Abstract
Evolutionary theories of aging predict the existence of certain genes that provide selective advantage early in life with adverse effect on lifespan later in life (antagonistic pleiotropy theory) or longevity insurance genes (disposable soma theory). Indeed, the study of human and animal genetics is gradually identifying new genes that increase lifespan when overexpressed or mutated: gerontogenes. Furthermore, genetic and epigenetic mechanisms are being identified that have a positive effect on longevity. The gerontogenes are classified as lifespan regulators, mediators, effectors, housekeeping genes, genes involved in mitochondrial function, and genes regulating cellular senescence and apoptosis. In this review we demonstrate that the majority of the genes as well as genetic and epigenetic mechanisms that are involved in regulation of longevity are highly interconnected and related to stress response.
Keywords: aging, epigenetics, evolution, genetics, longevity
Introduction
During aging, vital bodily functions such as regeneration and reproduction slowly decline. As a result, the organism loses its ability to maintain homeostasis and becomes more susceptible to stress, diseases, and injuries. A loss of essential body functions leads to age-associated pathologies, which ultimately cause death.
Traditionally, there have been many theories of aging, proposing underlying mechanisms of how aging evolved. The major evolutionary theories of aging are the theory of programmed death,1-3 the mutation accumulation theory of aging,4,5 the antagonistic pleiotropic theory of aging,6 and the evolutionary maintenance (see ref. 7 for a review). Weisman initiated the theoretical approach to the evolution of aging, arguing that natural selection inheritably “programs” death to limit individual lifespan and to clear space for new generations. His view was challenged by Haldane, Medawar, and Williams, who proposed that aging is more stochastic then programmed, because the forces of natural selection diminish with adult age, most rapidly after the peak of reproduction. Hamilton published theoretical work in 1966, deriving a mathematical equation that later became known as “Hamilton’s forces of natural selection” and showing that forces of natural selection indeed decline with age, which was later confirmed experimentally using Drosophila (see ref. 8 and references therein for a review). The mutation accumulation theory of aging postulates that the mechanism of aging evolved through the evolutionary accumulation of germinal mutations with small harmful effects, which do not appear until old age, and thus avoid the negative pressure of natural selection.4,5 The first theory that proposed gerontogenes was the theory of antagonistic pleiotropy.6 Williams postulated positive evolutionary selection of genes that have favorable effects in early life stages but adverse effects in late life (after reaching reproductive success). Indeed, now we know that mutations in many genes important for growth and development (e.g., PI3K, mTOR, see below) can prolong life of model organisms (yeasts, nematodes, flies, and mice). Disposable soma theory, a special case of the theory of antagonistic pleiotropy,9 predicts the existence of genes that control the redistribution of energy resources from body maintenance to growth and reproduction. According to this theory, repair of cellular damage requires energy, competing for energy needs with reproduction. Therefore, in favor to the growth and development conditions of existence, longevity-assurance genes reduce their activity or are temporarily turned off, and aging speed increases. As predicted by this theory, longevity assurance genes exist, as confirmed by experimental overexpression of some antioxidant, DNA-, protein-, and cellular-repair genes, which prolong the lifespan of model animals (fruit flies and mice).
Identification of dozens of genes with mutations that prolong life supports another evolutionary theory, “longevity program” theory.10-14 The longevity program could have arisen in the evolution so the organisms can survive in conditions of short-term extreme environmental stress (overheating, overcooling, overpopulation, reducing caloric intake). Under stress, the program allows the body to exceed its normal lifespan by entering “maintenance mode”. This is associated with such modifications as increased stress resistance, downregulation of the biosynthesis of structural proteins, suspension of growth, and reproduction. Indeed, the survival rate of offspring in circumstances of short-term adverse changes in the environment will be minimal, so it is to evolutionary advantage to reallocate resources to extended longevity of adults, which can start breeding after the improvement of the environmental conditions. For example, C. elegans is showing that genetic program by actively promoting longevity of adults at cold temperatures.15 Artificially induced pro-longevity mutations affect this program, so that individuals go into stress-resistant mode independently of the exogenous conditions. As we shall see, the analysis of large amount of experimental data shows that most of the molecular pathways of longevity are associated with increased stress tolerance.
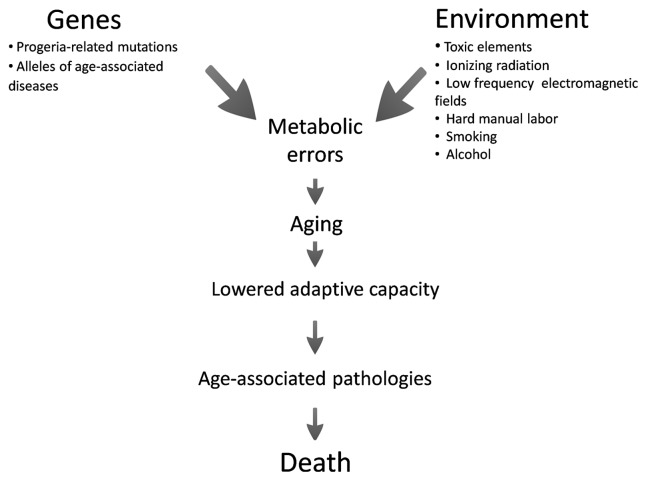
According to accumulation of the errors theory, aging has been viewed as a mechanical exhaustion and accumulation of errors. This model suggests that accidental errors and stress caused by environmental factors result in metabolic abnormalities, increase in free radical production and macromolecular damage at both cellular and tissue levels (Fig. 1).
Figure 1. The effect of environmental and genetic factors on aging and the formation of age-dependent diseases.
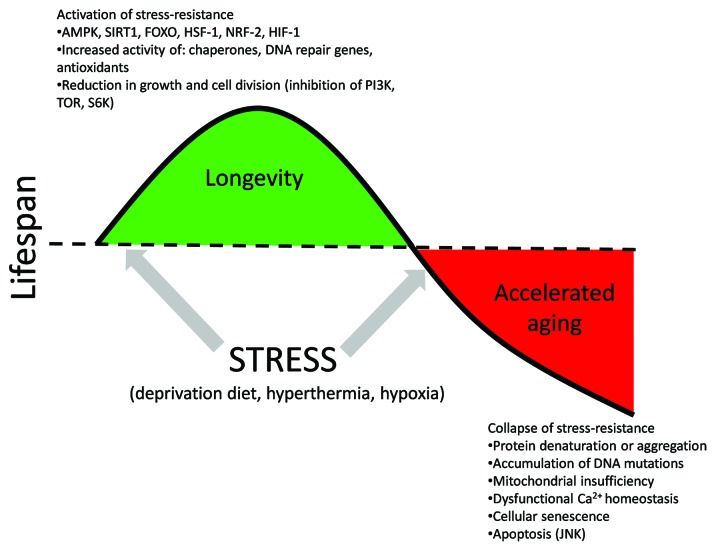
At the same time, it is known that moderate stress could have beneficial effects stimulating innate defense resources of the body, thereby boosting its ability to cope with higher stress levels and slowing down aging.16,17 This is the so-called lifespan hormesis effect.18,19 For instance, in our experiments, we observed the role of DNA repair genes and heat shock protein genes in radiation hormesis in fruit flies.20-22 Moderate stress stimulates expression of genes responsible for stress-resistance promoting prevention or elimination of genetic errors, including the novel and spontaneous ones, thereby delaying the aging process (Fig. 2). On the other hand, prolonged or severe stress exposure exhausts the defense mechanisms, causing drastic accumulation of errors and physiological abnormalities, accelerating the process of aging (Fig. 2).
Figure 2. Stresses of various magnitudes affect aging rate and lifespan through different mechanisms.
Aging research has undergone dramatic expansion in recent years, with the discovery of gerontological genes, or gerontogenes, members of conserved biological pathways across species that increase lifespan when overexpressed or mutated. This discovery led to a renewed interest in understanding how aging is regulated and opened up a new field developing pharmacological treatments that can extend healthy lifespan and slow down human aging process, pioneered by Cynthia Kenyon and Linda Partridge.23-27
Genetics of Aging and Longevity
Identification of gerontogenes—the genes controlling aging and longevity—typically involves model organisms to screen for mutant strains whose rate of aging differs significantly from that of a control group.
The two most efficient methods for identifying new genes are: (1) loss of function: lifespan increases when the gene is inactivated; (2) gain of function: lifespan increases in a mutant with an overexpressed candidate gene.
The phenotypic characteristics that are evaluated are increase in longevity, or emergence of functional aberrations associated with aging (e.g., the dynamics in behavioral responses, elevation of cellular levels of lipofuscin, etc.). In order to accelerate these studies, stress factors can be employed, typically a heat shock or oxidative stress, because stress resistance is frequently linked with life extension.28 Some of the genes, like LMNA, whose mutant version leads to decreased longevity, may be used to find clues for ameliorating age-related diseases.29 However, the most valuable gerontogenes that may ultimately lead to prospective drug candidates for life extension are the genes whose overexpression or polymorphisms lead to increased longevity of the organism.
Using various model organisms, hundreds of genes whose activity was altered in long-lived mutants have been identified. The following signaling pathways are involved in regulating the aging process: insulin/IGF-1, PI3K, TOR, MAPK, AMPK, PKC, NF-κB, TGF-β, Notch and WNT. Under favorable environmental conditions, these signaling cascades control energy balance, cellular plasticity, and the mechanisms supporting homeostasis, growth, and reproduction.30 However, under harsh conditions, the hormonal stimulation of growth is blocked, while stress-resistance proteins are activated. These pathways are evolutionary conserved from invertebrate to mammals.31
Lifespan regulators
The most studied pathway in the aging field is the insulin-like signaling pathway. Upon insulin-like growth factor (IGF-1) binding to its receptor, IGF-1 receptor (IGF-1R), the intracellular phosphoinositol-3-kinase (PI3K) is activated, leading to formation of the downstream intermediate phosphoinositide-3,4,5-triphosphate. The latter binds to 3-phosphoinositid-dependent kinase 1 (PDK-1), which, in turn, phosphorylates and activates the kinases Akt/PKB and SGK-1 that control regular growth processes in the cell. At the same time, the stress-resistance factors, such as FOXO transcriptional factor, are inactivated (see the ref. 28 for review).
It is known, that centenarians are more sensitive to insulin while maintaining low blood levels.32 Insulin-like signaling activity as well as the expression level of insulin-like peptides are reduced in long-lived nematodes, mice, and humans.33-36 Heterozygous mice and humans harboring mutation in a gene encoding receptor for IGF-1 live longer than usual.35,37 Mutations in genes encoding for substrates of insulin receptor 1 and 2 result in the extended lifespan in Drosophila and mouse.38-40 Mutations in genes encoding kinases PI3K, AKT/PKB, and PDK are associated with a prolonged life in animals.41-43 Activity of phosphatases such as PTEN, SHIP1, and SHIP2, counteracting the function of PI3K, also promote longevity.36 Insulin-like signaling inhibits the mechanisms of stress response regulated by FOXO transcription factor. FOXO activity, together with the activity of FOXO-dependent genes, including PEPCK, Hsps, and MnSod, results in life extension.44,45 Another FOXO-dependent gene, GADD45, when overexpressed, leads to a prolonged lifespan and stress resistance in Drosophila and is also associated with a number of age-dependent pathologies in humans.22,46,47
Mutation in a gene encoding kidney hormone Klotho leads to a shortened life in mice, while its overexpression promotes longevity. Klotho suppresses the effect of the insulin/IGF-1 signaling pathway, reinforcing the resistance to oxidative stress at the cellular and organismal levels, thereby promoting longevity.48
A characteristic feature of long-lived Drosophila with the reduced insulin signaling activity is high lipid level.49 Lipid metabolism is downregulated with time, leading to age-dependent diseases such as metabolic syndrome and atherosclerosis. Dyslipidemia is associated with altered activity in a number of genes. Hormones regulating lipid metabolism, such as adiponectin,50 leptin,51 ghrelin,52 and resistin53 play an important role in age-related diseases and longevity.
Lifespan mediators
Peroxisome proliferator-activated receptors (PPARs) are ligand-inducible transcription factors that belong to the nuclear hormone receptor superfamily. PPAR forms a heterodimer with its partner, the retinoic acid receptor X (RXR), which, upon ligand stimulation, binds target DNA sequences called peroxisome proliferator response element (PPRE) to induce gene transcription. PPAR ligands comprise fatty acids and their derivatives. PPARα is expressed in tissues, where a high level of mitochondrial oxidation of fatty acids is required, such as liver, kidney, heart, skeletal muscle, and in blood vessels. PPARα is activated by fatty acids, eicosanoids, 15-d prostaglandin, and oxidized fatty acids. PPARα regulates genes promoting lipid oxidation and metabolism of lipoproteins, such as main apo-lipoprotein of high density, Apo A-1. Through these activities, PPARα function antagonizes the metabolic syndrome and aging in general.54 Pparg-2 (Nr1c3) is activated by fatty acids in an adipose tissue and is known as one of the longevity genes in mammals.55 Pparg-2 plays central role in enhancing insulin sensitivity in the tissue, while at the same time, it stimulates adipogenesis and takes part in neoplastic processes such as intestinal cancer.
Lifespan effectors
The expression levels of lipogenesis controlling enzymes such as ATP-citrate and acetyl-CoA carbolase,56 as well as cytosolic phospholipase A2 and phospholipase C-y1,57,58 are reduced with age. On the contrary, overexpression of genes responsible for β-oxidation of fatty acids leads to life extension in Drosophila melanogaster.59
A characteristic feature of centenarians is the presence of large lipoprotein particles and raised level of high-density lipoproteins.60 Gene encoding proteins involved in triglyceride transport such as apolipoprotein E461 and apolipoprotein D62 are also associated with aging and longevity. It has been demonstrated in Drosophila that overexpression of human ApoD as well as its own homolog GLaz, lead to a longer life.63,64
In response to calorie restriction, the metabolic networks adjust by switching to an economy regime. Upon cellular energy deprivation, the NAD+-dependent deacetylases such as SIRT1 and HDAC1, 3, and 4, are activated, and it was shown that elevating levels of their expression prolongs lifespan.65 AMPK, the sensor of cellular AMP level, is another factor promoting longevity.66 Contrary to that, TOR kinase is activated in the presence of amino acids and accelerates aging; inhibiting TOR kinase activity leads to an extended lifespan in mice.67 Also, a knockout of RSK3/S6 protein kinase, which is activated by mTOR, resulted in long-lived mice.68 PHA-4/FOXA transcription factor serves as a mediator of calorie restriction effects and promotes life extension in C. elegans.69
Housekeeping genes
Excessive protein biosynthesis is toxic for cells and leads to stress in endoplasmic reticulum.70 Reduced expression of initiation factors eIF4E, eIF4G, eIF4E-BP resulted in extended lifespan in both worms and mice.71 The activities of several systems responsible for clearing up the damaged or excessive proteins, such as proteasome 20S C272 and the lysosomal and autophagy systems,73 are reduced with age. In model organisms, the overexpression of genes encoding proteins of regulatory proteasome subunit74 and autophagy proteins75 lead to life extension. Other enzymes involved in regulation of the lifespan are certain E3-ubiquitin ligases.76,77 Mitochondrial proteins are the most sensitive (susceptible) to oxidative damage. Overexpressing a mitochondrial chaperone Hsp22 in Drosophila resulted in life extension,78 and over-activation of mitochondrial protease LON in fungi Podospora anserina prolonged its lifespan.79
About 50% of proteins associated with aging and longevity are involved in signal transduction mechanisms.80 For example, TGF-β signaling pathway is reduced (downregulated) in long-lived worms.34 When muscles age, a pathological activation of Wnt signaling is observed.81 In long-living sea urchins, Notch signaling pathway activity is increased with age.82 The role of stress response associated with MAP kinase signaling cascade in regulating the lifespan has been recently elucidated: various small GTPases initiate MAP kinase signaling during stress and cellular aging.83 . Overexpression of p38MAP kinase extended Drosophila lifespan.84 The activity of kinases MEK1, MEK2, ERK1, and ERK2 was elevated (higher) in B-cell precursors in aged mice.85 In Drosophila, elevation in the levels of stress-activated protein kinase SAPK/JNK causes life extension,86 while GSK3 kinase inhibition leads to cellular aging.87
Genes involved in mitochondrial functions
Another group of genes playing an important role in aging are those regulating the free radical production. Some of these genes facilitate an extra production of free radicals. Mice lived longer when a gene, p66Shc, the mitochondrial target of p53 in response to oxidative stress, was eliminated from their genome.88 The same effect on extending the lifespan was achieved in nematodes carrying a mutation in Clk-1 gene regulating the biosynthesis of a component from electron transport chain in mitochondria and an antioxidant ubiquinone, as well as in mice heterozygous for the same gene.89 Mitochondrial uncoupling proteins UCP-1, -2, and -3 reduce the formation of active oxygen species in mitochondria.90 Oxidative stress sensors VDAC1 and VDAC3 play role in lifespan in different organisms.91
Enhanced activity in a number of proteins involved in antioxidant protection has also been proposed to promote longevity. In a course of cellular response to the oxidative stress through MAP kinase-signaling cascade, SKN-1 transcription factor is activated. SKN-1 activity is elevated in long-lived nematodes, mice, and flies.92 When genes encoding for peroxyredoxin II (Jafrac 1) and peroxyredoxin 5 (dPrx5), which are responsible for controlling peroxide levels in a cell, were overexpressed in Drosophila, the flies lived longer.93,94 Overexpression of Mn-SOD is also beneficial for life extension in flies and mice in number of cases.95Overexpressing Cu/Zn SOD in neurons extends life in Drosophila.96 Transgenic mice carrying a copy of mitochondrial catalase have shown delayed changes in aging markers in the heart in rodents.97
Genes regulating cellular senescence and apoptosis
In humans, a large number of genes undergo change in their expression with aging (Fig. 2). Some of these genes are downregulated as growth and development slow down, while other genes become activated in the course of pro-inflammatory and stress responses, which arise due to accumulation of damage and errors at the levels of cells and tissues.
Epigenetics of Aging and Longevity
Epigenetic marks on DNA and chromosomes
One of the main reasons for change in gene expression during aging is epigenetic regulation, which includes alterations in the methylated states of regulatory DNA sequences, covalent modifications of histone proteins, and the expression of regulatory non-coding RNAs. Epigenetic theory of aging is a rapidly developing modern concept postulating that non-adaptive epigenetic alterations are fundamental to aging. It is well established that epimutations accumulate with age, leading to activation of genes normally downregulated epigenetically.98,99 Genetically identical twins, as they age, exhibit significant differences in genome methylation pattern, leading to differences in gene expression and, ultimately, lifespan.100,101 Variations in epigenetic markers among different cells within the same tissue of an organism are increased with age.102 A global demethylation of DNA sequence repeats, such as mobile genetic elements, occurs with aging,103 as well as the local hypermethylation of promoters of genes transcribed by RNA polymerase II, such as rRNA.104,105 Senescence is accompanied by the formation of nuclear regions called senescence-associated heterochromatin foci (SAHF). These foci are determined by the recruitment of heterochromatin proteins and Rb protein to E2F-dependent promoters of proliferative genes, leading to the repression of E2F target genes.106 During aging, the activities of methyltransferases DNMT1 and DNMT3a107 as well as deacetylase SIRT1108 are reduced, while the activities of histone demethylases Jmjd3109 and Jarid1b,110 are enhanced. These changes result in non-adaptive alterations of epigenetic landscape, thereby changing gene expression and leading to aging.
Non-coding RNA
Non-coding RNAs include small RNAs, such as microRNAs and piwi-interacting RNAs, and a wide range of long non-coding RNAs (lnc RNAs).
MicroRNA
The aging process has become a potentially important target in cancer therapy after realization that cancer cells can be induce to undergo aging-type responses under stress of chemotherapeutics.111 In a search of appropriate age-related biomarkers, the role of microRNA (miRNA) in induction, regulation, and fine-tuning of the aging process has been discovered.112 miRNAs represent a class of small RNAs that play very important roles in various biological processes in health and in the development of human diseases through specific posttranscriptional downregulation of gene expression. One of the microRNAs, miR-34a, has been designated as an aging marker in several tissues and system. Boon et al. has shown that miR-34a is upregulated in the aging heart, and that miR-34a inhibition reduces cell death and fibrosis following acute myocardial infarction.113 The results of Boon et al. identified miR-34a and its target PNUTS as a key mechanism that regulates cardiac contractile function during aging by inducing DNA damage responses and telomere attrition. Klotho is an anti-aging protein in mice that regulates pathways classically associated with longevity, such as insulin/IGF-1 and Wnt signaling. Protein expression of Klotho decreases in normal aging of mice. In silico analysis has identified miRNA-339 and miRNA-556 to bind to 3′ untranslated region of Klotho mRNA. In vitro results confirmed that these miRNAs can directly decrease Klotho protein expression, indicating that these miRNAs might be playing a role in age-related downregulation of Klotho mRNA in vivo.114 In addition to intracellular miRNAs, there is a novel category of circulatory miRNAs that can be considered as a completely new intercellular and system level communication. Accumulated evidence suggests that circulatory miRNAs can exert 2 opposite roles, activating as well as inhibiting inflammatory pathways (inflamma-miRs). Several of the circulatory miRNAs seem to be common for the major age-related diseases that share a chronic, low-level proinflammatory status, such as cardiovascular disease, type 2 diabetes mellitus, Alzheimer disease, rheumatoid arthritis, and cancer.115
Long noncoding RNAs
The role of long non-coding RNAs (lncRNAs) in aging has been suggested in the work of Chang et al., in which he was studying gene expression changes of aged and rejuvenated human skin. He found that skin aging was associated with a significantly altered expression level of 2265 coding and noncoding RNAs, of which 1293 became “rejuvenated” after broadband light treatment. Rejuvenated genes (RGs) included several known key regulators of organismal longevity and their proximal long noncoding RNAs.116 Abdelmohsen et al. described identification of senescence-associated long non-coding RNAs (SAL-RNAs). He looked at the lncRNAs that are differentially expressed during replicative senescence of human diploid WI-38 fibroblasts by RNA-seq. SAL-RNA1 (XLOC_023166) has been identified as putative age-delaying lncRNA, since its reduction with small inhibitory RNAs (siRNA) induced rapid aging changes of the fibroblasts, such as large cell morphology, positive β-galactosidase activity, and upregulation of p53117
Pathway Analysis
The longevity genes described in this paper were separated into categories using Gene Ontology (GO), and their interactions were analyzed using GeneGo Metacore.
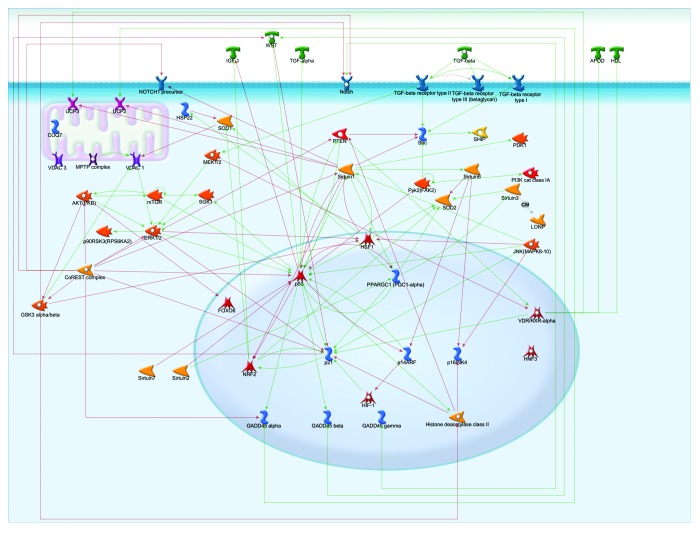
Most of the longevity genes described are related to stress response. The major regulatory hubs in stress response were P53, Sirtuin 1, P21, HSF1, and the CoREST and VDR/RXR-α complexes (Fig. 3).
Figure 3. Longevity genes involved in stress response. The relationship between proteins is depicted with arrows, where green and red represent activation and inhibition, respectively.
The VDR/RXR-α complex, a complex including over 20 elements, mainly PPAR and RXR, upregulates many proteins in the GADD45 family, P21, APOA1, APOD, WNT 4 UCP2, and UCP3. On the contrary, all of the interactions of the CoREST complex are downregulatory. It downregulates GADD45 α, P53, P21, ERK1/2, PTEN, AKT (PKB), GSK3 α/β complex, Notch pre-cursor, and NOTCH.
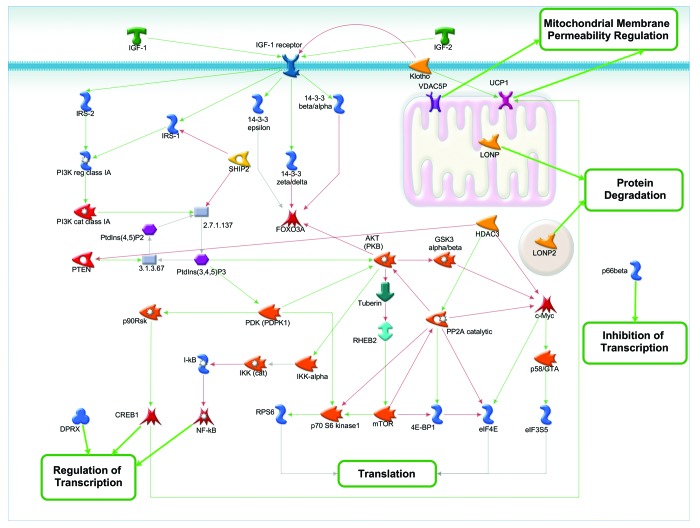
There are few genes that do not relate to stress response and are not classified as such in GO (Fig. 4). To get a deeper understanding of their action, we combined these genes with insulin-like growth factor signaling pathway. IGF binding, the tyrosine kinase activity of IGF-1 receptor, leads to the phosphorylation of several substrates, including the insulin receptor substrate family of proteins (such as insulin receptor substrate 1 and 2 [IRS-1 and IRS-2], SHC [Src homology 2 domain containing] transforming protein 1 (Shc), and some others).
Figure 4. IGF-1-mediated signaling combined with longevity proteins that are not directly involved in stress response.
After phosphorylation these proteins activate downstream signaling through the phosphatidylinositol 3-kinase (PI3K) or GRB2/SOS/H-Ras pathways. Activation of these pathways initiates metabolic cascades that result in the inhibition of apoptosis, activation of several transcription factors (CREB1, NK-κB), stimulation of protein synthesis via activation of ribosomal protein S6 kinase (p70 S6 kinase 1), mTOR, с-Myc, and also enhances glucose uptake, glycogen synthesis, and lipid storage. One of the anti-apoptotic pathways is mediated by 14-3-3 proteins. Three members of the 14-3-3 family of proteins (14-3-3 β/α, 14-3-3 zeta/delta, and 14-3-3 epsilon) interact with the IGF-1 receptor and in tandem with AKT (PKB) inhibit major stress response transcriptional factor FOXO3A. To simplify the schematic, we left out the GRB2/SOS/H-Ras and some of the anti-apoptotic pathways activated by IGF-1. Both HDAC3 and catalytic PP2A downregulate c-Myc, one of the major oncogenes, which may be one of the possible mechanisms for increased longevity in mammals. Both LON peptidases (mitochondrial LONP and peroxisomal LONP2) involved in protein degradation process, along with transcriptional repressor p66 beta and divergent paired-related homeobox protein (DPRX), have no direct interactions with other components of the network. Membrane protein Klotho directly activates only mitochondrial uncoupling protein 1 (UCP1), which facilitates the transfer of anions from the inner to the outer mitochondrial membrane and the return transfer of protons from the outer to the inner mitochondrial membrane. It’s known that Klotho overexpression extends lifespan, whereas loss of Klotho accelerates the development of aging-like phenotypes, but the exact mechanisms are still rather vague.
The effects of the interventions associated with the genes in Figures 3 and 4 on life extension of model organisms are summarized in Table 1.
Table 1. Life extension in model organisms.
| Gene | Human homolog | Organism | Wild-type lifespan | Life extension (%) | Mechanism | Gender | References |
|---|---|---|---|---|---|---|---|
| daf-2 | IGFR-1 | Caenorhabditis elegans | 14.9 ± 0.1 d | 83.0% | Gene inactivation leads to disruption of insulin signaling | N/A | 118 |
| age-1 | PI3K | Caenorhabditis elegans | 16 ± 2 d | ~1000% | Gene inactivation leads to disruption of insulin signaling | N/A | 41 |
| bec-1 | beclin | Caenorhabditis elegans | 22.4–31.1 d (mean lifespan) | −15–30% (across six trials) | Gene inactivation leads to disruption of autophagy | N/A | 119 |
| hsf-1 | HSF | Caenorhabditis elegans | 13.8 ± 0.5 d | 22.0% | Gene overexpression leads to activation of the heat shock promoter | N/A | 120 |
| daf-16 | FOXO | Caenorhabditis elegans | 23.2 ± 0.8 d | −27.0% | Gene inactivation leads to disregulation of stress response machinnery | N/A | 120 |
| let-363 | TOR | Caenorhabditis elegans | 10 d | 250.0% | Gene inactivation leads to disruption of insulin signaling | N/A | 121 |
| sgk-1 | SGK | Caenorhabditis elegans | 14.7 ± 0.3 d | 61.0% | Gene inactivation leads to disruption of insulin signaling (as sgk-1 acts in parallel with AKT kinases) and better stress response. | N/A | 122 |
| hcf-1 | HCFC1 | Caenorhabditis elegans | 14.3 ± 0.1 d | 28.0% | Gene inactivation leads to activation of stress response by daf-16/FOXO | N/A | 123 |
| jnk-1 | JNK | Caenorhabditis elegans | 16.8 ± 0.2 d | −21.7% | Gene inactivation leads to disruption of stress response by daf-16/FOXO | N/A | 124 |
| jkk-1 | JKK1 | Caenorhabditis elegans | 16.8 ± 0.2 d | −20.9% | Gene inactivation leads to disruption of stress response by daf-16/FOXO | N/A | 124 |
| akt-1 akt-2 | AKT1 AKT2 | Caenorhabditis elegans | 14.7 ± 0.3 d | 19.0% | Simultaneous inactivation of these genes leads to disruption of insulin signaling | N/A | 122 |
| sod1 | SOD1 | Caenorhabditis elegans | 18 d | 33% (averaged across trials 1 and 2) | Overexpression of sod1 activates longevity-promoting transcription factors. | N/A | 125 |
| sod2 | SOD2 | Caenorhabditis elegans | 19 d | 10% (averaged across trials 5 and 6) | Overexpression of sod2 activates longevity-promoting transcription factors. | N/A | 125 |
| dSir2 | SIRT1 | Drosophila melanogaster | 37 d | 57.0% | Overexpression of dSir2 enhances energy metabolism | female | 126 |
| dSir2 | SIRT1 | Drosophila melanogaster | 41 d | 32.0% | Overexpression of dSir2 enhances energy metabolism | male | 126 |
| chico | InRS | Drosophila melanogaster | 44 d | 47.7% | Gene inactivation leads to disruption of insulin signaling | female | 38 |
| InR | InR | Drosophila melanogaster | N/A | 85.0% | Gene inactivation leads to disruption of insulin signaling | female | 127 |
| dFOXO | FOXO | Drosophila melanogaster | Varies across trials | 19.4% (averaged across trials) | Overexpression of dFOXO leads to disruption of insulin signaling | female | 128 |
| dFOXO | FOXO | Drosophila melanogaster | Varies across trials | 15.5% (averaged across trials) | Overexpression of dFOXO leads to disruption of insulin signaling | male | 128 |
| dPTEN | PTEN | Drosophila melanogaster | 57 d | 17.4% | Overexpression of dPTEN leads to disruption of insulin signaling | female | 128 |
| dPTEN | PTEN | Drosophila melanogaster | 51 d | 19.6% | Overexpression of dPTEN leads to disruption of insulin signaling | male | 128 |
| hsp22 | HSP22 | Drosophila melanogaster | 60 ± 3 d | 32.0% | Overexpression of hsp22 increases cell-protection against oxidative injuries | male | 78 |
| sod2 | SOD2 | Drosophila melanogaster | 77.8 ± 5.7 d and 74.7 ± 5.1 d | −9.5% and −7.4% | Overexpression of SOD2 caused decrease of mitochondrial H2O2 release and enhancement of free methionine content essential for normal biological processes. | male | 129 |
| sod1 | SOD1 | Drosophila melanogaster | 27 d | >66% | Overexpression of sod1 in motorneurons enhances RO metabolism | male | 130 |
| mTOR | TOR | Drosophila melanogaster | N/A | 30.0% | Overexpression of dominant negative form of TOR alters stress responses translation and/or mitochondrial function | male | 131 |
| dS6K | S6K | Drosophila melanogaster | N/A | 29.0% | Overexpression of dominant negative form of S6 kinase alters stress responses translation and/or mitochondrial function | male | 131 |
| IGFR-1 | IGFR-1 | Mus musculus | 568 ± 49 d | 33.0% | Gene inactivation leads to disruption of insulin signaling | female | 37 |
| IGFR-1 | IGFR-1 | Mus musculus | 585 ± 69 d | 16.0% | Gene inactivation leads to disruption of insulin signaling | male | 37 |
| p66shc | p66 | Mus musculus | 761 ± 19.02 d | 30.0% | Disactivation of p66 contributes to increased cellular and organism oxidative stress resistance | male and female | 88 |
| Klotho | KLOTHO | Mus musculus | 715 ± 44 d | 20.0 and 30.8% (transgenic lines EFmKL46 and EFmKL48) | Gene inactivation leads to disruption of insulin signaling | male | 48 |
| Klotho | KLOTHO | Mus musculus | 697 ± 45 d | 18.8 and 19.0% (transgenic lines EFmKL46 and EFmKL48) | Gene inactivation leads to disruption of insulin signaling | female | 48 |
| Arf | p19 | Mus musculus | 113.8 ± 2.4 wk | 16.0% | Hypothetically activation of Arf/p53 module provides anti-cancer and anti-aging effect detecting cellular damage. | male and female | 132 |
| SIRT6 | SIRT6 | Mus musculus | 851.3 ± 24.9 and 724.0 ± 35.0 d (transgenic lines 55 and 108) | 14.8% and 16.9% (transgenic lines 55and 108) | Overexpression leads to higher levels of IGF-binding protein 1 and altered phosphorylation levels of major components of IGF1 signaling | male | 133 |
| p63 | p63 | Mus musculus | 121 wk (median lifespan) | −21.5% | p63 deficiency activates widespread cellular senescence with enhanced expression of senescent markers SA-β-gal PML and p16INK4a | male and female | 134 |
| Brca1 | Brca | Mus musculus | 713 ± 146 d | −8.0% | Gene inactivation leads to hypersensitivity to DNA damaging agents and consequently genomic instability of cells | female | 135 |
There are many genes in the pathways related to stress response where overexpression led to life extension. In Drosophila, overexpression of the stress response gene dGADD45 led to up to 73% increases in lifespan. In both C. elegans and Drosophila, inactivation of TOR signaling led to 250% and 30% increases, respectively. Major lifespan increases across all species were achieved by interventions into the interconnected IGFR-1 and TOR pathways, with IGFR-1 inactivation in mice resulting in up to 33% and Age-1 (PI3K) inactivation in C. elegans producing up to 1000% average life extension.
Aging Research Trends
To better understand the general trends in aging genetics, the funding and citation information for the longevity genes in Figures 3 and 4 was collected using the International Aging Research Portfolio (IARP) system136 as well as the NCBI PubMed system.
The names of human genes and animal homologs were used as search terms for the IARP system to produce the total funding amounts of grants with grant applications containing these search terms. The process was repeated using the gene name, “AND”, and “aging” as search terms. The same process was performed in PubMed to compile the number of citations for each gene. While the exact funding amounts and the number of published papers for each gene may differ, Table 2 illustrates the general trends.
Table 2. Summary of the available funding and citation data.
| Process | Gene | Funding | Citations | F/C | F/TF | AF | AF/TF** | YFC | YFCA | FC-A | A-T |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cellular response to stress | TP53 or P53 or Dmp53 | $4 027 210 538 | 68 834 | $58 506 | 46.97% | $195 599 425 | 4.86% | 1979 | 1987 | 32 | 8 |
| MAPK14 | $458 530 482 | 1706 | $268 775 | 5.35% | $23 968 444 | 5.23% | 2001 | 2006 | 10 | 5 | |
| MAPK8 | $424 571 226 | 1196 | $354 993 | 4.95% | $22 578 844 | 5.32% | 2000 | 2000 | 11 | 0 | |
| SOD1 or sod-1 | $274 749 561 | 4128 | $66 558 | 3.20% | $46 503 534 | 16.93% | 1975 | 1985 | 36 | 10 | |
| SOD2 or sod-2 | $203 094 775 | 1374 | $147 813 | 2.37% | $28 305 859 | 13.94% | 1973 | 1985 | 38 | 12 | |
| CDKN1A | $131 529 936 | 10 438 | $12 601 | 1.53% | $13 032 831 | 9.91% | 1993 | 1993 | 18 | 0 | |
| SIRT1 or sir2 or dSir2 | $116 967 665 | 3052 | $38 325 | 1.36% | $101 117 219 | 86.45% | 1984 | 1999 | 27 | 15 | |
| MAPK1 or mpk1 | $103 268 829 | 11 237 | $9190 | 1.20% | $8 223 868 | 7.96% | 1982 | 1993 | 29 | 11 | |
| HDAC6 | $101 832 683 | 474 | $214 837 | 1.19% | $- | 0.00% | 1999 | 2006 | 12 | 7 | |
| MAPK9 | $32 669 688 | 252 | $129 642 | 0.38% | $368 385 | 1.13% | 1994 | - | |||
| HDAC2 | $31 533 278 | 802 | $39 318 | 0.37% | $- | 0.00% | 1997 | 2001 | 14 | 4 | |
| RXRA | $30 032 747 | 356 | $84 362 | 0.35% | $1 848 693 | 6.16% | 1992 | 2011 | 19 | 19 | |
| WNT5A | $27 790 400 | 862 | $32 239 | 0.32% | $ - | 0.00% | 1993 | 2000 | 18 | 7 | |
| GSK3 or sgg | $27 250 742 | 1372 | $19 862 | 0.32% | $2 505 572 | 9.19% | 1980 | 1995 | 31 | 15 | |
| MAPK10 | $17 683 286 | 88 | $200 946 | 0.21% | $1 216 021 | 6.88% | 1991 | - | |||
| GADD45A | $12 777 259 | 482 | $26 509 | 0.15% | $1 296 966 | 10.15% | 1995 | 1999 | 16 | 4 | |
| GADD45G | $7 537 188 | 69 | $109 235 | 0.09% | $2 062 285 | 27.36% | 1998 | 2010 | 13 | 12 | |
| FOXA3 or HNF3G or TCF3G | $5 315 519 | 119 | $44 668 | 0.06% | $ - | 0.00% | 1990 | 1999 | 21 | 9 | |
| MAPK12 | $1 438 384 | 68 | $21 153 | 0.02% | $ - | 0.00% | 1992 | - | |||
| SIRT7 | $1766 | 51 | $35 | 0.00% | $1766 | 100% | 2000 | 2005 | 11 | 5 | |
| Total Stress Response | $6 035 785 952 | 106 960 | $56 430 | $448 629 712 | 7.43% | ||||||
| Insulin-like signaling | MTOR or TOR | $821 029 426 | 23 778 | $34 529 | 9.58% | $68 845 232 | 8.39% | 1975 | 1988 | 36 | 13 |
| PPARG | $213 853 403 | 10 059 | $21 260 | 2.49% | $15 193 855 | 7.10% | 1993 | 1994 | 18 | 1 | |
| AKT1 or akt-1 | $121 140 702 | 5408 | $22 400 | 1.41% | $7 246 805 | 5.98% | 1977 | 1999 | 34 | 22 | |
| PPARA | $74 581 825 | 750 | $99 442 | 0.87% | $1 120 366 | 1.50% | 1993 | 2002 | 18 | 9 | |
| AKT2 or akt-2 | $71 265 752 | 875 | $81 447 | 0.83% | $2 598 915 | 3.65% | 1987 | 1999 | 24 | 12 | |
| RXRA | $27 790 400 | 356 | $78 063 | 0.32% | $1 848 693 | 6.65% | 1992 | 2011 | 19 | 19 | |
| HDAC5 | $24 234 263 | 310 | $78 175 | 0.28% | $986 700 | 4.07% | 1999 | 2002 | 12 | 3 | |
| SHC1 | $16 912 072 | 898 | $18 833 | 0.20% | $1 238 936 | 7.33% | 1992 | 1997 | 19 | 5 | |
| HDAC9 | $5 844 910 | 106 | $55 141 | 0.07% | $- | 0.00% | 2001 | 2002 | 10 | 1 | |
| GSK3A | $2 120 564 | 150 | $14 137 | 0.02% | $- | 0.00% | 1995 | 2013 | 16 | 18 | |
| EIF4EBP1 or d4EBP | $608 042 | 818 | $743 | 0.01% | $- | 0.00% | 1994 | 2006 | 17 | 12 | |
| Total Insulin Stimulus | $1 379 381 359 | 43 508 | $31 704 | $99 079 502 | 7.18% | ||||||
| Regulation of translation | MTOR or TOR | $821 029 426 | 23 778 | $34 529 | 9.58% | $68 845 232 | 8.39% | 1975 | 1988 | 36 | 13 |
| AKT1 or akt-1 | $121 140 702 | 5408 | $22 400 | 1.41% | $7 246 805 | 5.98% | 1977 | 1999 | 34 | 22 | |
| MAPK1 | $96 943 128 | 11 067 | $8 760 | 1.13% | $8 223 868 | 8.48% | 1991 | 1993 | 20 | 2 | |
| EIF4E | $69 771 228 | 2392 | $29 169 | 0.81% | $3 935 384 | 5.64% | 1991 | 2001 | 20 | 10 | |
| PTK2B | $44 532 289 | 298 | $149 437 | 0.52% | $- | 0.00% | 1995 | 2010 | 16 | 15 | |
| EIF4G1 or EIF4G | $3 998 998 | 970 | $4123 | 0.05% | $911 650 | 22.80% | 1995 | 2001 | 16 | 6 | |
| EIF4EBP1 or d4EBP | $608 042 | 818 | $743 | 0.01% | $- | 0.00% | 1994 | 2006 | 17 | 12 | |
| Total reg. of translation | $1 158 023 813 | 44 731 | $25 889 | $89 162 939 | 7.70% | ||||||
| Total | $8 573 191 124 | 195 199 | $43 920 | $636 872 153 | 7.43% |
F/C, funding per citation; F/TF, funding for a specific gene as percentage of total funding; AF, funding for projects with the specific gene name and “aging” in the grant application; YFC, year of first citation; YFCA, year of first citation with “aging” in the abstract; FC-A, the time between first citation of the gene and citation with “aging”; A-T, the time between 2013 and the time of the first citation of the gene with “aging” in the abstract.
PubMed was also queried with the name of each gene and the name of each gene “AND” “aging” to identify the year of the first citation and the year of the first citation with “aging” in the abstract.
The science of aging genetics is a comparatively new field. P53 was discovered in 1979 and implicated in aging in 1987. On average, genes in Table 2 were discovered 21 years ago, and it took 9.7 years between the first citation and the first citation with “aging”.
The approximate amount of funding spent on genes related to aging is at over $8.5 billion, with over 195 thousand citations, with the most funding spent on genes involved in stress response. On average approximately 7.4% of the funding was spent on projects with “aging” in the grant application, and this was consistent across all 3 categories. The average amount of funding per citation was over $43.9 thousand.
The largest amount of funding spent on a single gene with “aging” in the grant abstract was $195 million, which represents fewer than 5% of the total funding spent on P53 research. SIRT1 and homologs is the only gene with over $100 million spent on analyzing its role in aging, with just under 14% of the funding spent on non-aging-related projects.
Most of the genes related to aging and longevity were associated with other biologic processes, and most of the funding and publications citing these genes are related to areas other than aging.
Conclusion
Based on the analysis of current knowledge on evolutionarily conserved genetic regulation of aging and longevity, it has been possible to generate a functional classification of genes controlling lifespan137
(1) Lifespan “regulators.” These act as switches of ontogenetic programs and are responsible for sensing and transmitting external environmental signals: synthesis, response, and transmission of hormones belonging to insulin-like pathway and secondary lipophilic hormones. A large fraction of these genes promote growth and reproduction while suppressing stress resistance. However, some of these genes stimulate stress response (see Klotho for an example).
(2) “Mediators” include kinases, protein deacetylases, and transcription factors. These genes are controlled by regulators and are responsible for switching stress response programs depending on the environmental signals, such as food availability, overpopulation (crowding), light and temperature conditions, and irradiation or endogenous oxidative stress. Mediators act either as tissue-specific regulators of effector genes or directly controlling protein activity or lifetime. Mediators also interact among themselves, stimulating or inhibiting one another’s activity.
(3) “Effectors” are stress-resistance genes, including heat shock proteins, antioxidants, protein and DNA damage repair proteins, proteasome components, calpains, autophagy proteins, innate immunity, detoxification of xenobiotics, and metabolic regulators. Overexpression of these genes is usually correlated with extended lifespan. Often, the effectors act in additive manner, becoming activated by distinct “mediators” and extending lifespan under stress conditions. However, a number of “mediators” suppress “effectors” activity.
(4) Housekeeping genes. These act ubiquitously at every stage of life and are responsible for supporting cellular structure, respiration, synthesis of amino acids, lipids, nucleotides, etc. Mutations in these genes are either lethal or result in pathologies. Under stress conditions, some of the housekeeping genes are temporarily repressed by “mediators”, which allows saving energy and resources for “effectors” and extending lifespan.
(5) Genes involved in mitochondrial function. These are components of electron transport chain, Krebs cycle, uncoupling proteins, clk-1 gene in nematodes. These genes regulate energy metabolism, the level of free radicals, and also apoptosis.
(6) Genes regulating cellular senescence and apoptosis (p53, p21, p16, pRB). These genes are responsible for cancer prevention, cell cycle regulation, and elimination of extra or malignant cells during early ontogenesis and maturity. Cellular senescence (replicative or stress-induced) of dividing cells or excessive elimination of postmitotic cells is a pleiotropic side effect of aging.
How do all these new developments in the new science of aging and discovery of genes that drastically alter longevity fit in with classical evolutionary theories? Which one is standing the test of time and new developments in the field of aging? At this point in time it appears that each of them is holding bits of truth, and each of them is explaining the evolution and mechanism of aging using dualistic principles (adaptive/nonadaptive, molecular/organismic, etc.). There is a newer theory proposed that offers an integrated theory of aging that helps us to better grasp similarity rather than differences among all these processes, the fractal theory of aging.138 The fractal theory is based, first, on the multilevel nature and complexity of aging, as well as self-similarity of those levels. Another important property of a fractal is a combination of stochastic and regular traits. The fractal principle of aging manifests in a combination of random (i.e., aging rates) and regular (i.e., sequence of geriatric changes) traits. Thus, according to this theory, aging can be defined as an age-dependent fractal increase in the number of deviations from homeostasis at the molecular, subcellular, cellular, tissue, and systemic levels. Actually, what would be highly desirable at this point in time is a unified theory of aging that would offer experimentally testable predictions. If we are able to mathematically describe the aging for one (e.g., cellular) level or one biological trait on a small interval of time, this model could be extrapolated to predict the aging at all other levels of organization of life, including individual lifespan. Substituting the model parameters with experimental measurements could lead to finding of biomarkers of aging rate and efficiency of anti-aging interventions.
Our pathway analysis shows that most of the gerontogenes are members of the stress response pathways and confirms the existence of genetics “longevity program”. As a rule, genes, regulators of the longevity program, which suppress mild stress response as well as mutations that make some of those pathways less efficient, provide life-extension benefits. Mild overexpression of effector longevity genes involved in stress response to DNA, protein, or other cellular damages (e.g., Hsps, Sod, GADD45, ATGs) prolongs lifespan. While moderate stress induces “longevity program” by stimulating expression of life-assurance genes and promoting prevention or elimination of errors, chronic or acute stress exposure exhausts the defense mechanisms and therefore accelerates aging. Pro-aging and anti-aging gene-determined processes exist on all levels of the organismal system—from molecules to systems (metabolic, endocrine, immune, inter-cellular communication). Their multi-level organization, the interpenetration of levels, a combination of regular and stochastic elements is what makes the process of aging a fractal process.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed
Acknowledgments
We would like to thank the editorial team and the reviewers for their valuable advice and comments pertaining to the content of this manuscript and Brian Kennedy, Jan Vijg, Yousin Suh, Viktoria Lunyak, Mark Tatar, and Charles Cantor for productive discussions on the genetics and epigenetics of aging and longevity. We would like to thank the UMA Foundation for their help in preparation of this manuscript. The work was supported by Grant of RFBR 14-04-01596 and Grant of President of Russian Federation MD-1090.2014.4.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/28433
References
- 1.Weismann A. Über die Dauer des Lebens G. Fisher: 1882. [Google Scholar]
- 2.Weismann A. Essays Upon Heredity and Kindred Biological Problems. Clarendon Press: Oxford, 1889. [Google Scholar]
- 3.Weismann A. Über Leben und Tod. Verlag von Gustav Fisher, Jena, Germany. Jena, Gustav Fischer: 1892. [Google Scholar]
- 4.Medawar PB. Old age and natural death. Modern Quarterly. 1946;1:30–56. [Google Scholar]
- 5.Medawar PB. An Unsolved Problem of Biology H.K.Lewis: London, 1952. [Google Scholar]
- 6.Williams GC. Pleiotropy, natural selection and the evolution of senescence. Evolution. 1957;11:398–411. doi: 10.2307/2406060. [DOI] [Google Scholar]
- 7.Gavrilov LA, Gavrilova NS. Evolutionary theories of aging and longevity. ScientificWorldJournal. 2002;2:339–56. doi: 10.1100/tsw.2002.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rose MR, Burke MK, Shahrestani P, Mueller LD. Evolution of ageing since Darwin. J Genet. 2008;87:363–71. doi: 10.1007/s12041-008-0059-6. [DOI] [PubMed] [Google Scholar]
- 9.Kirkwood TB. Evolution of ageing. Nature. 1977;270:301–4. doi: 10.1038/270301a0. [DOI] [PubMed] [Google Scholar]
- 10.Guarente L, Kenyon C. Genetic pathways that regulate ageing in model organisms. Nature. 2000;408:255–62. doi: 10.1038/35041700. [DOI] [PubMed] [Google Scholar]
- 11.Lithgow GJ, White TM, Melov S, Johnson TE. Thermotolerance and extended lifespan conferred by single-gene mutations and induced by thermal stress. Proc Natl Acad Sci U S A. 1995;92:7540–4. doi: 10.1073/pnas.92.16.7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Longo VD, Mitteldorf J, Skulachev VP. Programmed and altruistic ageing. Nat Rev Genet. 2005;6:866–72. doi: 10.1038/nrg1706. [DOI] [PubMed] [Google Scholar]
- 13.Murakami S, Johnson TE. A genetic pathway conferring life extension and resistance to UV stress in Caenorhabditis elegans. Genetics. 1996;143:1207–18. doi: 10.1093/genetics/143.3.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Partridge L, Gems D, Withers DJ. Sex and death: what is the connection? Cell. 2005;120:461–72. doi: 10.1016/j.cell.2005.01.026. [DOI] [PubMed] [Google Scholar]
- 15.Xiao R, Zhang B, Dong Y, Gong J, Xu T, Liu J, Xu XZ. A genetic program promotes C. elegans longevity at cold temperatures via a thermosensitive TRP channel. Cell. 2013;152:806–17. doi: 10.1016/j.cell.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaman L, Stoian I, Atanasiu V. Can ageing be slowed?: Hormetic and redox perspectives. J Med Life. 2011;4:346–51. [PMC free article] [PubMed] [Google Scholar]
- 17.Gems D, Partridge L. Stress-response hormesis and aging: “that which does not kill us makes us stronger”. Cell Metab. 2008;7:200–3. doi: 10.1016/j.cmet.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 18.Cornelius C, Perrotta R, Graziano A, Calabrese EJ, Calabrese V. Stress responses, vitagenes and hormesis as critical determinants in aging and longevity: Mitochondria as a “chi”. Immun Ageing. 2013;10:15. doi: 10.1186/1742-4933-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rattan SI. Hormesis in aging. Ageing Res Rev. 2008;7:63–78. doi: 10.1016/j.arr.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 20.Moskalev A. Radiation-induced lifespan alteration of Drosophila lines with genotype differences. Biogerontology. 2007;8:499–504. doi: 10.1007/s10522-007-9090-x. [DOI] [PubMed] [Google Scholar]
- 21.Moskalev AA, Plyusnina EN, Shaposhnikov MV. Radiation hormesis and radioadaptive response in Drosophila melanogaster flies with different genetic backgrounds: the role of cellular stress-resistance mechanisms. Biogerontology. 2011;12:253–63. doi: 10.1007/s10522-011-9320-0. [DOI] [PubMed] [Google Scholar]
- 22.Plyusnina EN, Shaposhnikov MV, Moskalev AA. Increase of Drosophila melanogaster lifespan due to D-GADD45 overexpression in the nervous system. Biogerontology. 2011;12:211–26. doi: 10.1007/s10522-010-9311-6. [DOI] [PubMed] [Google Scholar]
- 23.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Partridge L, Thornton J, Bates G. The new science of ageing. Philos Trans R Soc Lond B Biol Sci. 2011;366:6–8. doi: 10.1098/rstb.2010.0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fontana L, Partridge L, Longo VD. Extending healthy lifespan--from yeast to humans. Science. 2010;328:321–6. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–12. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 27.Partridge L. The new biology of ageing. Philos Trans R Soc Lond B Biol Sci. 2010;365:147–54. doi: 10.1098/rstb.2009.0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moskalev AA. [Prospective trends in genetics of aging and longevity] Adv Gerontol. 2009;22:92–103. [PubMed] [Google Scholar]
- 29.Zhavoronkov A, Smit-McBride Z, Guinan KJ, Litovchenko M, Moskalev A. Potential therapeutic approaches for modulating expression and accumulation of defective lamin A in laminopathies and age-related diseases. J Mol Med (Berl) 2012;90:1361–89. doi: 10.1007/s00109-012-0962-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barzilai N, Huffman DM, Muzumdar RH, Bartke A. The critical role of metabolic pathways in aging. Diabetes. 2012;61:1315–22. doi: 10.2337/db11-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim SK. Common aging pathways in worms, flies, mice and humans. J Exp Biol. 2007;210:1607–12. doi: 10.1242/jeb.004887. [DOI] [PubMed] [Google Scholar]
- 32.Cheng CL, Gao TQ, Wang Z, Li DD. Role of insulin/insulin-like growth factor 1 signaling pathway in longevity. World J Gastroenterol. 2005;11:1891–5. doi: 10.3748/wjg.v11.i13.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bartke A. Insulin and aging. Cell Cycle. 2008;7:3338–43. doi: 10.4161/cc.7.21.7012. [DOI] [PubMed] [Google Scholar]
- 34.Parrella E, Longo VD. Insulin/IGF-I and related signaling pathways regulate aging in nondividing cells: from yeast to the mammalian brain. ScientificWorldJournal. 2010;10:161–77. doi: 10.1100/tsw.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suh Y, Atzmon G, Cho MO, Hwang D, Liu B, Leahy DJ, Barzilai N, Cohen P. Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proc Natl Acad Sci U S A. 2008;105:3438–42. doi: 10.1073/pnas.0705467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tazearslan C, Ayyadevara S, Bharill P, Shmookler Reis RJ. Positive feedback between transcriptional and kinase suppression in nematodes with extraordinary longevity and stress resistance. PLoS Genet. 2009;5:e1000452. doi: 10.1371/journal.pgen.1000452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holzenberger M, Dupont J, Ducos B, Leneuve P, Géloën A, Even PC, Cervera P, Le Bouc Y. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421:182–7. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- 38.Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, Hafen E, Leevers SJ, Partridge L. Extension of lifespan by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292:104–6. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- 39.Selman C, Lingard S, Choudhury AI, Batterham RL, Claret M, Clements M, Ramadani F, Okkenhaug K, Schuster E, Blanc E, et al. Evidence for lifespan extension and delayed age-related biomarkers in insulin receptor substrate 1 null mice. FASEB J. 2008;22:807–18. doi: 10.1096/fj.07-9261com. [DOI] [PubMed] [Google Scholar]
- 40.Taguchi A, Wartschow LM, White MF. Brain IRS2 signaling coordinates lifespan and nutrient homeostasis. Science. 2007;317:369–72. doi: 10.1126/science.1142179. [DOI] [PubMed] [Google Scholar]
- 41.Ayyadevara S, Alla R, Thaden JJ, Shmookler Reis RJ. Remarkable longevity and stress resistance of nematode PI3K-null mutants. Aging Cell. 2008;7:13–22. doi: 10.1111/j.1474-9726.2007.00348.x. [DOI] [PubMed] [Google Scholar]
- 42.Narasimhan SD, Yen K, Tissenbaum HA. Converging pathways in lifespan regulation. Curr Biol. 2009;19:R657–66. doi: 10.1016/j.cub.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Puig O, Tjian R. Transcriptional feedback control of insulin receptor by dFOXO/FOXO1. Genes Dev. 2005;19:2435–46. doi: 10.1101/gad.1340505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanson RW, Hakimi P. Born to run; the story of the PEPCK-Cmus mouse. Biochimie. 2008;90:838–42. doi: 10.1016/j.biochi.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tower J. Hsps and aging. Trends Endocrinol Metab. 2009;20:216–22. doi: 10.1016/j.tem.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moskalev A, Plyusnina E, Shaposhnikov M, Shilova L, Kazachenok A, Zhavoronkov A. The role of D-GADD45 in oxidative, thermal and genotoxic stress resistance. Cell Cycle. 2012;11:4222–41. doi: 10.4161/cc.22545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moskalev AA, Smit-McBride Z, Shaposhnikov MV, Plyusnina EN, Zhavoronkov A, Budovsky A, Tacutu R, Fraifeld VE. Gadd45 proteins: relevance to aging, longevity and age-related pathologies. Ageing Res Rev. 2012;11:51–66. doi: 10.1016/j.arr.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kurosu H, Yamamoto M, Clark JD, Pastor JV, Nandi A, Gurnani P, McGuinness OP, Chikuda H, Yamaguchi M, Kawaguchi H, et al. Suppression of aging in mice by the hormone Klotho. Science. 2005;309:1829–33. doi: 10.1126/science.1112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Broughton SJ, Piper MD, Ikeya T, Bass TM, Jacobson J, Driege Y, Martinez P, Hafen E, Withers DJ, Leevers SJ, et al. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc Natl Acad Sci U S A. 2005;102:3105–10. doi: 10.1073/pnas.0405775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Atzmon G, Pollin TI, Crandall J, Tanner K, Schechter CB, Scherer PE, Rincon M, Siegel G, Katz M, Lipton RB, et al. Adiponectin levels and genotype: a potential regulator of lifespan in humans. J Gerontol A Biol Sci Med Sci. 2008;63:447–53. doi: 10.1093/gerona/63.5.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Naito M, Fujikura J, Ebihara K, Miyanaga F, Yokoi H, Kusakabe T, Yamamoto Y, Son C, Mukoyama M, Hosoda K, et al. Therapeutic impact of leptin on diabetes, diabetic complications, and longevity in insulin-deficient diabetic mice. Diabetes. 2011;60:2265–73. doi: 10.2337/db10-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Albarran-Zeckler RG, Sun Y, Smith RG. Physiological roles revealed by ghrelin and ghrelin receptor deficient mice. Peptides. 2011;32:2229–35. doi: 10.1016/j.peptides.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gharibeh MY, Al Tawallbeh GM, Abboud MM, Radaideh A, Alhader AA, Khabour OF. Correlation of plasma resistin with obesity and insulin resistance in type 2 diabetic patients. Diabetes Metab. 2010;36:443–9. doi: 10.1016/j.diabet.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 54.Schiff M, Bénit P, Jacobs HT, Vockley J, Rustin P. Therapies in inborn errors of oxidative metabolism. Trends Endocrinol Metab. 2012;23:488–95. doi: 10.1016/j.tem.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Argmann C, Dobrin R, Heikkinen S, Auburtin A, Pouilly L, Cock TA, Koutnikova H, Zhu J, Schadt EE, Auwerx J. Ppargamma2 is a key driver of longevity in the mouse. PLoS Genet. 2009;5:e1000752. doi: 10.1371/journal.pgen.1000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nogalska A, Pankiewicz A, Goyke E, Swierczynski J. The age-related inverse relationship between ob and lipogenic enzymes genes expression in rat white adipose tissue. Exp Gerontol. 2003;38:415–22. doi: 10.1016/S0531-5565(02)00210-3. [DOI] [PubMed] [Google Scholar]
- 57.Gentili C, Morelli S, de Boland AR. PTH and phospholipase A2 in the aging process of intestinal cells. J Cell Biochem. 2004;93:312–26. doi: 10.1002/jcb.20158. [DOI] [PubMed] [Google Scholar]
- 58.Utsuyama M, Wakikawa A, Tamura T, Nariuchi H, Hirokawa K. Impairment of signal transduction in T cells from old mice. Mech Ageing Dev. 1997;93:131–44. doi: 10.1016/S0047-6374(96)01837-4. [DOI] [PubMed] [Google Scholar]
- 59.Lee SH, Lee SK, Paik D, Min KJ. Overexpression of fatty-acid-β-oxidation-related genes extends the lifespan of Drosophila melanogaster. Oxid Med Cell Longev. 2012;2012:854502. doi: 10.1155/2012/854502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Atzmon G, Rincon M, Schechter CB, Shuldiner AR, Lipton RB, Bergman A, Barzilai N. Lipoprotein genotype and conserved pathway for exceptional longevity in humans. PLoS Biol. 2006;4:e113. doi: 10.1371/journal.pbio.0040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bonomini F, Filippini F, Hayek T, Aviram M, Keidar S, Rodella LF, Coleman R, Rezzani R. Apolipoprotein E and its role in aging and survival. Exp Gerontol. 2010;45:149–57. doi: 10.1016/j.exger.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 62.Muffat J, Walker DW, Apolipoprotein D. Apolipoprotein D: an overview of its role in aging and age-related diseases. Cell Cycle. 2010;9:269–73. doi: 10.4161/cc.9.2.10433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muffat J, Walker DW, Benzer S, Human Apo D. Human ApoD, an apolipoprotein up-regulated in neurodegenerative diseases, extends lifespan and increases stress resistance in Drosophila. Proc Natl Acad Sci U S A. 2008;105:7088–93. doi: 10.1073/pnas.0800896105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walker DW, Muffat J, Rundel C, Benzer S. Overexpression of a Drosophila homolog of apolipoprotein D leads to increased stress resistance and extended lifespan. Curr Biol. 2006;16:674–9. doi: 10.1016/j.cub.2006.01.057. [DOI] [PubMed] [Google Scholar]
- 65.Virtue S, Vidal-Puig A. It’s not how fat you are, it’s what you do with it that counts. PLoS Biol. 2008;6:e237. doi: 10.1371/journal.pbio.0060237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Greer EL, Banko MR, Brunet A. AMP-activated protein kinase and FoxO transcription factors in dietary restriction-induced longevity. Ann N Y Acad Sci. 2009;1170:688–92. doi: 10.1111/j.1749-6632.2009.04019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–5. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Selman C, Tullet JM, Wieser D, Irvine E, Lingard SJ, Choudhury AI, Claret M, Al-Qassab H, Carmignac D, Ramadani F, et al. Ribosomal protein S6 kinase 1 signaling regulates mammalian lifespan. Science. 2009;326:140–4. doi: 10.1126/science.1177221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–5. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- 70.Brown MK, Naidoo N. The endoplasmic reticulum stress response in aging and age-related diseases. Front Physiol. 2012;3:263. doi: 10.3389/fphys.2012.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hipkiss AR. On why decreasing protein synthesis can increase lifespan. Mech Ageing Dev. 2007;128:412–4. doi: 10.1016/j.mad.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 72.Rodriguez KA, Gaczynska M, Osmulski PA. Molecular mechanisms of proteasome plasticity in aging. Mech Ageing Dev. 2010;131:144–55. doi: 10.1016/j.mad.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cuervo AM. Autophagy and aging: keeping that old broom working. Trends Genet. 2008;24:604–12. doi: 10.1016/j.tig.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tonoki A, Kuranaga E, Tomioka T, Hamazaki J, Murata S, Tanaka K, Miura M. Genetic evidence linking age-dependent attenuation of the 26S proteasome with the aging process. Mol Cell Biol. 2009;29:1095–106. doi: 10.1128/MCB.01227-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4:176–84. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- 76.Carrano AC, Liu Z, Dillin A, Hunter T. A conserved ubiquitination pathway determines longevity in response to diet restriction. Nature. 2009;460:396–9. doi: 10.1038/nature08130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li W, Gao B, Lee SM, Bennett K, Fang D. RLE-1, an E3 ubiquitin ligase, regulates C. elegans aging by catalyzing DAF-16 polyubiquitination. Dev Cell. 2007;12:235–46. doi: 10.1016/j.devcel.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 78.Morrow G, Samson M, Michaud S, Tanguay RM. Overexpression of the small mitochondrial Hsp22 extends Drosophila lifespan and increases resistance to oxidative stress. FASEB J. 2004;18:598–9. doi: 10.1096/fj.03-0860fje. [DOI] [PubMed] [Google Scholar]
- 79.Luce K, Osiewacz HD. Increasing organismal healthspan by enhancing mitochondrial protein quality control. Nat Cell Biol. 2009;11:852–8. doi: 10.1038/ncb1893. [DOI] [PubMed] [Google Scholar]
- 80.Wolfson M, Budovsky A, Tacutu R, Fraifeld V. The signaling hubs at the crossroad of longevity and age-related disease networks. Int J Biochem Cell Biol. 2009;41:516–20. doi: 10.1016/j.biocel.2008.08.026. [DOI] [PubMed] [Google Scholar]
- 81.Brack AS, Conboy MJ, Roy S, Lee M, Kuo CJ, Keller C, Rando TA. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 2007;317:807–10. doi: 10.1126/science.1144090. [DOI] [PubMed] [Google Scholar]
- 82.Loram J, Bodnar A. Age-related changes in gene expression in tissues of the sea urchin Strongylocentrotus purpuratus. Mech Ageing Dev. 2012;133:338–47. doi: 10.1016/j.mad.2012.03.012. [DOI] [PubMed] [Google Scholar]
- 83.DeNicola GM, Tuveson DA. RAS in cellular transformation and senescence. Eur J Cancer. 2009;45(Suppl 1):211–6. doi: 10.1016/S0959-8049(09)70036-X. [DOI] [PubMed] [Google Scholar]
- 84.Vrailas-Mortimer A, del Rivero T, Mukherjee S, Nag S, Gaitanidis A, Kadas D, Consoulas C, Duttaroy A, Sanyal S. A muscle-specific p38 MAPK/Mef2/MnSOD pathway regulates stress, motor function, and lifespan in Drosophila. Dev Cell. 2011;21:783–95. doi: 10.1016/j.devcel.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.King AM, Van der Put E, Blomberg BB, Riley RL. Accelerated Notch-dependent degradation of E47 proteins in aged B cell precursors is associated with increased ERK MAPK activation. J Immunol. 2007;178:3521–9. doi: 10.4049/jimmunol.178.6.3521. [DOI] [PubMed] [Google Scholar]
- 86.Wang MC, Bohmann D, Jasper H. JNK signaling confers tolerance to oxidative stress and extends lifespan in Drosophila. Dev Cell. 2003;5:811–6. doi: 10.1016/S1534-5807(03)00323-X. [DOI] [PubMed] [Google Scholar]
- 87.Kim YM, Seo YH, Park CB, Yoon SH, Yoon G. Roles of GSK3 in metabolic shift toward abnormal anabolism in cell senescence. Ann N Y Acad Sci. 2010;1201:65–71. doi: 10.1111/j.1749-6632.2010.05617.x. [DOI] [PubMed] [Google Scholar]
- 88.Migliaccio E, Giorgio M, Mele S, Pelicci G, Reboldi P, Pandolfi PP, Lanfrancone L, Pelicci PG. The p66shc adaptor protein controls oxidative stress response and lifespan in mammals. Nature. 1999;402:309–13. doi: 10.1038/46311. [DOI] [PubMed] [Google Scholar]
- 89.Lapointe J, Hekimi S. Early mitochondrial dysfunction in long-lived Mclk1+/- mice. J Biol Chem. 2008;283:26217–27. doi: 10.1074/jbc.M803287200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Caldeira da Silva CC, Cerqueira FM, Barbosa LF, Medeiros MH, Kowaltowski AJ. Mild mitochondrial uncoupling in mice affects energy metabolism, redox balance and longevity. Aging Cell. 2008;7:552–60. doi: 10.1111/j.1474-9726.2008.00407.x. [DOI] [PubMed] [Google Scholar]
- 91.Messina A, Reina S, Guarino F, De Pinto V. VDAC isoforms in mammals. Biochim Biophys Acta. 2012;1818:1466–76. doi: 10.1016/j.bbamem.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 92.Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–38. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee KS, Iijima-Ando K, Iijima K, Lee WJ, Lee JH, Yu K, Lee DS. JNK/FOXO-mediated neuronal expression of fly homologue of peroxiredoxin II reduces oxidative stress and extends lifespan. J Biol Chem. 2009;284:29454–61. doi: 10.1074/jbc.M109.028027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Radyuk SN, Michalak K, Klichko VI, Benes J, Rebrin I, Sohal RS, Orr WC. Peroxiredoxin 5 confers protection against oxidative stress and apoptosis and also promotes longevity in Drosophila. Biochem J. 2009;419:437–45. doi: 10.1042/BJ20082003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hu D, Cao P, Thiels E, Chu CT, Wu GY, Oury TD, Klann E. Hippocampal long-term potentiation, memory, and longevity in mice that overexpress mitochondrial superoxide dismutase. Neurobiol Learn Mem. 2007;87:372–84. doi: 10.1016/j.nlm.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Phillips JP, Parkes TL, Hilliker AJ. Targeted neuronal gene expression and longevity in Drosophila. Exp Gerontol. 2000;35:1157–64. doi: 10.1016/S0531-5565(00)00117-0. [DOI] [PubMed] [Google Scholar]
- 97.Dai DF, Santana LF, Vermulst M, Tomazela DM, Emond MJ, MacCoss MJ, Gollahon K, Martin GM, Loeb LA, Ladiges WC, et al. Overexpression of catalase targeted to mitochondria attenuates murine cardiac aging. Circulation. 2009;119:2789–97. doi: 10.1161/CIRCULATIONAHA.108.822403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bennett-Baker PE, Wilkowski J, Burke DT. Age-associated activation of epigenetically repressed genes in the mouse. Genetics. 2003;165:2055–62. doi: 10.1093/genetics/165.4.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Salpea P, Russanova VR, Hirai TH, Sourlingas TG, Sekeri-Pataryas KE, Romero R, Epstein J, Howard BH. Postnatal development- and age-related changes in DNA-methylation patterns in the human genome. Nucleic Acids Res. 2012;40:6477–94. doi: 10.1093/nar/gks312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bell JT, Tsai PC, Yang TP, Pidsley R, Nisbet J, Glass D, Mangino M, Zhai G, Zhang F, Valdes A, et al. MuTHER Consortium Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS Genet. 2012;8:e1002629. doi: 10.1371/journal.pgen.1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suñer D, Cigudosa JC, Urioste M, Benitez J, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604–9. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Anisimov VN, Bartke A, Barzilai N, Batin MA, Blagosklonny MV, Brown-Borg H, Budovskaya Y, Campisi J, Friguet B, Fraifeld V, et al. The second international conference “genetics of aging and longevity”. Aging (Albany NY) 2012;4:305–17. doi: 10.18632/aging.100458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Barbot W, Dupressoir A, Lazar V, Heidmann T. Epigenetic regulation of an IAP retrotransposon in the aging mouse: progressive demethylation and de-silencing of the element by its repetitive induction. Nucleic Acids Res. 2002;30:2365–73. doi: 10.1093/nar/30.11.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Machwe A, Orren DK, Bohr VA. Accelerated methylation of ribosomal RNA genes during the cellular senescence of Werner syndrome fibroblasts. FASEB J. 2000;14:1715–24. doi: 10.1096/fj.99-0926com. [DOI] [PubMed] [Google Scholar]
- 105.Swisshelm K, Disteche CM, Thorvaldsen J, Nelson A, Salk D. Age-related increase in methylation of ribosomal genes and inactivation of chromosome-specific rRNA gene clusters in mouse. Mutat Res. 1990;237:131–46. doi: 10.1016/0921-8734(90)90019-N. [DOI] [PubMed] [Google Scholar]
- 106.Narita M, Nũnez S, Heard E, Narita M, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–16. doi: 10.1016/S0092-8674(03)00401-X. [DOI] [PubMed] [Google Scholar]
- 107.Casillas MA, Jr., Lopatina N, Andrews LG, Tollefsbol TO. Transcriptional control of the DNA methyltransferases is altered in aging and neoplastically-transformed human fibroblasts. Mol Cell Biochem. 2003;252:33–43. doi: 10.1023/A:1025548623524. [DOI] [PubMed] [Google Scholar]
- 108.Marton O, Koltai E, Nyakas C, Bakonyi T, Zenteno-Savin T, Kumagai S, Goto S, Radak Z. Aging and exercise affect the level of protein acetylation and SIRT1 activity in cerebellum of male rats. Biogerontology. 2010;11:679–86. doi: 10.1007/s10522-010-9279-2. [DOI] [PubMed] [Google Scholar]
- 109.Jung JW, Lee S, Seo MS, Park SB, Kurtz A, Kang SK, Kang KS. Histone deacetylase controls adult stem cell aging by balancing the expression of polycomb genes and jumonji domain containing 3. Cell Mol Life Sci. 2010;67:1165–76. doi: 10.1007/s00018-009-0242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nijwening JH, Geutjes EJ, Bernards R, Beijersbergen RL. The histone demethylase Jarid1b (Kdm5b) is a novel component of the Rb pathway and associates with E2f-target genes in MEFs during senescence. PLoS One. 2011;6:e25235. doi: 10.1371/journal.pone.0025235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cairney CJ, Bilsland AE, Evans TR, Roffey J, Bennett DC, Narita M, Torrance CJ, Keith WN. Cancer cell senescence: a new frontier in drug development. Drug Discov Today. 2012;17:269–76. doi: 10.1016/j.drudis.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 112.Bilsland AE, Revie J, Keith W. MicroRNA and senescence: the senectome, integration and distributed control. Crit Rev Oncog. 2013;18:373–90. doi: 10.1615/CritRevOncog.2013007197. [DOI] [PubMed] [Google Scholar]
- 113.Boon RA, Iekushi K, Lechner S, Seeger T, Fischer A, Heydt S, Kaluza D, Tréguer K, Carmona G, Bonauer A, et al. MicroRNA-34a regulates cardiac ageing and function. Nature. 2013;495:107–10. doi: 10.1038/nature11919. [DOI] [PubMed] [Google Scholar]
- 114.Mehi SJ, Maltare A, Abraham CR, King GD. MicroRNA-339 and microRNA-556 regulate Klotho expression in vitro. Age (Dordr) 2014;36:141–9. doi: 10.1007/s11357-013-9555-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Olivieri F, Rippo MR, Procopio AD, Fazioli F. Circulating inflamma-miRs in aging and age-related diseases. Front Genet. 2013;4:121. doi: 10.3389/fgene.2013.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chang AL, Bitter PH, Jr., Qu K, Lin M, Rapicavoli NA, Chang HY. Rejuvenation of gene expression pattern of aged human skin by broadband light treatment: a pilot study. J Invest Dermatol. 2013;133:394–402. doi: 10.1038/jid.2012.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Abdelmohsen K, Panda A, Kang MJ, Xu J, Selimyan R, Yoon JH, Martindale JL, De S, Wood WH, Becker KG, et al. SAL-RNAs: Senescence-associated long non-coding RNAs. Aging Cell. 2013 doi: 10.1111/acel.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Halaschek-Wiener J, Khattra JS, McKay S, Pouzyrev A, Stott JM, Yang GS, Holt RA, Jones SJ, Marra MA, Brooks-Wilson AR, et al. Analysis of long-lived C. elegans daf-2 mutants using serial analysis of gene expression. Genome Res. 2005;15:603–15. doi: 10.1101/gr.3274805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Morley JF, Morimoto RI. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell. 2004;15:657–64. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Müller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- 122.Hertweck M, Göbel C, Baumeister R. C. elegans SGK-1 is the critical component in the Akt/PKB kinase complex to control stress response and lifespan. Dev Cell. 2004;6:577–88. doi: 10.1016/S1534-5807(04)00095-4. [DOI] [PubMed] [Google Scholar]
- 123.Rizki G, Iwata TN, Li J, Riedel CG, Picard CL, Jan M, Murphy CT, Lee SS. The evolutionarily conserved longevity determinants HCF-1 and SIR-2.1/SIRT1 collaborate to regulate DAF-16/FOXO. PLoS Genet. 2011;7:e1002235. doi: 10.1371/journal.pgen.1002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Oh SW, Mukhopadhyay A, Svrzikapa N, Jiang F, Davis RJ, Tissenbaum HA. JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc Natl Acad Sci U S A. 2005;102:4494–9. doi: 10.1073/pnas.0500749102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cabreiro F, Ackerman D, Doonan R, Araiz C, Back P, Papp D, Braeckman BP, Gems D. Increased lifespan from overexpression of superoxide dismutase in Caenorhabditis elegans is not caused by decreased oxidative damage. Free Radic Biol Med. 2011;51:1575–82. doi: 10.1016/j.freeradbiomed.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A. 2004;101:15998–6003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS. A mutant Drosophila insulin receptor homolog that extends lifespan and impairs neuroendocrine function. Science. 2001;292:107–10. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- 128.Hwangbo DS, Gershman B, Tu MP, Palmer M, Tatar M. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature. 2004;429:562–6. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- 129.Bayne AC, Mockett RJ, Orr WC, Sohal RS. Enhanced catabolism of mitochondrial superoxide/hydrogen peroxide and aging in transgenic Drosophila. Biochem J. 2005;391:277–84. doi: 10.1042/BJ20041872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Parkes TL, Elia AJ, Dickinson D, Hilliker AJ, Phillips JP, Boulianne GL. Extension of Drosophila lifespan by overexpression of human SOD1 in motorneurons. Nat Genet. 1998;19:171–4. doi: 10.1038/534. [DOI] [PubMed] [Google Scholar]
- 131.Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 2004;14:885–90. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Matheu A, Maraver A, Klatt P, Flores I, Garcia-Cao I, Borras C, Flores JM, Viña J, Blasco MA, Serrano M. Delayed ageing through damage protection by the Arf/p53 pathway. Nature. 2007;448:375–9. doi: 10.1038/nature05949. [DOI] [PubMed] [Google Scholar]
- 133.Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, Nahum L, Bar-Joseph Z, Cohen HY. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483:218–21. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- 134.Keyes WM, Wu Y, Vogel H, Guo X, Lowe SW, Mills AA. p63 deficiency activates a program of cellular senescence and leads to accelerated aging. Genes Dev. 2005;19:1986–99. doi: 10.1101/gad.342305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jeng YM, Cai-Ng S, Li A, Furuta S, Chew H, Chen PL, Lee EY, Lee WH. Brca1 heterozygous mice have shortened lifespan and are prone to ovarian tumorigenesis with haploinsufficiency upon ionizing irradiation. Oncogene. 2007;26:6160–6. doi: 10.1038/sj.onc.1210451. [DOI] [PubMed] [Google Scholar]
- 136.Zhavoronkov A, Cantor CR. Methods for structuring scientific knowledge from many areas related to aging research. PLoS One. 2011;6:e22597. doi: 10.1371/journal.pone.0022597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Moskalev A. Aging and genes Science: St Petersburg, 2008; p 358. [Google Scholar]
- 138.Moskalev AA. Evolutionary ideas on the nature of aging. Adv Gerontol. 2011;1:112–21. doi: 10.1134/S207905701102010X. [DOI] [Google Scholar]