MicroRNAs (miRNAs) are full of surprises. The more we study these ~22-nucleotide non-coding RNAs, the more we realize how little is understood about this mode of gene regulation.1 Case in point: lin-4, the founding member of the miRNA superfamily discovered over 20 years ago. Thousands of miRNAs have since been discovered in plant, animal, and viral genomes. They direct important cellular processes, such as proliferation, apoptosis, differentiation, metabolic and immune responses. Mounting evidence supports the canonical view that miRNAs function to block expression of their targets by associating in a sequence-specific manner to messenger RNA (mRNA) transcripts, causing mRNA degradation and/or translational repression. A recent report by Turner et al. offers an expanded function for miRNAs in regulating gene expression.2 Their work, focusing on lin-4, suggests that miRNAs can bind to their own promoters to induce their transcription—a process referred to as RNA activation (RNAa).
Our fundamental understanding of miRNA mechanisms arose from studying lin-4 in the simple nematode Caenorhabditis elegans (C. elegans). Early work by the Ambros and Ruvkun laboratories revealed lin-4 as the first gene in a cascade of factors required to direct post-embryonic proliferation and differentiation of hypodermal stem cells into specialized skin cells and egg-laying structures in the worm.3 lin-4 is turned “on” at the end of the first of 4 larval stages (late L1) and binds to complementary regions in the 3′UTR of lin-14 and lin-28 to suppress their expression. Proper timing of lin-4 expression is critical to control the transition between the first and second larval stages during the nematode life cycle. Mutant worms lacking lin-4 reiterate cell fates normally seen in L1, which result in seam cell defects, absence of egg-laying structures, long body shape, and uncoordinated movement in adults.
The question is, then, who is regulating the expression of lin-4, a master regulator of developmental timing in the worm? The control of lin-4 expression is complicated, involving both negative and positive factors (Fig. 1). The RNA binding protein RBM-28 promotes lin-4 activity by directing the processing of lin-4 precursor transcripts into biologically active mature miRNAs.4 FLYWCH Zn-finger transcription factors FLH-1 and FLH-2 bind directly to the lin-4 promoter and repress lin-4 transcription during embryogenesis, but somehow this repression is lifted in the larva at late L1.5 Work in this recent article by Turner et al. implicates lin-4 as the elusive positive trans-acting regulatory factor that activates its own expression during early larval development.2
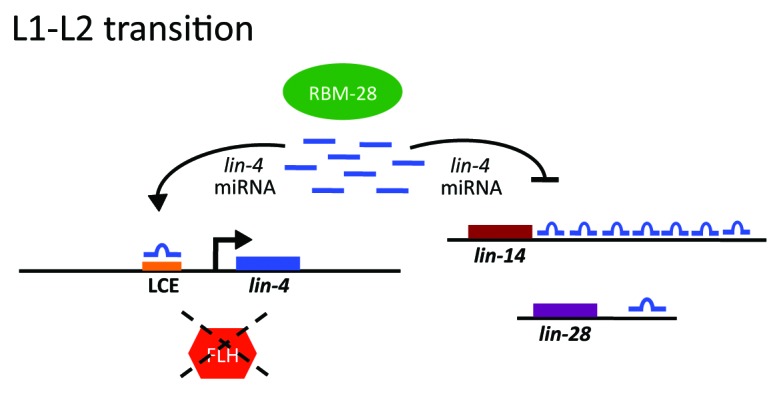
Figure 1. A model of micromanaging gene regulation. In C. elegans, lin-4 controls the transition between the first and second larval stages (L1 and L2) by suppressing lin-14 and lin-28 in a 3′UTR-dependent manner. lin-4 expression at late L1 is tightly regulated at its promoter by positive (lin-4 autoactivation) and negative (embryonically-expressed FLH-1,-2) factors as well as factors promoting lin-4 biogenesis (RBM-28).
The authors used a transcriptional fusion of the lin-4 promoter with green fluorescent protein (GFP) to identify the key element required to turn “on” lin-4 at late L1. This 56-nucleotide DNA region mapped 50 bases upstream of the lin-4 transcriptional start site. Interestingly, a binding site for lin-4 was noted in this region. The lin-4-complementary element (LCE) is conserved in other nematodes (C. remanei and C. vulgaris), implying its biological importance. The authors experimentally verified that lin-4 was essential for activating its own expression, which required an intact LCE and association of RNA polymerase II to the lin-4 promoter. Deletion of the LCE blocked lin-4 activity in worms, resulting in abnormal seam cell development and egg-laying defects.
The exact mechanism used by lin-4 for autoregulation is unknown. Electromobility shift assays indicated that the LCE recruits an RNA/protein complex to the lin-4 promoter.2 Unfortunately, factors within this complex were not identified. It is also unclear if lin-4 autoactivation is due to direct recruitment of the general transcription machinery to its promoter and/or activation of RNA polymerase II at the transcriptional start site, or if this is accomplished indirectly through chromatin or histone modifications.
The ability of a miRNA to induce (rather than block) gene expression has been documented for other small RNAs. The C. elegans let-7 miRNA autoregulates its expression by recruiting Argonaute to the 3′end of the let-7 primary transcript to promote its biogenesis.6 miR-373 can bind to the promoters of human E-cadherin and CSDC2 to activate transcription and increase their protein levels.7 However, this recent Cell Cycle report is novel, because it shows that lin-4 binds to its own promoter to autoactivate expression.2
MiRNAs are more complex than initially predicted and direct important functions in the nucleus as well as the cytoplasm to modulate gene expression in negative and positive ways. Lessons learned in C. elegans imply that the 3 human lin-4 homologs (miR-125a, -125b-1, and -125b-2) likely use similar strategies for their regulation. More work is required to determine how widely miRNA autoregulation is employed across the superfamily. Misexpression of the lin-4 family is closely associated with human diseases such as cancer, and therefore these findings could lead to innovative therapeutics in the clinic.8 Stay tuned—these tiny RNAs likely have bigger surprises in store for us!
Turner MJ, et al. Cell Cycle. 2014;13:772–781. doi: 10.4161/cc.27679.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/28384
References
- 1.Breving K, et al. Int J Biochem Cell Biol. 2010;42:1316–29. doi: 10.1016/j.biocel.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 2.Turner MJ, et al. Cell Cycle. 2014;13 doi: 10.4161/cc.27679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambros V. Curr Opin Genet Dev. 2000;10:428–33. doi: 10.1016/S0959-437X(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 4.Bracht JR, et al. Dev Biol. 2010;348:210–21. doi: 10.1016/j.ydbio.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ow MC, et al. Genes Dev. 2008;22:2520–34. doi: 10.1101/gad.1678808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zisoulis DG, et al. Nature. 2012;486:541–4. doi: 10.1038/nature11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Place RF, et al. Proc Natl Acad Sci U S A. 2008;105:1608–13. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esquela-Kerscher A, et al. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]


