. [DOI] [PubMed] [Google Scholar]
- 56.Mencarelli A, et al. The bile acid sensor farnesoid X receptor is a modulator of liver immunity in a rodent model of acute hepatitis. J. Immunol. 2009;183:6657–6666. doi: 10.4049/jimmunol.0901347. [DOI] [PubMed] [Google Scholar]
- 57.Diao H, et al. Osteopontin as a mediator of NKT cell function in T cell-mediated liver diseases. Immunity. 2004;21:539–550. doi: 10.1016/j.immuni.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 58.Lenz K. Bile acid metabolism and vitamin B12 absorption in ulcerative colitis. Scand. J. Gastroenterol. 1976;11:769–775. [PubMed] [Google Scholar]
- 59.Rutgeerts P, Ghoos Y, Vantrappen G. Bile acid studies in patients with Crohn’s colitis. Gut. 1979;20:1072–1077. doi: 10.1136/gut.20.12.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 62.Cani PD, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 63.Kobayashi M, et al. Prevention and treatment of obesity, insulin resistance, and diabetes by bile acid-binding resin. Diabetes. 2007;56:239–247. doi: 10.2337/db06-0353. [DOI] [PubMed] [Google Scholar]
- 64.Henao-Mejia J, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qin J, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 66.Karlsson FH, et al. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat. Commun. 2012;3:1245. doi: 10.1038/ncomms2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koren O, et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc. Natl. Acad. Sci. USA. 2011;108:4592–4598. doi: 10.1073/pnas.1011383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sobhani I, et al. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS ONE. 2011;6:e16393. doi: 10.1371/journal.pone.0016393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abt MC, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37:158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ganal SC, et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. 2012;37:171–186. doi: 10.1016/j.immuni.2012.05.020. References 69 and 70 demonstrate that commensal bacteria-derived signals regulate antiviral immunity.
- 71.Renga B, et al. The acid sensor FXR is required for immune-regulatory activities of TLR-9 in intestinal inflammation. PLoS ONE. 2013;8:e54472. doi: 10.1371/journal.pone.0054472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nijmeijer RM, et al. Farnesoid X receptor (FXR) activation and FXR genetic variation in inflammatory bowel disease. PLoS ONE. 2011;6:e23745. doi: 10.1371/journal.pone.0023745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Devkota S, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. This reference demonstrates that at least some bile acids promote outgrowth of a pathogenic bacterial species in IL-10-deficient mice.
- 74.Chang KO, et al. Bile acids are essential for porcine enteric calicivirus replication in association with down-regulation of signal transducer and activator of transcription 1. Proc. Natl. Acad. Sci. USA. 2004;101:8733–8738. doi: 10.1073/pnas.0401126101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chang KO, George DW. Bile acids promote the expression of hepatitis C virus in replicon-harboring cells. J. Virol. 2007;81:9633–9640. doi: 10.1128/JVI.00795-07. References 74 and 75 demonstrate that bile acids regulate viral replication.
- 76.Miller TL, Wolin MJ. Fermentations by saccharolytic intestinal bacteria. Am. J. Clin. Nutr. 1979;32:164–172. doi: 10.1093/ajcn/32.1.164. [DOI] [PubMed] [Google Scholar]
- 77.Cummings JH. Fermentation in the human large intestine: evidence and implications for health. Lancet. 1983;1:1206–1209. doi: 10.1016/s0140-6736(83)92478-9. [DOI] [PubMed] [Google Scholar]
- 78.Cummings JH, Macfarlane GT. The control and consequences of fermentation in the human colon. J. Appl. Bacteriol. 1991;70:443–459. doi: 10.1111/j.1365-2672.1991.tb02739.x. [DOI] [PubMed] [Google Scholar]
- 79.Wong JMW, de Souza R, Kendall CWC, Emam A, Jenkins DJA. Colonic health: fermentation and short chain fatty acids. J. Clin. Gastroenterol. 2006;40:235–243. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- 80.Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short chain fatty acids in human large intestine, portal, hepatic, and venous blood. Gut. 1987;28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proc. Nutr. Soc. 2003;62:67–72. doi: 10.1079/PNS2002207. [DOI] [PubMed] [Google Scholar]
- 82.Smiricky-Tjardes MR, et al. In vitro fermentation characteristics of selected oligosaccharides by swine fecal microflora. J. Anim. Sci. 2003;81:2505–2514. doi: 10.2527/2003.81102505x. [DOI] [PubMed] [Google Scholar]
- 83.Høverstad T, Midtvedt T. Short-chain fatty acids in germfree mice and rats. J. Nutr. 1986;116:1772–1776. doi: 10.1093/jn/116.9.1772. [DOI] [PubMed] [Google Scholar]
- 84.Donohoe DR, et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011;13:517–526. doi: 10.1016/j.cmet.2011.02.018. This article demonstrates that commensal bacteria-derived butyrate, an SCFA, is critical for maintaining metabolic homeostasis and regulating autophagy in colonocytes.
- 85.Maslowski KM, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. This article demonstrates that commensal bacteria-derived SCFAs have an anti-inflammatory role in a model of IBD.
- 86.Bjursell M, et al. Improved glucose control and reduced body fat mass in free fatty acid receptor 2-deficient mice fed a high-fat diet. Am. J. Physiol. Endocrinol. Metab. 2011;300:E211–E220. doi: 10.1152/ajpendo.00229.2010. [DOI] [PubMed] [Google Scholar]
- 87.Bellahcene M, et al. Male mice that lack the G-protein-coupled receptor GPR41 have low energy expenditure and increased body fat content. Br. J. Nutr. 2012;109:1755–1764. doi: 10.1017/S0007114512003923. [DOI] [PubMed] [Google Scholar]
- 88.Kimura I, et al. Short-chain fatty acids and ketones directly regulate sympathetic nervous system via G protein-coupled receptor 41 (GPR41) Proc. Natl. Acad. Sci. USA. 2011;108:8030–8035. doi: 10.1073/pnas.1016088108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sina C, Jiang H-P, Li J, Schreiber S, Rosenstiel P. G protein-coupled receptor 43 is essential for neutrophil recruitment during intestinal inflammation. J. Immunol. 2009;183:7514–7522. doi: 10.4049/jimmunol.0900063. [DOI] [PubMed] [Google Scholar]
- 90.Vinolo MA, et al. SCFAs induce mouse neutrophil chemotaxis through the GPR43 receptor. PLoS ONE. 2011;6:e21205. doi: 10.1371/journal.pone.0021205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Brown AJ, et al. The orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain fatty acids. J. Biol. Chem. 2003;278:11312–11319. doi: 10.1074/jbc.M211609200. [DOI] [PubMed] [Google Scholar]
- 92.Le Poul E, et al. Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J. Biol. Chem. 2003;278:25481–25489. doi: 10.1074/jbc.M301403200. [DOI] [PubMed] [Google Scholar]
- 93.Nilsson NE, Kotarsky K, Owman C, Olde B. Identification of a free fatty acid receptor, FFA2R, expressed on leukocytes and activated by short-chain fatty acids. Biochem. Biophys. Res. Commun. 2003;303:1047–1052. doi: 10.1016/s0006-291x(03)00488-1. References 91-93 provide comprehensive pharmacologic characterizations of SCFA-GPR41 and SCFA-GPR43 interactions and demonstrate that SCFAs regulate immune cells.
- 94.Cousens LS, Gallwitz D, Alberts BM. Different accessibilities in chromatin to histone acetylase. J. Biol. Chem. 1979;254:1716–1723. [PubMed] [Google Scholar]
- 95.Donohoe DR, et al. The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol. Cell. 2012;48:612–626. doi: 10.1016/j.molcel.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hinnebusch BF, Meng S, Wu JT, Archer SY, Hodin RA. The effects of short-chain fatty acids on human colon cancer cell phenotype are associated with histone hyperacetylation. J. Nutr. 2002;132:1012–1017. doi: 10.1093/jn/132.5.1012. [DOI] [PubMed] [Google Scholar]
- 97.Waldecker M, Kautenburger T, Daumann H, Busch C, Schrenk D. Inhibition of histone-deacetylase activity by short-chain fatty acids and some polyphenol metabolites formed in the colon. J. Nutr. Biochem. 2008;19:587–593. doi: 10.1016/j.jnutbio.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 98.Virgin HW, Levine B. Autophagy genes in immunity. Nat. Immunol. 2009;10:461–470. doi: 10.1038/ni.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hudson BD, Tikhonova IG, Pandey SK, Ulven T, Milligan G. Extracellular ionic locks determine variation in constitutive activity and ligand potency between species orthologs of the free fatty acid receptors FFA2 and FFA3. J. Biol. Chem. 2012;287:41195–41209. doi: 10.1074/jbc.M112.396259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cox MA, et al. Short-chain fatty acids act as anti-inflammatory mediators by regulating prostaglandin E(2) and cytokines. World J. Gastroenterol. 2009;15:5549–5557. doi: 10.3748/wjg.15.5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Venkatraman A, et al. Amelioration of dextran sulfate colitis by butyrate: role of heat shock protein 70 and NF-κB. Am. J. Physiol. Gastroenterol. Liver Physiol. 2003;285:G177–G184. doi: 10.1152/ajpgi.00307.2002. [DOI] [PubMed] [Google Scholar]
- 102.Berndt BE, et al. Butyrate increases IL-23 production by stimulated dendritic cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2012;303:G1384–G1392. doi: 10.1152/ajpgi.00540.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu L, et al. Butyrate interferes with the differentiation and function of human monocyte-derived dendritic cells. Cell Immunol. 2012;277:66–73. doi: 10.1016/j.cellimm.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 104.Eftimiadi C, et al. Divergent effect of the anaerobic bacteria by-product butyric acid on the immune response: suppression of T-lymphocyte proliferation and stimulation of interleuking-1 beta production. Oral Microbiol. Immunol. 1991;6:17–23. doi: 10.1111/j.1399-302x.1991.tb00446.x. [DOI] [PubMed] [Google Scholar]
- 105.Gilbert KM, DeLoose A, Valentine JL, Fifer EK. Structure-activity relationship between carboxylic acids and T cell cycle blockade. Life Sci. 2006;78:2159–2165. doi: 10.1016/j.lfs.2005.09.047. [DOI] [PubMed] [Google Scholar]
- 106.Bailón E, et al. Butyrate in vitro immune-modulatory effects might be mediated through a proliferation-related induction of apoptosis. Immunobiology. 2010;215:863–873. doi: 10.1016/j.imbio.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 107.Zimmerman MA, et al. Butyrate suppresses colonic inflammation through HDAC1-dependent Fas upregulation and Fas-mediated apoptosis of T cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2012;302:G1405–G1415. doi: 10.1152/ajpgi.00543.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Huang N, Katz JP, Martin DR, Wu GD. Inhibition of IL-8 gene expression in Caco-2 cells by compounds which induce histone hyperacetylation. Cytokine. 1997;9:27–36. doi: 10.1006/cyto.1996.0132. [DOI] [PubMed] [Google Scholar]
- 109.Patel KK, Stappenbeck TS. Autophagy and intestinal homeostasis. Annu. Rev. Physiol. 2012;75:241–262. doi: 10.1146/annurev-physiol-030212-183658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shakespear MR, Halili MA, Irvine KM, Fairlie DP, Sweet MJ. Histone deacetylases as regulators of inflammation and immunity. Trends Immunol. 2011;32:335–343. doi: 10.1016/j.it.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 111.Scheppach W, et al. Effect of butyrate enemas on the colonic mucosa in distal ulcerative colitis. Gastroenterology. 1992;103:51–56. doi: 10.1016/0016-5085(92)91094-k. [DOI] [PubMed] [Google Scholar]
- 112.Segain JP, et al. Buytrate inhibits inflammatory responses through NFkappaB inhibition: implications for Crohn’s disease. Gut. 2000;47:397–403. doi: 10.1136/gut.47.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Resta SC. Effects of probiotics and commensals on intestinal epithelial physiology: implications for nutrient handling. J. Physiol. (Lond.) 2009;587:4169–4174. doi: 10.1113/jphysiol.2009.176370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bhaskaram P. Micronutrient malnutrition, infection, and immunity: an overview. Nutr. Rev. 2002;60:S40–S45. doi: 10.1301/00296640260130722. [DOI] [PubMed] [Google Scholar]
- 115.Cheng CH, Chang SJ, Lee BJ, Lin KL, Huang YC. Vitamin B6 supplementation increases immune responses in critically ill patients. Eur. J. Clin. Nutr. 2006;60:1207–1213. doi: 10.1038/sj.ejcn.1602439. [DOI] [PubMed] [Google Scholar]
- 116.Meydani SN, et al. Vitamin E supplementation and in vivo immune response in healthy elderly subjects: a randomized controlled trial. J. Am. Med. Assoc. 1997;277:1380–1386. doi: 10.1001/jama.1997.03540410058031. [DOI] [PubMed] [Google Scholar]
- 117.Tamura J, et al. Immunomodulation by vitamin B12: augmentation of CD8+ T lymphocytes and natural killer (NK) cell activity in vitamin B12-deficient patients by methyl-B12 treatment. Clin. Exp. Immunol. 1999;116:28–32. doi: 10.1046/j.1365-2249.1999.00870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cantorna MT, Zhu Y, Froicu M, Wittke A. Vitamin D status, 1,25-dihydroxyvitamin D3, and the immune system. Am. J. Clin. Nutr. 2004;80:1717S–1720S. doi: 10.1093/ajcn/80.6.1717S. [DOI] [PubMed] [Google Scholar]
- 119.Hashimoto T, et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487:477–481. doi: 10.1038/nature11228. This article suggests that commensal bacteria may regulate intestinal inflammation by influencing absorption of amino acids.
- 120.Kunisawa J, Hashimoto E, Ishikawa I, Kiyono H. A pivotal role of vitamin B9 in the maintenance of regulatory T cells in vitro and in vivo. PLoS ONE. 2012;7:e32094. doi: 10.1371/journal.pone.0032094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Spencer SP, Belkaid Y. Dietary and commensal derived nutrients: shaping mucosal and systemic immunity. Curr. Opin. Immunol. 2012;24:379–384. doi: 10.1016/j.coi.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kjer-Nielsen L, et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 2012;491:717–723. doi: 10.1038/nature11605. This article demonstrates that B-vitamin metabolites bind MR1 and promote mucosa-associated invariant T cell activation.
- 123.Dusseaux M, et al. Human MAIT cells are xenobiotic resistant, tissue-targeted, CD161hi IL-17 secreting T cells. Blood. 2011;117:1250–1259. doi: 10.1182/blood-2010-08-303339. [DOI] [PubMed] [Google Scholar]
- 124.Walker LJ, et al. Human MAIT cells and CD8alphaalpha cells develop from a pool of type-17 precommitted CD8+ T cells. Blood. 2012;119:422–433. doi: 10.1182/blood-2011-05-353789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Le Bourhis L, et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat. Immunol. 2010;11:701–708. doi: 10.1038/ni.1890. [DOI] [PubMed] [Google Scholar]
- 126.Le Bourhis L, Mburu YK, Lantz O. MAIT cells, surveyors of a new class of antigen: development and functions. Curr. Opin. Immunol. 2013;25:174–180. doi: 10.1016/j.coi.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 127.Smith MI, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339:548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Trehan I, et al. Antibiotics as part of the management of severe acute malnutrition. N. Engl. J. Med. 2013;368:425–435. doi: 10.1056/NEJMoa1202851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mestdagh R, et al. Gut microbiota modulate the metabolism of brown adipose tissue in mice. J. Proteome Res. 2012;11:620–630. doi: 10.1021/pr200938v. [DOI] [PubMed] [Google Scholar]
- 130.Tannahill GM, et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 2013;496:238–242. doi: 10.1038/nature11986. This article demonstrates that glucose oxidation and amounts of the citric acid cycle intermediate succinate regulate production of IL-1β.
- 131.Matsumoto M, et al. Impact of intestinal microbiota on intestinal luminal metabolome. Scientific Reports. 2012;2:233. doi: 10.1038/srep00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Whitt DD, Demoss RD. Effect of microflora on the free amino acid distribution in various regions of the mouse gastrointestinal tract. Appl. Microbiol. 1975;30:609–615. doi: 10.1128/am.30.4.609-615.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.McGaha TL, et al. Amino acid catabolism: a pivotal regulator of innate and adaptive immunity. Immunol. Rev. 2012;249:135–157. doi: 10.1111/j.1600-065X.2012.01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Morris SM., Jr. Arginases and arginine deficiency syndromes. Curr. Opin. Clin. Nutr. Metab. Care. 2012;15:64–70. doi: 10.1097/MCO.0b013e32834d1a08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Puccetti P, Grohmann U. IDO and regulatory T cells: a role for reverse signaling and non-canonical NF-κB activation. Nat. Rev. Immunol. 2007;7:817–823. doi: 10.1038/nri2163. [DOI] [PubMed] [Google Scholar]
- 136.Das P, Lahiri A, Lahiri A, Chakravortty D. Modulation of the arginase pathway in the context of microbial pathogenesis: a metabolic enzyme moonlighting as an immune modulator. PLoS Pathog. 2010;6:e1000899. doi: 10.1371/journal.ppat.1000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Munn DH, et al. Inhibition of T cell proliferation by macrophage tryptophan catabolism. J. Exp. Med. 1999;189:1363–1372. doi: 10.1084/jem.189.9.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Nowak EC, et al. Tryptophan hydroxylase-1 regulates immune tolerance and inflammation. J. Exp. Med. 2012;209:2127–2135. doi: 10.1084/jem.20120408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rodriguez PC, et al. Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Res. 2004;64:5839–5849. doi: 10.1158/0008-5472.CAN-04-0465. [DOI] [PubMed] [Google Scholar]
- 140.Cobbold SP, et al. Infectious tolerance via the consumption of essential amino acids and mTOR signaling. Proc. Natl. Acad. Sci. USA. 2009;106:12055–12060. doi: 10.1073/pnas.0903919106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Scrimshaw NS, Wilson D, Bressani R. Infection and kwaszhiorkor. J. Trop. Pediatr. 1960;6:37–43. doi: 10.1093/oxfordjournals.tropej.a057567. [DOI] [PubMed] [Google Scholar]
- 142.Müller O, Krawinkel M. Malnutrition and health in developing countries. CMAJ. 2005;173:279–286. doi: 10.1503/cmaj.050342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Black RE, et al. Maternal and child undernutrition: global and regional exposures and health consequences. Lancet. 2008;371:243–260. doi: 10.1016/S0140-6736(07)61690-0. [DOI] [PubMed] [Google Scholar]
- 144.Rice AL, Sacco L, Hyder A, Black RE. Malnutrition as an underlying cause of childhood deaths associated with infectious diseases in developing countries. Bull. World Health Organ. 2000;78:1207–1221. [PMC free article] [PubMed] [Google Scholar]
- 145.Pretorius PJ, De Villers LS. Antibody response in children with protein malnutrition. Am. J. Clin. Nutr. 1962;10:379–383. doi: 10.1093/ajcn/10.5.379. [DOI] [PubMed] [Google Scholar]
- 146.Savy M, et al. Landscape analysis of interactions between nutrition and vaccine responses in children. J. Nutr. 2009;139:2154S–2218S. doi: 10.3945/jn.109.105312. [DOI] [PubMed] [Google Scholar]
- 147.Dumas ME, et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc. Natl. Acad. Sci. USA. 2006;103:12511–12516. doi: 10.1073/pnas.0601056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Rossjohn J, Pellicci DG, Patel O, Gapin L, Godfrey DI. Recognition of CD1d-restricted antigens by natural killer T cells. Nat. Rev. Immunol. 2012;12:845–857. doi: 10.1038/nri3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wei B, et al. Commensal microbiota and CD8+ T cells shape the formation of invariant NKT cells. J. Immunol. 2010;184:1218–1226. doi: 10.4049/jimmunol.0902620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Olszak T, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kidani Y, Bensinger SJ. Liver X receptor and peroxisome proliferator-activated receptor as integrators of lipid homeostasis and immunity. Immunol. Rev. 2012;249:72–83. doi: 10.1111/j.1600-065X.2012.01153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hong C, et al. Coordinate regulation of neutrophil homeostasis by liver X receptors in mice. J. Clin. Invest. 2012;122:337–347. doi: 10.1172/JCI58393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Odegaard JI, et al. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007;447:1116–1120. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Odegaard JI, et al. Alternative M2 activation of Kupffer cells by PPARdelta ameliorates obesity-induced insulin resistance. Cell Metab. 2008;7:496–507. doi: 10.1016/j.cmet.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mukundan L, et al. PPAR-δ senses and orchestrates clearance of apoptotic cells to promote tolerance. Nat. Med. 2009;15:1266–1272. doi: 10.1038/nm.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kelly D, et al. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-γ and RelA. Nat. Immunol. 2004;5:104–112. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- 157.Are A, et al. Enterococcus faecalis from newborn babies regulate endogenous PPARgamma activity and IL-10 levels in colonic epithelial cells. Proc. Natl. Acad. Sci. USA. 2008;105:1943–1948. doi: 10.1073/pnas.0711734105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gerriets VA, Rathmell JC. Metabolic pathways in T cell fate and function. Trends Immunol. 2012;33:168–173. doi: 10.1016/j.it.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pearce EL, et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–107. doi: 10.1038/nature08097. This article suggests that metabolism of fatty acids is critical for formation of CD8+ memory T cells.
- 160.Ito K, et al. PML-PPAR-δ pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat. Med. 2012;18:1350–1358. doi: 10.1038/nm.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]