Abstract
The recessive ataxia-telangiectasia (A-T) syndrome is characterized by cerebellar degeneration, immunodeficiency, cancer susceptibility, premature aging, and insulin-resistant diabetes and is caused by loss of function of the ATM kinase, a member of the phosphoinositide 3-kinase–like protein kinases (PIKKs) family. ATM plays a crucial role in the DNA damage response (DDR); however, the complexity of A-T features suggests that ATM may regulate other cellular functions. Here we show that ATM affects proper bipolar mitotic spindle structure independently of DNA damage. In addition, we find that in mitosis ATM forms a complex with the poly(ADP)ribose (PAR) polymerase Tankyrase (TNKS) 1, the spindle pole protein NuMA1, and breast cancer susceptibility protein BRCA1, another crucial DDR player. Our evidence indicates that the complex is required for efficient poly(ADP)ribosylation of NuMA1. We find further that a mutant NuMA1 version, non-phosphorylatable at potential ATM-dependent phosphorylation sites, is poorly PARylated and induces loss of spindle bipolarity. Our findings may help to explain crucial A-T features and provide further mechanistic rationale for TNKS inhibition in cancer therapy.
Keywords: ATM, BRCA1, NuMA1, Tankyrase1, spindle structure, poly(ADP)ribosylation
Introduction
ATM is a crucial DDR regulator particularly relevant for double-strand break repair.1,2 Although DDR defects have been directly linked to cancer, sterility, and radiosensitivity,1,3 how DDR deficit causes other A-T syndrome features, like premature aging, insulin-resistant diabetes, and neurological deficit, is less clear. Premature aging may derive from premature replicative cellular senescence, a phenomenon linked to accelerated telomere shortening.4 Although ATM has been shown to help mounting a DDR at shortened telomeres,2 how ATM deficit translates into telomere shortening is unclear. DDR deficit has also been involved in the pathogenesis of ataxia, lack of movement coordination, a major neurological A-T feature. It has been suggested that ataxia arises from malfunctioning, or death, of DNA-damaged cerebellar neurons.1,5 However, recent findings from both A-T patients and mouse models suggest a cause–effect relationship between neurological deficit and higher numbers of aneuploid neurons present in A-T brains.6,7 Indeed, A-T cells show chromosomal instability (CIN) with both structural (deletions, fusions, inversions, translocations, etc.) and numerical (gain or loss of entire chromosomes) chromosomal abnormalities (aneuploidy).6,7 Structural aneuploidy is mechanistically linked to DDR defects, while numerical aneuploidy is rather linked to defects in execution of mitosis.8 Conversely, imprecise chromosome segregation may contribute to structural aneuploidy by causing DNA damage.9 These observations suggest that ATM may play a role in mitosis. Indeed, recent evidence indicated that ATM is activated in mitosis by Aurora B kinase and helps the spindle assembly checkpoint (SAC), a safeguard mechanism that delays mitosis exit until spindle assembly completion, to prevent chromosome segregation errors.10 Thus, ATM might have DDR-independent functions in mitosis. Importantly, the breast cancer susceptibility gene product BRCA1, another crucial DDR player that cooperates with ATM in DDR, appears to have relevant, DDR-independent mitotic roles.11,12 BRCA1-deficient cells show structural deficit in mitotic spindle assembly, and BRCA1 has been shown to interact with spindle pole proteins like the spindle pole organizer protein NuMA1.11-13 These observations suggest that DDR and mitosis control share common players. To date, however, little is known about whether and how ATM might affect mitosis, apart from its role in SAC. In search for potential mitotic functions of ATM, in the present study we unveiled a molecular pathway composed by ATM, BRCA1, TNKS1, and NuMA1 that controls NuMA1 PARylation and correct bipolar spindle assembly. We discuss how the ATM–TNKS1 relationship helps to explain several A-T features and provides further mechanistic rationale for TNKS and ATM inhibition in cancer treatment.
Results
ATM affects mitotic spindle structure
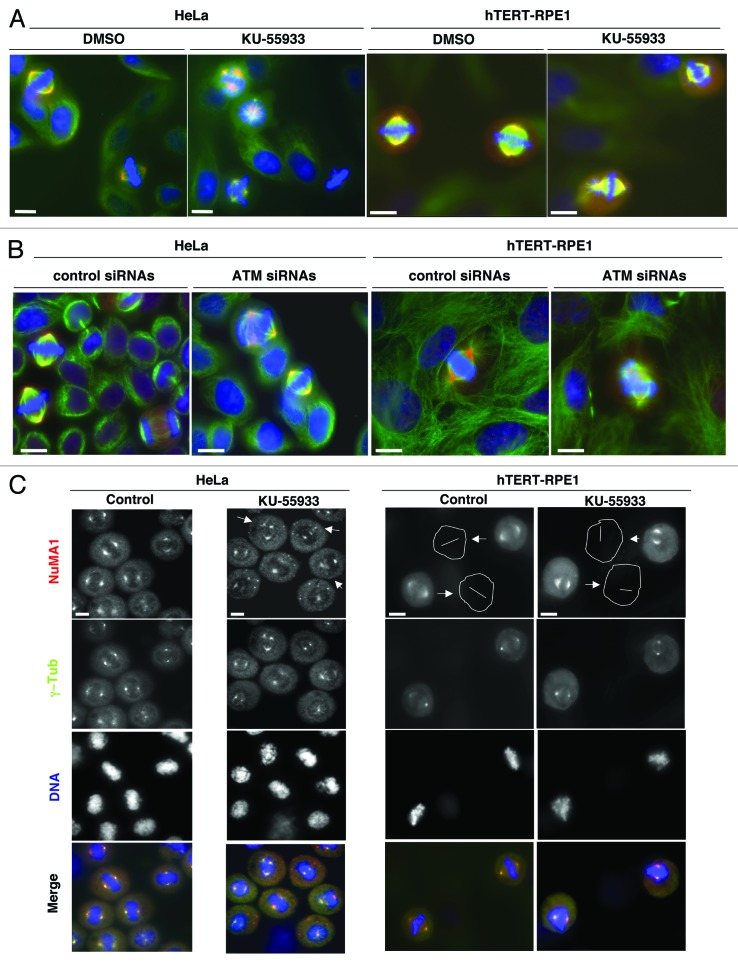
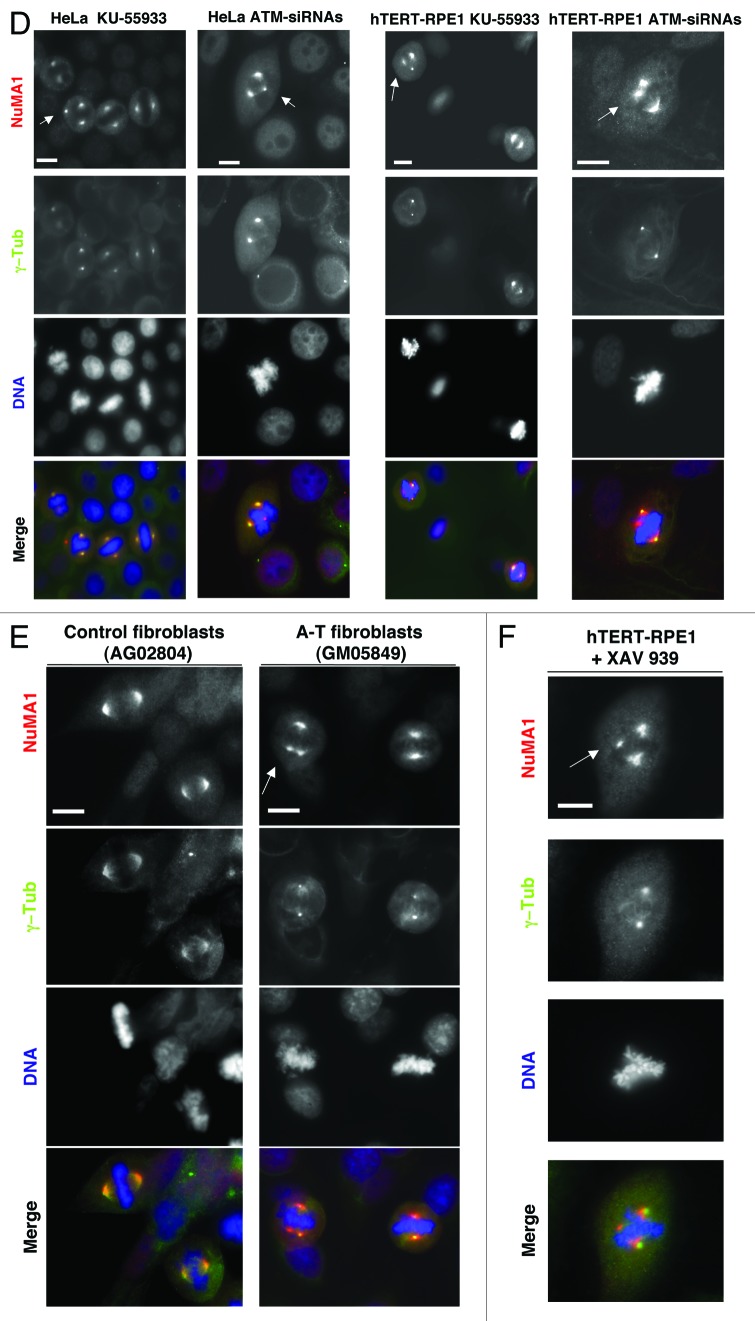
In search for mitotic effects of ATM, transformed HeLa and non-transformed, telomerase-immortalized, retinal pigment epithelial (hTERT-RPE1) cells were challenged for 24 h with selective ATM chemical inhibitor KU-55933, and mitotic cells were analyzed by staining NuMA1, α-tubulin, and DNA. Treatment significantly increased frequency of aberrant mitosis in both cell lines (Fig. 1A). Mitotic abnormalities included defective metaphase chromosome alignment, multipolar spindles, derangement of astral microtubules, and spindle displacement from cell center. Treatment increased chromosome misalignment in both cell lines, induced prominent higher number of multipolar spindles in HeLa cells, and spindle displacement from cell center and astral microtubules derangement in about 20% of mitosis in hTERT-RPE1 cells, features never observed in control cells (Fig. 1A; Fig. S1A and B). Increased frequency of similar mitotic abnormalities was also induced by treatment with ATM-small interfering RNAs (siRNAs, Fig. 1B; Figs. S1C, S2A, and 2B). Increase in spindle abnormalities was detected, as well, in cells arrested at the G2–M transition prior to release and treatment with KU-55933 (Fig. 1C), indicating genuine mitotic defects, rather than consequence of previous cell cycle stage perturbations. ATM-downregulated cells also showed increased frequency of spindles in which one extranumerary pole stained positive for NuMA1 but not for the centrosomal marker γ-tubulin (Fig. 1D; see also Fig. S2E), and of anaphases with lagging chromosomes (Fig. S1D and E), perhaps as a consequence of increased merotelic attachments that associate with deranged spindle geometry.14 Immortalized fibroblasts from an A-T patient also showed increased mitotic defects relatively to control immortalized fibroblasts, and, importantly, cells with NuMA1 positive and γ-tubulin negative spindle poles were scored in A-T but not in control cells (Fig. 1E; Fig. S1I).
Figure 1A–C. Mitotic defects upon chemical or genetic ATM downregulation. (A) HeLa or hTERT-RPE1 cells treated 24 h with vehicle (dimethylsulfoxide; DMSO) or with ATM inhibitor KU-55933. (B) HeLa or hTERT-RPE1 cells treated 72 h with control or ATM small interfering RNAs (siRNA). Cells were fixed, permeabilized, and stained for NuMA1 (red), α-tubulin (green), and DNA (blue). (C) HeLa and hTERT-RPE1 cells were treated 20 h with Cdk1 inhibitor RO-3306 and released into fresh medium; 10 min post-release, either DMSO (control) or KU-55933 were added. HeLa and hTERT-RPE1 cells, fixed, respectively, 30 and 50 min after release from RO-3306, were permeabilized and stained for NuMA1 (red), γ-tubulin (green), and DNA (blue). (D) HeLa and hTERT-RPE1 cells were treated 24 h with KU-55933 or ATM-siRNAs, fixed, permeabilized, and stained for NuMA1 (red), γ-tubulin (green), and DNA (blue). (E) Control (AG02804) and A-T (GM05849) fibroblasts were fixed, permeabilized, and stained for NuMA1 (red), γ-tubulin (green), and DNA (blue). (F) hTERT-RPE1 cells treated 6 h with TNKS inhibitor XAV939 were fixed, permeabilized, and stained for NuMA1 (red), γ-tubulin (green), and DNA (blue). For quantitations see Figure S1. Images are representative of 4–6 independent experiments. For each experiment around 200 mitotic cells were scored. Treatment with DMSO or control siRNA did not increase abnormal spindles number relatively to untreated cells. Arrowheads indicate spindle abnormalities, and in (C) cell contours and inter-poles axis have been marked to show spindle displacement from cell center. Scale bars, 10 μm.
Figure 1D–F. See legend for Figure 1A–C.
NuMA1 PARylation requires ATM
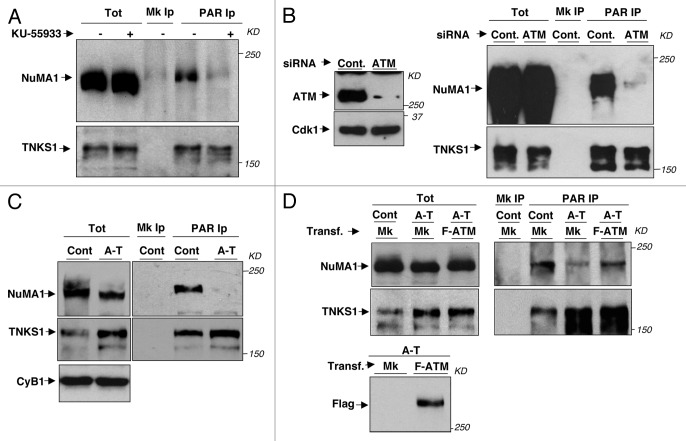
Although at low penetrance, mitotic abnormalities observed in ATM-downregulated cells were reminiscent of those induced by genetic downregulation of the telomere maintenance protein and PAR polymerase (PARP) Tankyrase (TNKS) 1.15,16 In addition, treating cells with the TNKS1 enzymatic inhibitor XAV939, at concentrations reported to upset the β-catenin pathway and that significantly inhibited global PARylation in mitotic cells (see also Fig. S3A),16,17 increased the frequency of similar mitotic abnormalities to ATM downregulation, albeit to a slightly lower extent (Fig. 1F; Fig. S1F, G, and H). XAV939-induced spindle abnormalities, however, were substantially lower in frequency than that reported for genetic TNKS1 downregulation.15,16 Considering that catalytic inactive TNKS1 was reported to be unable to complement mitotic defects in genetically TNKS1-downregulated cells,15,16 it is possible that spindle derangement at high frequency requires a strong, near complete, inhibition of TNKS1 activity, difficult to reach with available chemical inhibitors (without the risk of upsetting other functions or pathways).16,17 A major mitotic TNKS1 target is the spindle pole organizer protein NuMA1, whose PARylation has been suggested to be relevant for spindle bipolarization.16,18 Thus, we asked if ATM affected mitotic PARylation of NuMA1. HeLa cells were arrested in mitosis by a 14-h treatment with the microtubule polymerization inhibitor nocodazole, a condition in which TNKS1-dependent NuMA1 PARylation is at a maximum,19 and further treated with KU-55933 for 60 min. Treatment did not induce mitosis exit, indicated by persistence of mitotic specific MPM2 epitope signals (Fig. S3B), but reduced NuMA1 recovered in anti-PAR immunoprecipitates (IPs, Fig. 2A). PARylation of TNKS1, which can auto-PARylate,15 was not significantly affected (Fig. 2A). Similarly, ATM-siRNAs-treated HeLa cells arrested as efficiently as controls in mitosis (Fig. S3C) but contained much less PARylated NuMA1. A-T fibroblasts showed higher levels of TNKS1 and similar levels of NuMA1 compared with control cells, but significantly reduced levels of PARylated NuMA1 (Fig. 2C; see also Fig. S3D). Importantly, higher levels of NuMA1 PARylation could be restored in A-T fibroblasts upon re-expression of wild-type ATM by transfection of a Flag-tagged-ATM expression vector (Fig. 2D). Low NuMA1 PARylation was also detected in mitotic A-T lymphoblasts (Fig. S3E). Together, these observations suggest that ATM promotes NuMA1 PARylation, a reaction likely required for proper bipolar spindle assembly.16-19
Figure 2. ATM requirements for NuMA1 PARylation. (A) Mitotic HeLa cells, detached after 14-h treatment with nocodazole, were treated with vehicle or KU-55933 (−/+) for 60 min in nocodazole. (B) Detached HeLa cells treated with control or ATM siRNAs for 60 h prior to nocodazole for 14 h were collected. (C) Control and A-T fibroblasts, detached after 14 h-treatment with nocodazole, were collected. (D) Control and A-T fibroblasts were mock-transfected (Mk) along with A-T fibroblasts transfected with a Flag-ATM expression vector (F-ATM) 48 h prior to nocodazole treatment (Transf.), then, detached cells were collected. Total cytosolic extracts (Tot), mock (Mk) or anti-Poly(ADP)ribose (PAR) immunoprecipitates (IP), from total cytosolic extracts, were probed for NuMA1, TNKS1. F-ATM was also probed from total cytosolic extract samples with anti-Flag antibody. The levels of expression of TNKS1 and NuMA1 were not affected by mock transfection in control and A-T fibroblasts. The data shown are representative of at least 3 independent experiments per type.
ATM, BRCA1, TNKS1, and NuMA1 form a complex required for efficient NuMA1 PARylation
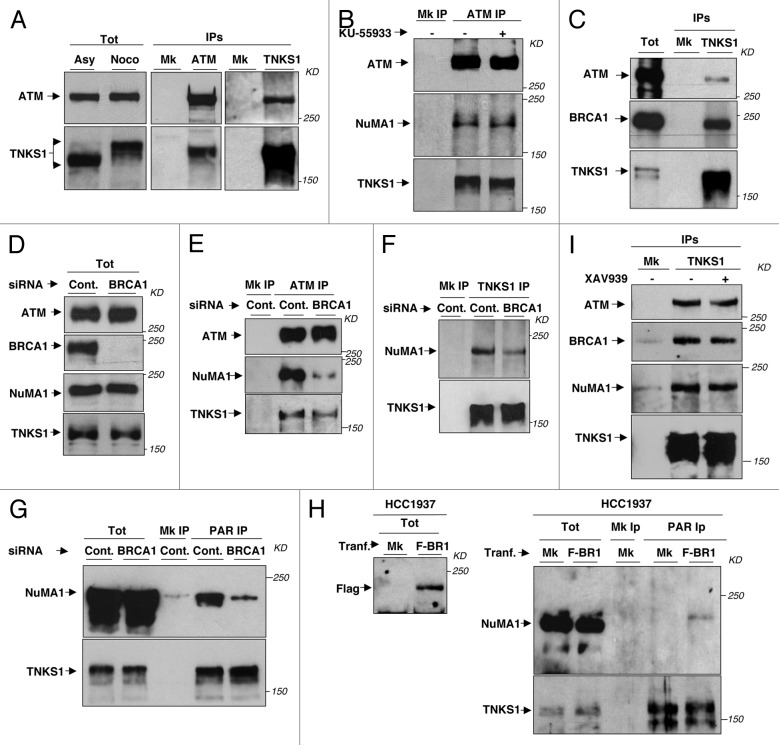
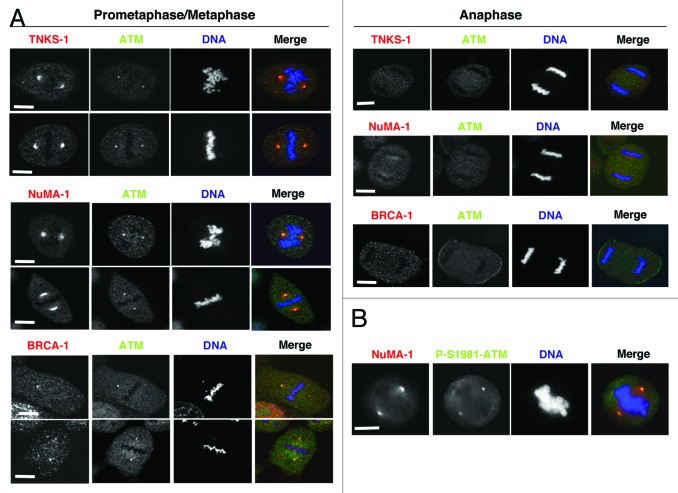
Although it cannot be excluded that other PARPs, in addition to TNKS1,20 regulated mitotic NuMA1 PARylation, we asked whether ATM physically interacted with TNKS1. By reciprocal co-IPs, we found that ATM and TNKS1 interacted in mitotic HeLa cells (Fig. 3A) and other cell types (Fig. S4A). Analysis of TNKS1 deletion mutants indicated that its ankyrin repeats domain was crucial for ATM–TNKS1 interaction (Fig. S4B and C). NuMA1 was also present in ATM IPs from mitotic cells, and the interaction of ATM with TNKS1 and NuMA1 was not significantly affected by treatment with KU-55933 (Fig. 3B). Since ATM inhibition impairs NuMA1 PARylation (Fig. 2A and B), these data may suggest that an ATM-TNKS1-NuMA1 complex forms in mitosis, in which ATM activity affects NuMA1 PARylation. To gain further insight into how ATM regulated NuMA1 PARylation, we took under consideration the potential contribution of BRCA1, another crucial DDR player, physically and functionally interacting with ATM in DDR, that has also been shown to interact with NuMA1 and affect mitotic spindle assembly.11,12 We first determined if BRCA1 interacted with TNKS1 in mitotic cells, and found that BRCA1 could be detected in TNKS1 IPs (Fig. 3C). Next, BRCA1 downregulation, via siRNAs (Fig. 3D; see also Fig. S2C), significantly lowered the amounts of NuMA1 in ATM IPs and also reduced NuMA1 amounts in TNKS1 IPs (Fig. 3E and F). Importantly, BRCA1 downregulation strongly inhibited NuMA1, rather than TNKS1, PARylation (Fig. 3G) and induced increased spindle abnormalities (Fig. S1J). To determine whether BRCA1 contributed to control NuMA1 PARylation in naturally BRCA1-deficient cells, we assessed NuMA1 and TNKS1 PARylation in HCC1937 cells, a BRCA1-deficient breast cancer cell line that shows various spindle aberrations, including NuMA1-positive and γ-tubulin-negative extranumerary poles (Fig. S2D), and in these cells after Flag-tagged-wild type BRCA1 expression vector transfection, in complementation experiments. Poor NuMA1, rather than TNKS1, PARylation was detected in mitotic HCC1937 cells, but detectable levels of PARylated NuMA1 could be retrieved upon wild-type BRCA1 re-expression (Fig. 3H). The ATM-TNKS1-NuMA1-BRCA1 complex may not crucially depend on TNKS1 activity, since treatment of mitotic HeLa cells with XAV939 only slightly reduced ATM, BRCA1, and NuMA1 retrieved in TNKS1 IPs (Fig. 3I). ATM, TNKS1, NuMA1, and BRCA1 analysis by confocal microscopy revealed the 4 proteins localized at spindle poles in prometaphase/metaphase and dissociated from this structure in anaphase (Fig. 4A). In addition, in mitotic HeLa cells, an antibody anti-P-Ser1981-ATM, a phosphorylation that marks active ATM, stained spindle poles (Fig. 4B) in agreement with previously reported data.21 Together these data suggest that ATM, TNKS1, NuMA1, and BRCA1 form a complex in mitosis that promotes efficient spindle assembly and NuMA1 PARylation. In ATM-downregulated, as well as in XAV939-treated cells, ATM, like BRCA1, always co-localized with γ-tubulin, while TNKS1 showed staining at γ-tubulin-negative poles with similar frequency to NuMA1 (Fig. S2F and G). Possibly, poor NuMA1 PARylation favors formation of NuMA1 aggregates that form extranumerary poles and that can bind TNKS1. ATM and BRCA1, instead, can only localize to centromeric, γ-tubulin-positive, spindle pole structures.
Figure 3. ATM and BRCA1 interact with TNKS1 and control NuMA1 PARylation. (A) Total cytosolic extracts from asynchronous (Asy) and nocodazole-treated (Noco) HeLa cells and mock (Mk), ATM, and TNKS1 IPs from extracts of nocodazole-treated cells were probed for ATM and TNKS1 (B) HeLa cells were treated as in Figure 2A; ATM, NuMA1 and TNKS1 were probed in mock (Mk) or ATM IPs. (C) Mock (Mk) or TNKS1 IPs from total cytosolic extracts (Tot) of mitotic-arrested HeLa cells probed for ATM, BRCA1, and TNKS1. (D–G) HeLa cells were treated with control or BRCA1 siRNAs 60 h prior to nocodazole treatment: (D) ATM, BRCA1, NuMA1, and TNKS1 in total cytosolic extracts (Tot); (E) ATM, NuMA1, and TNKS1 in ATM IPs; (F) NuMA1 and TNKS1 in TNKS1 IPs and (G) NuMA1 and TNKS1 in total cytosolic extracts (Tot) and anti-PAR IPs. (H) HCC1937 cells were either mock-transfected or transfected with a Flag-BRCA1 expression vector (F-BR1) 60 h prior to nocodazole treatment; then, left panel, total cytosolic extracts (Tot) probed with anti-Flag antibody, and, right panels, NuMA1 and TNKS1 probed in total cytosolic extracts (Tot) and anti-PAR IPs. (I) Mitotic HeLa cells, that detached after 14-h treatment with nocodazole, were further treated with vehicle or XAV939 (−/+) for 60 min in nocodazole. ATM, BRCA1, NuMA1, and TNKS1 probed in mock (Mk) and TNKS1 IPs. The data shown are representative of 3 independent experiments per type.
Figure 4. (A) Mitotic localization of ATM, BRCA1, NuMA1, and TNKS1. HeLa cells were fixed and processed for immunofluorescence and confocal microscopy analysis. BRCA1, NuMA1, and TNKS1 were stained in red, ATM in green, and DNA in blue. Mitotic phases are indicated. (B) HeLa cells were fixed and processed for immunofluorescence. NuMA1 was stained in red, P-Ser1981-ATM in green, and DNA in blue. Scale bars, 10 μm.
NuMA1 phosphorylation is required for its efficient PARylation and correct spindle assembly
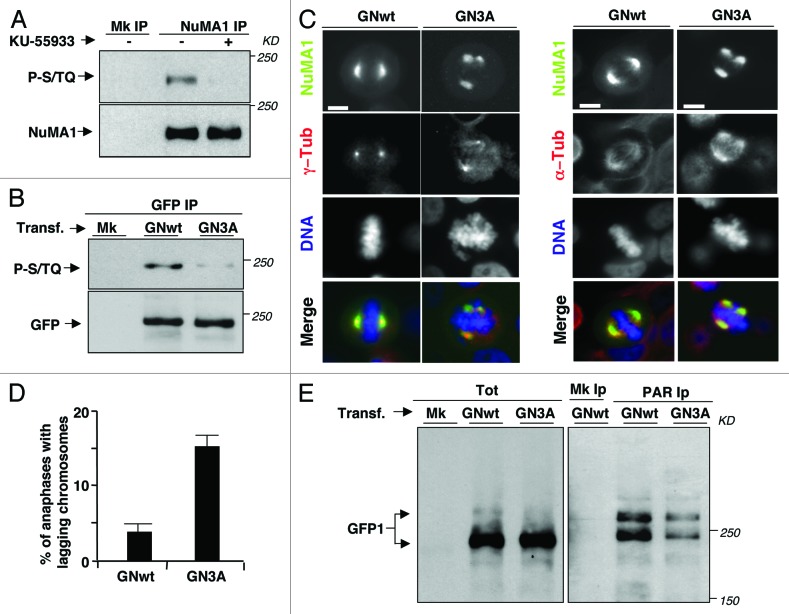
ATM kinase inhibition depressed NuMA1, rather than TNKS1, PARylation (Fig. 2A). We hypothesized that ATM kinase activity rendered NuMA1 a better PARylation substrate, rather than stimulated TNKS1 activity. NuMA1 has been shown to be phosphorylated at Ser395 by ATM upon DNA damage;22 however, NuMA1-Ser395 was not significantly phosphorylated in mitosis (Fig. S5A). Nevertheless, mitotic NuMA1 reacted with an anti-phosphoS/TQ (the basal ATM consensus phosphorylation target) antibody in a KU-55933-sensitive manner (Fig. 5A), and mitotic NuMA1 could be further phosphorylated in vitro by ATM isolated from mitotic cells (Fig. S5B). In the search for potential ATM-dependent, mitotic, phosphorylation sites in NuMA1, through a bioinformatic software at high stringency for ATM phosphorylation consensus (NetPhos 2.0),23we focused our attention on 3 human NuMA1 serine residues (Ser1262, Ser1601, Ser1887). Importantly, phosphorylated Ser1262 and Ser1601 have been detected in NuMA1 from mitotic cells via proteomic assays.24,25 These residues were mutated (ser-to-ala) in a green fluorescent protein (GFP)-tagged wild-type human NuMA1 expression vector (GNwt) to produce an ATM-phosphorylation-resistant NuMA1 version (GN3A). Indeed, when retrieved from transfected and mitotic-arrested HeLa cells, GN3A reacted poorly with anti-phosphoS/TQ relatively to GNwt (Fig. 5B). To determine whether GN3A affected spindle structure, we analyzed spindle morphology 24 h post-transfection of GNwt and GN3A in HeLa cells, when exogenous NuMA1 level was about 1-fold over endogenous level for both constructs (Fig. S5C), to limit spindle derangement potentially induced by NuMA1 overexpression per se.13,26 GN3A-expressing cells showed increased abnormal spindle frequency over GNwt-expressing cells, including higher frequency of NuMA1-positive/γ-tubulin-negative extranumerary spindle poles and increased frequency of anaphases with lagging chromosomes (Fig. 5C and D). To determine whether deranged mitosis correlated with decreased PARylation of GN3A, HeLa cells expressing GNwt or GN3A were arrested in mitosis 74 h post-transfection, and PARylated NuMA1 amounts determined. Significantly less GN3A than GNwt could be recovered in anti-PAR IPs (Fig. 5E, in total lysates of transfected cells, the faint anti GFP-reacting bands migrating above the 250 kD marker perhaps represent hyper-PARylation or other modifications of PARylated GFP-NuMA1, since these are enriched in PAR IPs, especially from the GNwt-transfectants, but never detected in non- or mock-transfected cells). Not excluding that ATM might affect other PARPs activity that may PARylate NuMA1 in mitosis nor TNKS1 activity directly,20 these data suggest that ATM-dependent phosphorylation of NuMA1 is required for efficient NuMA1 PARylation. Being poorly PARylated, an ATM-phosphorylation-resistant NuMA1 facilitates formation of abnormal aggregates of itself distorting normal spindle structure.
Figure 5. NuMA1 phosphorylations affecting spindle assembly and PARylation. (A) HeLa cells were treated as described in Figure 2A; mock (Mk) or NuMA1 IPs probed for phospho-S/TQ sites (P-S/TQ; top panel), NuMA1 (bottom panel). (B) HeLa cells were either mock (Mk) or transfected with a GFP-NuMA1-wild type (GNwt) or with GFP-NuMA1–3-ser-to-ala mutant (GN3A) fusion protein expression vectors 24 h prior to a 14 h nocodazole treatment. GFP IPs probed for phospho-S/TQ sites (P-S/TQ; top panel) and for GFP (bottom panel). (C) HeLa cells were transfected with GNwt or GN3A expression vectors. Twenty-four hours post-tranfections, cells were fixed and immunostained for (left panels) γ-tubulin (red), or (right panels) α-tubulin (red), the GFP-NuMA1 fusion proteins were visualized by autofluorescence (green) and DNA by Hoechst 33258 staining (blue). Scale bars, 10 μm. (D) Histogram: percent of anaphases with lagging chromosomes in GNwt- and GN3A-transfected HeLa cells. Error bars indicate s.d. (E) HeLa cells were either mock transfected (Mk) or transfected with GNwt or GN3A expression vectors 60 h prior to 14 h-nocodazole treatment. Detached cells were isolated, and total cytosolic extracts (Tot), mock (Mk), and anti-PAR IPs were probed with an anti-GFP antibody. In transfected cells, the faint bands recognized by the anti-GFP antibody migrating above the 250-kD marker in total lysates perhaps represent hyper-PARylation or other modifications of PARylated GFP-NuMA1, since these appear enriched in PAR IPs (especially in the GNwt-transfectants) and are never detected in non- or mock-transfected cells. The data shown are representative of 3 independent experiments per type.
Discussion
Our data indicate that ATM affects mitotic spindle architecture by helping enforcing spindle bipolarity and positioning, and that ATM forms a complex with TNKS1, NuMA1, and BRCA1 in mitosis that promotes efficient NuMA1 PARylation. The exact role for NuMA1 PARylation in spindle assembly is not known at present, nor whether it only depends on TNKS activity. However, it has been described that genetic TNKS1 downregulation depressed NuMA1 PARylation and increased the frequency of abnormal spindle structures, including spindles with extranumerary, γ-tubulin-negative spindle poles formed by NuMA1 aggregates, similarly to what we found by downregulating ATM.16,18-20 The mitotic phenotypes appear substantially more penetrant in the case of genetic TNKS1 knockdown relative to ATM deficit or TNKS PARP activity inhibition.16,18-20 Nevertheless, the contribution of ATM, and possibly full NuMA1 PARylation, to spindle assembly and structure may be particularly relevant during the logarithmic growth rate phase of neural development, and defects in bipolar spindle assembly may help to explain increased aneuploidy in neurons of A-T patients and animal models.6,7,27 In addition, defects in spindle assembly and positioning in stem cell districts, where control of spindle structure and positioning is critical for stemness maintenance, may contribute to reduced regenerative potential and premature organ aging observed in A-T patients.28 Given the role for TNKS1 in telomere length maintenance and glucose transport, the relationship we unveiled between ATM and TNKS1 may also be relevant for other A-T manifestations, like telomere shortening and diabetes.29,30 ATM is aided by BRCA1 in the NuMA1 PARylation control. BRCA1 and the checkpoint kinase Chk2 have been described to affect the Ran-dependent spindle assembly pathway.11,12 Our data show that BRCA1 forms a mitotic complex with ATM, TNKS1, and NuMA1, in which, possibly, NuMA1 is phosphorylated by ATM to undergo TNKS1-dependent PARylation. In addition to provide evidence for a novel pathway by which ATM and BRCA1 contribute to bipolar spindle architecture, these observations offer a mechanistic explanation for why TNKS1 inhibition, by exacerbating mitotic defects in BRCA1-deficient cells, may be beneficial for BRCA1-associated cancers treatment and suggest that targeting both ATM and TNKS could be a wider approach to reduce cancer cell fitness.31,32
Materials and Methods
Cell cultures
HeLa and hTERT-RPE1 cells were maintained essentially as previously described.33,34 SV40-transformed control fibroblasts (AG02804) (Coriell Cell Repository), and SV40-transformed AT-fibroblasts (GM05849) (Coriell Cell Repository) were maintained in Eagle Minimum Essential Medium (Sigma Aldrich) with Earle salts supplemented with 10% fetal bovine serum (FBS) (Hyclone), 1% penicillin/streptomycin (pen/strep) (Sigma Aldrich), 2 mM L-Glutamine (Sigma Aldrich), and 1% non-essential amino acids (Sigma Aldrich). HCC1937 (ATCC) were grown in Iscove Modified Dulbecco Medium (Sigma Aldrich) supplemented with 20% FBS, 1% pen/strep, 2 mM L-Glutamine. EBV-transformed B-lymphocytes from a healthy subject (AHH1) and EBV-transformed B-lymphocytes from an A-T patient (GM03189) (Coriell Cell Repository) were grown in RPMI-1640 (Sigma Aldrich) supplemented with 15% FBS, 1% pen/strep, 2 mM L-Glutamine. HCT116 and NIH3T3 (ATCC) cells were grown in Dulbecco modified Eagle medium with 4.5 g/L glucose (Sigma Aldrich) supplemented with 10% FBS and 1% pen/strep.
siRNAs, vectors, site-directed mutagenesis, and transfections
For control (non-targeting), ATM, and BRCA1 siRNAs, ON-TARGETplus SMART-pools of 4 siRNAs were purchased from Dharmacon Research and transfected with DharmaFect (Dharmacon) according to the manufacturer’s instructions. pCMV 4XFlag-BRCA1 was a gift of Dr Richard Baer (Columbia University);35 pGFP-C1-NuMA1 was provided by AddGene (plasmid 28238); pcDNA3.1-Flag-ATM WT has been previously described.36 Site-directed mutagenesis of human NuMA1 at Serine-1262 (5′-gccgagtcag agaaggccca gaagctggag ga-3′ and 3′-cggctcagtc tcttccgggt cttcgacctc ct-5′), Serine-1601 (5′-ctgcaagccc agttggccca gaaggagcag gc-3′ and 3′-gacgttcggg tcaaccgggt cttcctcgtc cg-5′) and Serine-1887 (5′-cagttctgct cgtcgtgccc aggccg-3′ and 3′-gtcaagacga gcagcacggg tccggc-5′) into alanine was performed using pGFP-C1-NuMA1 vector as template by QuickChange II XL Site-Directed Mutagenesis Kit according to the manufacturer’s instructions (Agilent Technology). N-terminal-3XFlag-TNKS1-HPS and N-terminal-3XFlag-TNKS1-ANK were cloned in pReceinver-M12 (GeneCopeia Inc). Transfections were performed using Linear Polyethylenimine (PEI) (Polysciences Inc).
Chemicals for cell treatments
Nocodazole (Calbiochem), for mitotic synchronization was used at 100 ng/ml for HeLa, hTERT-RPE1, HCC1937, AHH1, and GM03189 and at 800 ng/ml for AG02804 and GM05849. KU-55933, XAV939, RO-3306 and Etoposide (all from Calbiochem) were used at 10, 5, 10, and 50 μM final respective concentrations.
Immunoprecipitations
Cells lysed in lysis buffer, 0.1% NP-40-PBS, plus phosSTOP and Complete Protease inhibitor (Roche) and PAR glycohydrolase inhibitor ADP-HPD (5 μM; Calbiochem), were put on ice for 10 min, centrifuged twice at 14 000 g for 10 min, and supernatants were collected. For PAR IPs only, to disrupt non covalent interactions, the supernatants were added with 1% SDS and incubated 10 min at room temperature then diluted 5-fold with lysis buffer. For PAR and NuMA1 IPs, 2 μg per sample of anti-PAR clone 10H (Tulip Biolabs), anti-NuMA1 (Novus Biological), or control non-immune mouse or rabbit IgG (Santa Cruz Biotech) were pre-adsorbed to 20 μl of 50% protein A+G-agarose conjugated slurry (AC; Santa Cruz Biotech.) by rotation overnight (O.N.) at 4 °C. Cell lysates were pre-cleared using 2 μg of control mouse or rabbit IgGs and 20 μl of 50% protein A+G-AC slurry on rotation for 120 min at 4 °C. Supernatants were incubated with the preadsorbed antibodies O.N. at 4 °C. For IPs with agarose- or sepharose-conjugated (AC, SC) antibodies, cell lysates were pre-cleared using 20 μl of a 50% slurry of relative heat-inactivated conjugated antibody (anti-ATM AC, 2 μg Novus Biologicals; anti-Tankyrase 1/2 AC, 1,5 μg Santa Cruz Biotech; anti-GFP SC, 2 μg Abcam; anti-Flag M2 AC, 2μg Sigma) on rotation for 120 min at 4 °C. Supernatants were incubated with heat-inactivated, mock, or active antibody beads O.N. at 4 °C. Beads were washed 3 times with lysis buffer, boiled in sample buffer, and separated on SDS-PAGE. Boiling was omitted for anti-PAR immunoblotting. For in vitro NuMA1 phosphorylation experiments, NuMA1 IPs from mitotic HeLa cells were incubated with Flag Ips from either mock- or Flag-ATM expression vector-transfected, mitotic, HeLa cells in a kinase reaction mixture (10 mM HEPES, pH 7.4; 10 mM MgCl2; 50 mM NaCl; 10 mM MnCl2), including Flag peptide (Sigma), 10 μCi γ-32P-ATP, and KU-55933 where indicated, for 15 min at 30 °C. NuMA1 IPs were washed with lysis buffer, boiled in sample buffer, separated on SDS/PAGE, and autoradiographed. One aliquot of the reactions was SDS/PAGE-separated and immunoblotted for NuMA1 as loading control.
Immunoblotting
Immunoblots were incubated separately with the following primary antibodies: mouse monoclonal anti-ATM (1:1000; 2C1; Novus Biologicals), rabbit polyclonal anti-pS(T)Q ATM/ATR substrates (1:1000; Cell Signaling), rabbit polyclonal anti-BRCA1 (1:350; C-20; Santa Cruz Biotechnology), mouse monoclonal anti-Cdc2 (1:1000; Cdk1; Santa Cruz Biotechnology), rabbit polyclonal anti-Cyclin B1 (1:1000; Santa Cruz Biotechnology), mouse monoclonal anti-Flag (1:1000; Sigma Aldrich), rabbit polyclonal anti-GFP (1:350; Santa Cruz Biotechnology), mouse monoclonal anti-phospho-Ser/Thr-Pro MPM-2 (1:1000; Millipore), rabbit polyclonal anti-NuMA1 (1:1500; Novus Biological), rabbit polyclonal anti-NuMA1 (1:350; H-300; Santa Cruz Biotechnology), rabbit polyclonal anti-pSer395-NuMA1 (1:1000; Cell Signaling), rabbit polyclonal anti-Poly(ADP-Ribose) (1:5000; BD Biosciences), rabbit polyclonal anti-TNKS1/2 (1:350; H-350; Santa Cruz Biotechnology), followed by horseradish peroxidase-conjugated donkey anti-rabbit or anti-mouse IgG (1:2500; Amersham). To extract total NuMA1 for anti NuMA1-Ser395 experiment, cells were collected and lysed in the following ice-cold buffer: 50 mM TRIS-HCl pH 7.4, 1 mM EDTA, 400 mM NaCl, 1% Triton X-100, 0.1% SDS, 1 mM DTT plus phosSTOP (Roche) and Complete Protease inhibitor (Roche).
Immunofluorescence and microscopy
Cells were grown on glass coverslips (Corning) pretreated with Poly-L-Lysine (Sigma Aldrich). After treatments, cells were fixed with 4% paraformaldehyde (Sigma Aldrich) for 10 min at room temperature (R.T.) and permeabilized for 10 min in PBS plus 0.25% Triton X-100. For γ-tubulin staining, the coverslips were then incubated for 15 min at −20 °C with ice-cold methanol. The coverslips were washed in PBS, blocked for 1 h with 3% BSA in PBS, and incubated for 3 h at R.T. with the mix of the following antibodies: mouse monoclonal anti-ATM (1:250; 2C1; Novus Biologicals), mouse monoclonal anti P-Ser1981-ATM (1:250; 4526; Cell Signaling), rabbit polyclonal anti-BRCA (1:150; C20; Santa Cruz Biotechnology), rabbit polyclonal anti-NuMA1 (1:150; H-300; Santa Cruz Biotechnology), rabbit polyclonal anti-TNKS1/2 (1:150; H-350; Santa Cruz Biotechnology), mouse monoclonal anti-γ-tubulin (1:1500; Sigma Aldrich), mouse monoclonal anti-α-tubulin (1:1000; Sigma Aldrich) antibodies. Secondary antibodies were conjugated to Alexa Fluor-488/-594 (1:300; Invitrogen). DNA was stained with Hoechst 33258 in PBS (10 μg/mL; Sigma) or with DRAQ5 (1 μM; Cell Signaling) for confocal microscopy. Samples were analyzed using an Axiovert 200M inverted microscope equipped with Apotome slider module (Carl Zeiss, Inc.), 63× objective oil immersion and Axiovert software (Carl Zeiss, Inc) or using a Leica TCS SP5 II confocal microscope, 63× objective NA 1.4 oil immersion (Leica Microsystems).
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank VE Avvedimento, ME Gottesman, R Baer, and Ceinge Dynamic Imaging facility. Supported by Italian Ministero dell’Università e della Ricerca -PRIN 2010–2011- and Associazione Italiana per la Ricerca sul Cancro (AIRC); L.P. is a Fondazione Italiana per la Ricerca sul Cancro (FIRC) fellowship recipient.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/27945
References
- 1.McKinnon PJ. ATM and ataxia telangiectasia. EMBO Rep. 2004;5:772–6. doi: 10.1038/sj.embor.7400210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14:197–210. doi: 10.1038/nrm3546. [DOI] [PubMed] [Google Scholar]
- 3.Humar B, Müller H, Scott RJ. Cell cycle dependent DNA break increase in ataxia telangiectasia lymphoblasts after radiation exposure. Mol Pathol. 2001;54:347–50. doi: 10.1136/mp.54.5.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armanios M. Syndromes of telomere shortening. Annu Rev Genomics Hum Genet. 2009;10:45–61. doi: 10.1146/annurev-genom-082908-150046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herzog KH, Chong MJ, Kapsetaki M, Morgan JI, McKinnon PJ. Requirement for Atm in ionizing radiation-induced cell death in the developing central nervous system. Science. 1998;280:1089–91. doi: 10.1126/science.280.5366.1089. [DOI] [PubMed] [Google Scholar]
- 6.Shen KC, Heng H, Wang Y, Lu S, Liu G, Deng CX, Brooks SC, Wang YA. ATM and p21 cooperate to suppress aneuploidy and subsequent tumor development. Cancer Res. 2005;65:8747–53. doi: 10.1158/0008-5472.CAN-05-1471. [DOI] [PubMed] [Google Scholar]
- 7.Yurov YB, Vorsanova SG, Iourov IY. GIN’n’CIN hypothesis of brain aging: deciphering the role of somatic genetic instabilities and neural aneuploidy during ontogeny. Mol Cytogenet. 2009;2:23. doi: 10.1186/1755-8166-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat Rev Genet. 2012;13:189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- 9.Ganem NJ, Pellman D. Linking abnormal mitosis to the acquisition of DNA damage. J Cell Biol. 2012;199:871–81. doi: 10.1083/jcb.201210040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang C, Tang X, Guo X, Niikura Y, Kitagawa K, Cui K, Wong ST, Fu L, Xu B. Aurora-B mediated ATM serine 1403 phosphorylation is required for mitotic ATM activation and the spindle checkpoint. Mol Cell. 2011;44:597–608. doi: 10.1016/j.molcel.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joukov V, Groen AC, Prokhorova T, Gerson R, White E, Rodriguez A, Walter JC, Livingston DM. The BRCA1/BARD1 heterodimer modulates ran-dependent mitotic spindle assembly. Cell. 2006;127:539–52. doi: 10.1016/j.cell.2006.08.053. [DOI] [PubMed] [Google Scholar]
- 12.Stolz A, Ertych N, Bastians H. Loss of the tumour-suppressor genes CHK2 and BRCA1 results in chromosomal instability. Biochem Soc Trans. 2010;38:1704–8. doi: 10.1042/BST0381704. [DOI] [PubMed] [Google Scholar]
- 13.Radulescu AE, Cleveland DW. NuMA after 30 years: the matrix revisited. Trends Cell Biol. 2010;20:214–22. doi: 10.1016/j.tcb.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silkworth WT, Cimini D. Transient defects of mitotic spindle geometry and chromosome segregation errors. Cell Div. 2012;7:19. doi: 10.1186/1747-1028-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith S, Giriat I, Schmitt A, de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 1998;282:1484–7. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- 16.Chang P, Coughlin M, Mitchison TJ. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat Cell Biol. 2005;7:1133–9. doi: 10.1038/ncb1322. [DOI] [PubMed] [Google Scholar]
- 17.Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–20. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 18.Chang W, Dynek JN, Smith S. NuMA is a major acceptor of poly(ADP-ribosyl)ation by tankyrase 1 in mitosis. Biochem J. 2005;391:177–84. doi: 10.1042/BJ20050885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang P, Coughlin M, Mitchison TJ. Interaction between Poly(ADP-ribose) and NuMA contributes to mitotic spindle pole assembly. Mol Biol Cell. 2009;20:4575–85. doi: 10.1091/mbc.E09-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boehler C, Dantzer F. PARP-3, a DNA-dependent PARP with emerging roles in double-strand break repair and mitotic progression. Cell Cycle. 2011;10:1023–4. doi: 10.4161/cc.10.7.15169. [DOI] [PubMed] [Google Scholar]
- 21.Oricchio E, Saladino C, Iacovelli S, Soddu S, Cundari E. ATM is activated by default in mitosis, localizes at centrosomes and monitors mitotic spindle integrity. Cell Cycle. 2006;5:88–92. doi: 10.4161/cc.5.1.2269. [DOI] [PubMed] [Google Scholar]
- 22.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–6. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 23.Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–62. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 24.Nousiainen M, Silljé HH, Sauer G, Nigg EA, Körner R. Phosphoproteome analysis of the human mitotic spindle. Proc Natl Acad Sci U S A. 2006;103:5391–6. doi: 10.1073/pnas.0507066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dephoure N, Zhou C, Villén J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A. 2008;105:10762–7. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quintyne NJ, Reing JE, Hoffelder DR, Gollin SM, Saunders WS. Spindle multipolarity is prevented by centrosomal clustering. Science. 2005;307:127–9. doi: 10.1126/science.1104905. [DOI] [PubMed] [Google Scholar]
- 27.Yurov YB, Iourov IY, Vorsanova SG, Liehr T, Kolotii AD, Kutsev SI, Pellestor F, Beresheva AK, Demidova IA, Kravets VS, et al. Aneuploidy and confined chromosomal mosaicism in the developing human brain. PLoS One. 2007;2:e558. doi: 10.1371/journal.pone.0000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamashita YM. Regulation of asymmetric stem cell division: spindle orientation and the centrosome. Front Biosci (Landmark Ed) 2009;14:3003–11. doi: 10.2741/3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metcalfe JA, Parkhill J, Campbell L, Stacey M, Biggs P, Byrd PJ, Taylor AM. Accelerated telomere shortening in ataxia telangiectasia. Nat Genet. 1996;13:350–3. doi: 10.1038/ng0796-350. [DOI] [PubMed] [Google Scholar]
- 30.Chi NW, Lodish HF. Tankyrase is a golgi-associated mitogen-activated protein kinase substrate that interacts with IRAP in GLUT4 vesicles. J Biol Chem. 2000;275:38437–44. doi: 10.1074/jbc.M007635200. [DOI] [PubMed] [Google Scholar]
- 31.McCabe N, Cerone MA, Ohishi T, Seimiya H, Lord CJ, Ashworth A. Targeting Tankyrase 1 as a therapeutic strategy for BRCA-associated cancer. Oncogene. 2009;28:1465–70. doi: 10.1038/onc.2008.483. [DOI] [PubMed] [Google Scholar]
- 32.Gilardini Montani MS, Prodosmo A, Stagni V, Merli D, Monteonofrio L, Gatti V, Gentileschi MP, Barilà D, Soddu S. ATM-depletion in breast cancer cells confers sensitivity to PARP inhibition. J Exp Clin Cancer Res. 2013;32:95. doi: 10.1186/1756-9966-32-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Visconti R, Palazzo L, Della Monica R, Grieco D. Fcp1-dependent dephosphorylation is required for M-phase-promoting factor inactivation at mitosis exit. Nat Commun. 2012;3:894. doi: 10.1038/ncomms1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Visconti R, Palazzo L, Grieco D. Requirement for proteolysis in spindle assembly checkpoint silencing. Cell Cycle. 2010;9:564–9. doi: 10.4161/cc.9.3.10581. [DOI] [PubMed] [Google Scholar]
- 35.Choudhury AD, Xu H, Baer R. Ubiquitination and proteasomal degradation of the BRCA1 tumor suppressor is regulated during cell cycle progression. J Biol Chem. 2004;279:33909–18. doi: 10.1074/jbc.M403646200. [DOI] [PubMed] [Google Scholar]
- 36.Trenz K, Smith E, Smith S, Costanzo V. ATM and ATR promote Mre11 dependent restart of collapsed replication forks and prevent accumulation of DNA breaks. EMBO J. 2006;25:1764–74. doi: 10.1038/sj.emboj.7601045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.