Abstract
Microtubules are structural components of the cell cytoskeleton and key factors for mitosis and ciliogenesis in eukaryotes. The regulation of MT dynamics requires non-motor MAPs. We previously showed that, in human cells in culture, MAP9 (also named ASAP) is involved in MT dynamics and is essential for mitotic spindle formation and mitosis progression. Indeed, misexpression of MAP9 leads to severe mitotic defects and cell death. Here, we investigated the in vivo role of map9 during zebrafish development. Map9 is expressed mainly as a maternal gene. Within cells, Map9 is associated with the MT network of the mitotic spindle and with centrosomes. Morpholino-mediated depletion of map9 leads to early development arrest before completion of epiboly. Map9 localizes to the MT array of the YSL. This MT network is destroyed in Map9-depleted embryos, and injection of anti-map9 morpholinos directly in the nascent YSL leads to arrest of epiboly/gastrulation. Finally, map9 knockdown deregulates the expression of genes involved in endodermal differentiation, dorso–ventral and left–right patterning, and other MT-based functions. At low morpholino doses, the surviving embryos show dramatic developmental defects, spindle and mitotic defects, and increased apoptosis. Our findings suggest that map9 is a crucial factor in early zebrafish development by regulating different MT-based processes.
Keywords: microtubule-associated protein, microtubules, zebrafish, knockdown, mitosis, yolk syncytial layer, epiboly, gastrulation, development
Introduction
MTs are fibrillar structures that are present in all eukaryotic cells. The centrosome is the primary MT-organizing center of animal cells and is involved in regulating cell motility and polarity in interphase as well as in organizing the spindle poles during mitosis. The centriole is the core centrosomal component. During development, and also in cultured cells, centrioles are involved in the formation of the mitotic spindle poles and the basal body of primary cilia. Cell divisions require coordination between chromosome segregation by the mitotic spindle (a bipolar MT-based structure) and cell cleavage by the cytokinetic apparatus. Spindle assembly and chromosome segregation depend on many factors1,2 that regulate MT dynamics,3-5 including also non-motor MAPs. In fish, the YSL is an extraembryonic tissue that forms at the surface of the yolk and that drives epiboly, during which the blastoderm spreads over the yolk cell toward the vegetal pole.6-8 YSL movements rely heavily on a specific and dense MT network, and during epiboly/gastrulation, YSL emits Nodal/TGFβ signals that specify the endoderm and mesoderm cell fate.6
We have recently characterized a novel human spindle MAP named ASAP or MAP9.9 MAP9 overexpression induces aberrant spindles in mitosis, and its depletion by RNA interference results in severe mitotic defects that lead to aneuploidy and/or cell death. These findings indicate that MAP9 has a crucial role in the organization of the bipolar mitotic spindle and in mitotic progression. MAP9 is phosphorylated by the mitotic kinases Aurora A10 and PLK111 to regulate its role in bipolar spindle assembly and centrosome integrity. TP53 is a key factor that controls the response to DNA damage, and we have shown that in response to DNA damage, MAP9 transiently accumulates and interacts with and stabilizes TP53.12
As MAP9 is thought to be involved in various aspects of MT dynamics, we decided to investigate, in vivo, the role of map9 during embryogenesis using zebrafish as a model. In addition to the aforementioned works to support this hypothesis, large-scale studies identified MAP9 and PLK1 as ciliopathy candidate genes,13-16 and Plk1 inhibition induces growth defects in developing zebrafish embryos.17,18
Here we report that map9 is expressed during early zebrafish development and localizes to the MTs of the mitotic spindle and to centrosomes. Inhibition of map9 expression using oligonucleotide morpholinos blocks zebrafish development during the early steps of embryogenesis, leading to embryo death or to dramatic developmental defects. Morphant embryos display mitotic defects and increased apoptosis. Injection of anti-map9 morpholino oligomers is associated with reduction of the MT network of the YSL, and injection of morpholino directly in the nascent YSL of early embryos blocks epiboly/gastrulation. Finally, map9 knockdown deregulates the expression of a number of genes involved mainly in the nodal pathway.
These in vivo results confirm the previous in vitro observations about ASAP/MAP9’s role in regulating MT dynamics, and suggest that Map9 is a key factor during the early steps of zebrafish development.
Results
Zebrafish map9 is expressed early during embryo development
To obtain a full-length zebrafish map9 cDNA clone, we screened the IMAGE library using the sequence of the predicted ENSDART00000133937 transcript (Ensembl), which encodes a 678-aa protein, and the partial sequence BC124374 (GenBank), which was identified as zebrafish map9. The clone IMAGp998O0412053Q (IMAGE ID 5413083) was ordered and fully sequenced. This 2520-bp cDNA sequence (GenBank accession number JQ768417) includes a 2037 nucleotide-long ORF that encodes a 678-aa protein with very few amino acid changes compared with the ENSDART00000133937 sequence. We also cloned and sequenced the map9 cDNA from the zebrafish golden mutant. The predicted protein sequence was identical to the one encoded by the JQ768417 clone. Protein sequence comparisons show that the zebrafish Map9 protein is ~50% similar (~27% protein identity) to the human and mouse MAP9 proteins.19
A previous microarray analysis20 established that, in zebrafish embryos, the level of map9 mRNA increases between 0 and 3 hpf (~1000-cell stage21), then decreases quickly until 24 hpf (pharyngula, prim-5 stage) and remains stable up to the swimming larva stage (5 d post-fertilization). We confirmed these findings by quantifying map9 mRNA expression in unfertilized eggs (0 hpf) and in embryos up to 24 hpf by qPCR (with normalization to β-actin, Fig. 1A). Specifically, maternal map9 mRNA was high in unfertilized eggs (0 hpf). Map9 level increased between 1 and 2 hpf (64-cell stage) and then gradually decreased and remained stable from 4 hpf (sphere period) to 24 hpf. This observation suggests that, like for many maternal gene transcripts,22 high map9 level is required in the early steps of embryogenesis that are characterized by high mitosis rate, while a basal level is then sufficient for normal cell function.
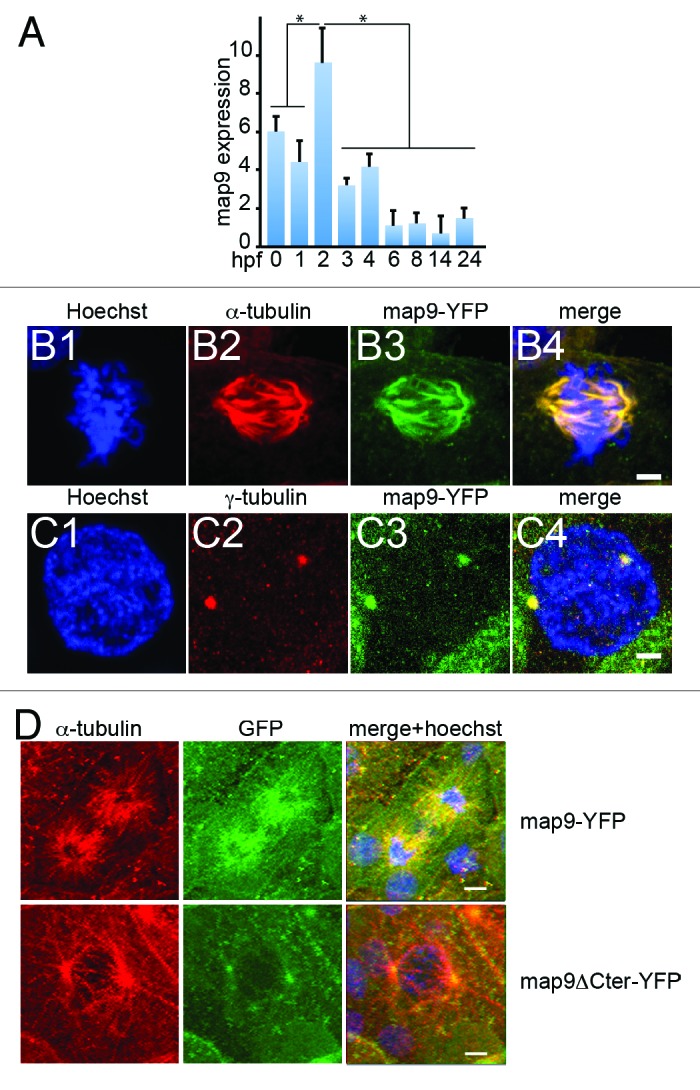
Figure 1. Expression of map9 in early zebrafish embryos. (A) Relative quantification of map9 mRNA by qPCR in 0 hpf (unfertilized eggs) to 24 hpf embryos. Data are the mean ± SD from 3 independent experiments, with n = 25–35 embryos per time point (*P < 0.005 by Student t test). (Band C) Subcellular localization of Map9 during mitosis in 24 hpf embryos following expression of map9-YFP. Two hundred pg of map9-YFP RNA was injected at the 1-cell stage, embryos fixed at 24 hpf, and stained with an anti-α-tubulin antibody (red) and Hoechst 33258. Confocal microscopy images show that Map9-YFP (green) co-localizes with α-tubulin on the microtubules of the mitotic spindle (B). In interphase cells (C), Map9-YFP co-localizes with γ-tubulin (red) at centrosomes. (Scale bars, 10 μm). (D) Loss of the fiber-like distribution of Map9 lacking its C-terminal MAP domain in mitotic cells. Map9-YFP or map9ΔCter-YFP RNA (200 pg) was injected in 1-cell stage embryos. Embryos were fixed at 24 hpf and stained with anti-α-tubulin, anti-GFP antibodies, and Hoechst 33258. Confocal microscopy images show that Map9-YFP but not Map9ΔCter-YFP co-localizes with α-tubulin on the microtubules of the mitotic spindle. (Scale bars, 10 μm).
Map9 is expressed in the nervous system and is associated with the mitotic spindle
In situ hybridization analysis of zebrafish embryos23 showed that map9 is expressed in the ventral spinal cord (from the 1–4 somite to the Prim-5 stage, i.e., from 10–24 hpf), the diencephalon, the olfactory placode and the tegmentum at the Prim-5 stage, but it was not detected before 10 hpf or after 24 hpf. We tried to study the expression pattern of map9 on whole embryos during epiboly (before 10 hpf) and in the adult, but we also did not get a significant labeling using in situ hybridization.
To follow Map9 expression in individual cells, we injected in vitro transcribed map9-YFP RNA in 1–2 cell stage embryos. Individual mitoses were observed at 24 hpf by confocal microscopy (Fig. 1B and C). Although Map9 localization to cytoplasmic MTs was not obvious over background, Map9 co-localized with α-tubulin in the MT network of the mitotic spindle (Fig. 1B), as we previously reported in other species.9,10,19 Specifically, zebrafish Map9 co-localized with MTs along the entire length of the mitotic spindle, from the centrosome to the distal ends. Like in other species, zebrafish Map9 was also localized to centrosomes, as revealed by co-localization with γ-tubulin (Fig. 1C). Although this experiment relies on an overexpression procedure that may produce overexpression phenotypes, it provides valuable information about the localization of zebrafish Map9 to the mitotic spindle, as we previously observed in other species using the same protocol or antibody detection.9,19 The MAP domain that spans the C-terminal part of the protein (amino acids 451–678 in zebrafish) is well-conserved in distantly related species (~38% identity and >65% similarity between fish and human), suggesting a conserved function. Also, the MIT-like domain19 (microtubule interacting and trafficking domain) is conserved in fish (amino acids 455–533) with most of its characteristic features. In addition, experiments using a YFP-ΔCterMap9 construct lacking the MAP domain showed a loss of fiber-like distribution of Map9 in mitotic cells, suggesting that this domain is required for zebrafish Map9 to localize to microtubules, as we previously showed in human9 (Fig. 1D). The nuclear localization signals (NLS) found in the human MAP domain are not conserved in fish. However another bipartite NLS was found in the same region using the pSORTII software (amino acids 570–587 and 598–614).
Map9 knockdown or overexpression leads to major developmental defects
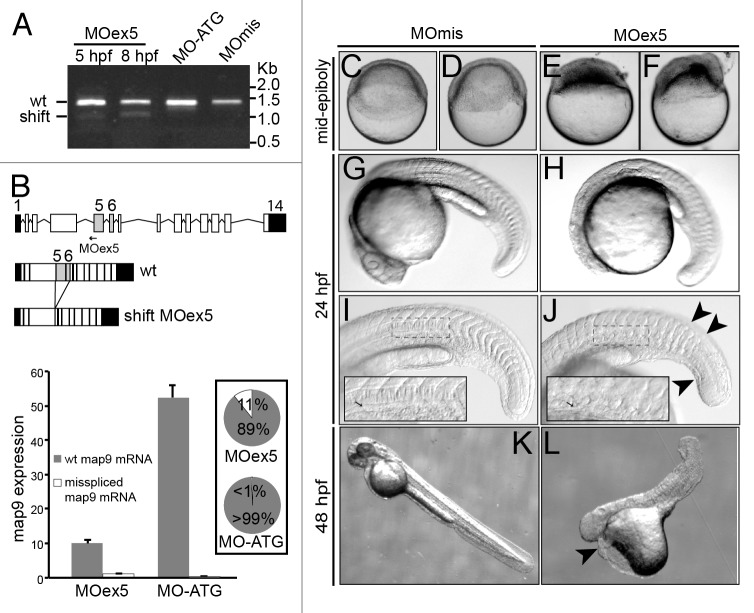
We previously showed9 that MAP9 expression must be tightly regulated in normal cells, as MAP9 depletion or overexpression leads to severe mitotic defects. To understand the role of Map9 in zebrafish development, we knocked map9 down using morpholino oligomers (MO) that block map9 splicing (MOex5) or translation (MO-ATG). These 2 MOs were injected separately into the yolk of 1–2-cell stage embryos. Although MO-ATG blocked the expression of maternal and zygotic map9 mRNAs, comparable effects were obtained with both MOs. We tested a range of concentrations (0.25, 0.50, 0.75, and 1 pmol per embryo) and found that most of the injected embryos died before 24 hpf (70% following injection of 0.25 pmol and 100% with 0.75–1.0 pmol MOs). Conversely, non-injected embryos or embryos injected with the control MOmis MO (same sequence than MOex5 except 5 mismatches) were relatively unaffected (death rate = 7% and 18%, respectively). In the rest of the study we used only MOex5. To ascertain the specificity of MOex5, we assessed map9 expression by RT-PCR using RNA from 5 and/or 8 hpf embryos injected with MOex5, MO-ATG, or control MOmis and the map9-specific primers ZF1/ZF8. A lower map9 band was detected on agarose gel in MOex5 morphants at 5 hpf, suggesting that the MO could start blocking map9 RNA splicing much earlier, but not in MO-ATG or MOmis-injected embryos (Fig. 2A). The ratio between the 2 bands increased up to 8 hpf and then remained stable. Partial blocking by MOs are frequent,24,25 and we have previously shown that mis-expression of MAP9 led to drastic effects.9 Sequencing of these 2 fragments revealed that the upper band corresponded to wild-type map9 (newly transcribed zygotic mRNAs and maternal mRNAs left) and the lower band to a mis-spliced form in which exons 5 and 6 were deleted (Fig. 2B), leading to the loss of 292 bp in the mRNA and 97 aa in the protein sequence. However, to evaluate the efficacy of MOex5, we quantified the 2 map9 mRNA species by qPCR using specific primer pairs, in 8 hpf MOex5 morphants (splicing blocked) and MO-ATG morphants (translation blocked) as a control (Fig. 2B, bottom panel). In MOex5 embryos, the wt map9 mRNA is only 19% of that in MO-ATG control embryos (similar results were obtained with MOmis embryos, not shown) and the misspliced form represents 11% of total map9 mRNA vs. background level in MO-ATG embryos. These results indicate that MOex5 injection leads to a decrease >80% of map9 transcripts, probably as a result of abnormal misspliced mRNAs that are degraded.
Figure 2. MO-mediated depletion of map9 leads to early defects and/or embryo death and growth failure. (A) Injection of splice-blocking MOex5 leads to the production of a mis-spliced map9 mRNA. RT-PCR was performed using map9 specific primers and RNA from embryos injected at the 1-cell stage with MOex5 (5 and 8 hpf), translation-blocking MO-ATG (8 hpf), or the control MOmis (8 hpf). PCR products were separated on 0.8% agarose gels. MOex5 PCR products show 2 bands that differ by ~300 bp in size. (B) Top: subcloning and sequencing of these 2 fragments revealed that the upper band corresponds to wild-type (wt) map 9, whereas the lower band corresponds to an mRNA in which exon 5 and 6 are deleted. mRNA from MO-ATG and MOmis-injected embryos show only wt map9. The position of MOex5 is indicated by an arrow on the acceptor splice junction of exon 5. Exons 5 and 6 are shaded in gray, and black boxes represent untranslated regions. Bottom: measure by qPCR of the expression of the 2 map9 mRNA forms in 8 hpf MOex5 and MO-ATG morphants. In MOex5 morphants the expression of map9 is ~5 times lower than in MO-ATG morphants, and the misspliced forms represents 11% of map9 mRNA vs. background level < 1% in MO-ATG morphants (inset). (C–F) One-cell stage zebrafish embryos were injected with 1 pmol of anti-map9 MOex5 or control MOmis and imaged at mid-epiboly. Growth and epiboly in MOex5 morphants are arrested very early. (G–L) Embryos at 24 and 48 hpf after injection of 0.25 pmol of the 2 MOs. Map9 morphants show complex malformations, including underdeveloped nervous system, absence of the eyes, abnormal yolk, notochord (I and J, arrow in the insets), somites and tail (J, arrowheads), and pericardial edema (L, arrowhead). In (I and J), the insets are enlargements of the dotted boxes.
Based on these results, we decided to use 1 pmol of MOs to detect early defects (<10 hpf, Fig. 2 C–F) and 0.25 pmol of MOs for analysis at 24 and 48 hpf (Fig. 2G–L). Following injection of 1 pmol MOex5, embryo development was blocked at mid-epiboly (5–6 hpf), and most embryos died around 9–10 hpf, because they could not complete epiboly. Few embryos injected with 0.25 pmol MOex5 survived up to 24 hpf (27%) and 48 hpf (5%), but they showed a range of complex defects, including possible embryo axis defects, large yolk, short body size, curved tail, flat head with small brain often associated with eye absence or almost no brain, abnormal somites, pericardial edema, and other features that were reminiscent of gastrulation defects (Fig. 2G–L). Moreover, in MOex5 morphants the notochord was abnormal (Fig. 2J, arrow in the inset; compare with Fig. 2I) and the chevron structure of somites disorganized. Similar defects were observed in embryos injected with 1 pmol or 0.25 pmol of MO-ATG (data not shown). Although these morphants do not represent the main effect of map9 knockdown (arrest of development at mid-epiboly), they provide information about the multiple effects of map9 misexpression.
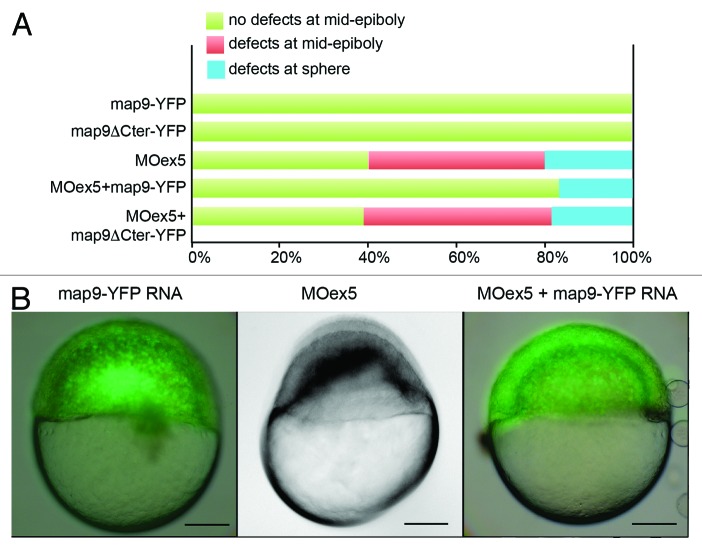
To ascertain the specificity of MOex5 effect on zebrafish embryo development, we co-injected map9-YFP mRNA and MOex5. However, map9-YFP (200 pg) only partially rescued the MOex5 phenotypes (Fig. 3). About 40% of MOex5-injected embryos showed defects at mid-epiboly (red bar) and died at ~9–10 hpf and 20% were blocked at the sphere stage (blue bars). Conversely, about 80% of embryos co-injected with MOex5 and map9-YFP RNA developed normally up to mid-epiboly (green bars), but died between 9–10 and 24 hpf (similar results were obtained by co-injecting MOex5 and map9 RNA, not shown). The percentage of embryos blocked at the sphere stage was slightly reduced. Control map9-YFP embryos developed normally up to mid-epiboly and also to later stages, and co-injection of map9ΔCter-YFP RNA (lacking the MAP domain) did not rescue the morphant phenotype. Injection of lower amounts of map9-YFP RNA (45, 90 pg) did not rescue morphant phenotype (not shown), and injection of higher amounts of map9-YFP RNA (400 pg) resulted in overexpression phenotypes, as shown in Figure S1 and in Saffin et al.,9 and thus affected the rescue of the MOex5 phenotypes. This experiment shows that only the initiation of epiboly could be rescued but not the embryo lethality phenotype, further confirming that the Map9 level in the cells should be tightly regulated for the development to proceed correctly.
Figure 3.Map9-YFP RNA can partially rescue the phenotypes of MOex5 morphants. (A) Embryos were injected at the 1-cell stage with 1 pmol MOex5, 200 pg map9-YFP RNA or map9ΔCter-YFP RNA, or co-injected with MOex5 and either RNA. Map9-YFP- and map9ΔCter-YFP-injected embryos developed normally up to mid-epiboly and later stages (green). Forty % of MOex5 morphants showed defects at mid-epiboly (red), 20% were blocked at the sphere stage (blue), and all of them died at 9–10 hpf. Following co-injection of MOex5 and map9-YFP RNA, about 80% of embryos developed normally up to mid-epiboly but died before 24 hpf, whereas co-injection with map9ΔCter-YFP did not rescue the morphant phenotype (n = 64 for map9ΔCter-YFP-injected embryos; n = 54 for embryos co-injected with MOex5 and map9ΔCter-YFP RNA). (B) Representative images of embryos injected with map9-YFP RNA (at mid-epiboly, n = 102), with MOex5 (showing developmental delay and defects before mid-epiboly, n = 85), or with both MOex5 and map9-YFP RNA (showing partial rescue of the phenotype at mid-epiboly, n = 153). Scale bars, 200 μm.
As overexpression of MAP9 in cultured human cells leads to severe mitotic defects and cell death,9 we overexpressed Map9 also in zebrafish embryos by injecting in vitro transcribed map9-YFP mRNA in 1-cell embryos. Abnormal phenotypes (Fig. S1) were observed following injection of 400 pg map9-YFP mRNA with a death rate of ~10% at 24 hpf (Fig. S1C), and similar results were obtained by injecting map9 mRNA (not shown). About 40% of embryos displayed complex malformations, such as brain and tail underdevelopment, abnormal yolk, and, in some cases, severe phenotypes (Fig. S1B) caused by abnormal gastrulation, similar to what was observed in MOex5 morphants.
Map9 inhibition induces apoptosis/cell death
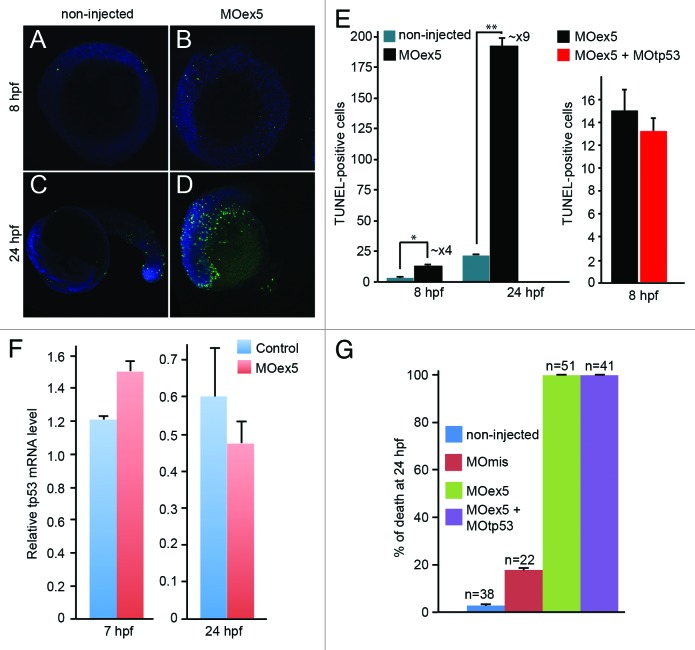
As depletion of human MAP9 in cultured cells is associated with high cell death rate,9 we quantified apoptosis in zebrafish embryos at 8 hpf (75% epiboly) and 24 hpf (Prim-5 stage) by TUNEL assay (Fig. 4A–D). Due to the high death rate of morphants, 1-cell embryos were injected with 1 pmol MOex5 and observed at 8 hpf, or with 0.25 pmol MOex5 and observed at 24 hpf (Fig. 4A–D). Although apoptosis is instrumental for proper embryo development,26 we detected a 4-fold increase in TUNEL-positive cells at 8 hpf and a ~9-fold increase at 24 hpf (Fig. 4E, left panel) in map9 morphants in comparison to non-injected controls. These results indicate that developmental defects in map9 morphants are correlated with increased apoptosis. We also verified that inhibition of tp53 was not able to rescue apoptosis in MOex5 morphants (Fig. 4E, right panel). To rule out the possibility that map9 depletion and/or morpholino injection might induce tp53 and, therefore, tp53-dependent cell death, we measured the relative tp53 mRNA level by qPCR using RNA from 7 and 24 hpf control embryos and morphants, and found only a slight variation of its expression (Fig. 4F). By comparison, MO depletion of plk1 or Aurora A, 2 mitotic kinases that phosphorylate Map9, induces tp53-dependent cell death by enhancing tp53 expression by 7- to 10-fold.27,28 In addition, depletion of tp53 by co-injection of MO-tp53 with MOex5 did not rescue the embryo mortality observed in MOex5-injected embryos (Fig. 4G).
Figure 4. Morpholino-mediated depletion of map9 leads to increased apoptosis. (A–D) Zebrafish embryos were injected, or not (control), at the 1-cell stage with 1 pmol of MOex5 for observation at 8 hpf, or with 0.25 pmol MOex5 for observation at 24 hpf. TUNEL-positive cells (green) were counted in injected (B and D) and control embryos (A and C) (n = 10/assay). (E) Left panel, number of TUNEL-positive cells in the control and MOex5-injected embryos depicted in (A–D), n = 5 embryos per assay, total cell count ~1070 (*P < 0.005, **P < 0.02); right panel, tp53 inhibition does not rescue apoptosis induced by MOex5. Number of TUNEL-positive cells in MOex5- and MOex5 + MOtp53-injected embryos (n = 20 embryos per assay, total cell count ~564). (F) Map9 depletion does not induce tp53 expression. Embryos were injected at the 1-cell stage with 1 pmol of MOex5. RNA from 7 hpf map9 morphants and control embryos was used to quantify tp53 gene expression by RT-qPCR. Expression data were normalized to β-actin. Experiments were made in quadruplicate, n = 42 control embryos, and 50 MOex5 morphants at 7 hpf, and n = 39/44 embryos at 24 hpf. (G) Co-injection of MOex5 (1 pmol) and MOtp53 (0.5 pmol) does not rescue MOex5 effects. Embryo mortality was not rescued after tp53 depletion by morpholino. Embryo mortality after injection of a control MO (MOmis) was not significantly different compared with non-injected embryos. This experiment was performed in duplicate.
Map9 inhibition disrupts mitosis
As knockdown of MAP9 by RNA interference in human cells leads to incomplete mitosis9 and, consequently, to the death of more than 40% of cells, we asked whether the higher apoptosis rate observed in MOex5 morphants in comparison to controls could be linked to mitotic defects.
To test this hypothesis we first counted the mitotic cells in embryos fixed at 5 and 8 hpf after injection of MOex5 or MOmis (control) and immunostained with an anti-phosphohistone H3 antibody (pH3, a mitosis marker)29 (Fig. 5A–C). The number of pH3-positive cells was reduced by 35–40% in MOex5-injected embryos, suggesting an early mitotic blockade. Moreover, in MOex5 morphants, cells displayed mitotic infidelity (such as irregular spindle assembly, hypercondensed chromosomes, irregular microtubule spindles and metaphase plates, as well as congression defects) (Fig. 5D–G) and higher percentage of aberrant mitoses in comparison to cells from MOmis-injected control embryos (44% vs. ~11%, respectively). Similarly, a strong reduction of cells in prophase (~1% vs. 25%) and a diminution of cells in anaphase/telophase (~25% vs. 35%, Fig. 5H) were observed in MOex5 morphants in comparison to control embryos. These results are in agreement with previous observation in human cells in culture showing that MAP9 inhibition led to abnormal spindles and nuclei as well as chromosome congression and segregation defects,9 and indicate that Map9 is critical for mitosis regulation during zebrafish embryo development.
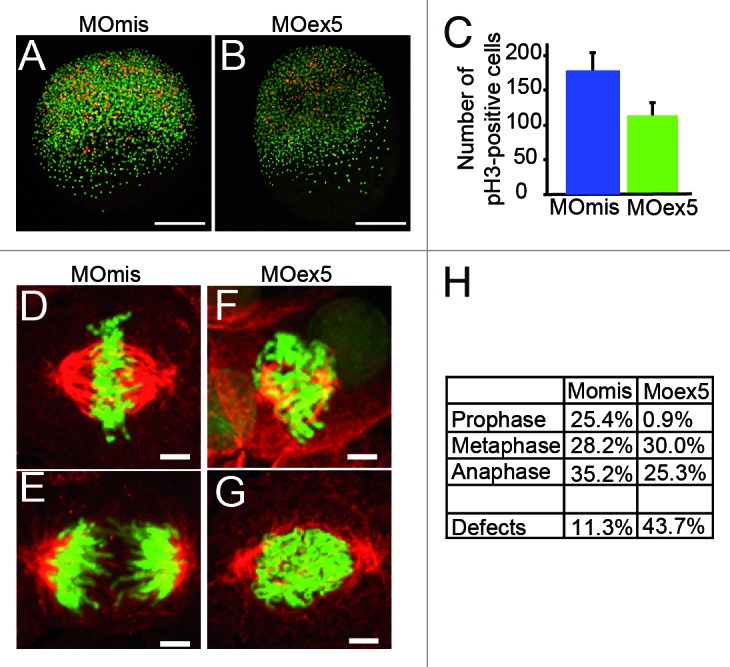
Figure 5. Morpholino-mediated depletion of map9 leads to a reduction in the number of mitotic cells and to mitotic defects. (AandB) Embryos were injected at the 1-cell stage with 1 pmol MOex5 or MOmis (control) and stained with the anti-phosphorylated histone H3 (pH3) antibody (red). DNA was labeled with Hoechst 33258 (blue). Scale bars, 200 μm. (C) pH3-positive mitotic cells were counted in injected embryos at 5 hpf (n = 10/group). In MOex5 morphants the number of mitotic cells was reduced by ~40% in comparison to controls. Similar results were obtained in 8 hpf embryos (not shown). (D–G) Embryos were injected at the 1-cell stage with 1 pmol MOmis (control) (D and E) or MOex5 (F and G) and mitotic cells were imaged in 8 hpf embryos by confocal microscopy (scale bars, 10 μm). DNA was labeled with Hoechst (green) and MTs of the mitotic spindles with an anti-α-tubulin antibody (red). (H) Percentage of cells at different phases of mitosis. Mitotic cells were scored in map9 morphants and control embryos (n = 10 embryos/assay).
Map9 is required for YSL function, epiboly and gastrulation
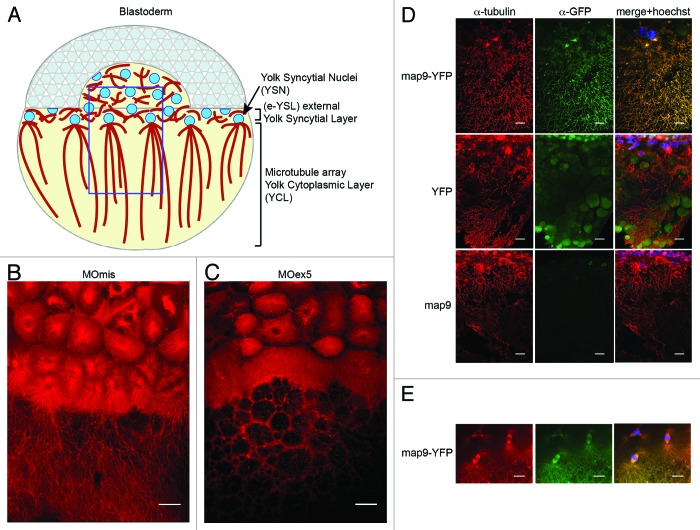
Based on the findings that map9 morphants display gastrulation/epiboly defects (Fig. 2), and that mammalian MAP9 binds to MTs and is essential for the correct formation of bipolar mitotic spindles and mitosis completion,9 we asked whether map9 could be involved in YSL dynamics.30 Indeed, just before the midblastula transition, the zebrafish MT network is necessary for the fusion of the marginal blastomeres with the yolk to form the YSL that will then drive epiboly and gastrulation.31,32 During epiboly, YSL nuclei, followed by the blastoderm, spread toward the vegetal pole31,33 guided by a MT array oriented along the animal–vegetal axis (Fig. 6A).7,31,32
Figure 6. Microtubules in the YSL are disorganized in map9 morphants. (A) Schematic representation of the organization of the YSL and yolk cell MT array in early zebrafish embryos (adapted from ref. 43). The portion boxed in blue represents the region analyzed in (B– D). (B andC) Embryos were injected at the 1-cell stage with 1 pmol of control MOmis (B) or MOex5 (C) and imaged by confocal microscopy at ~4.3 hpf (beginning of epiboly). The MT spindles/array are dense and clearly identified in MOmis-injected embryos, whereas they are depleted in MOex5 morphants. (D) Map9 localizes on the MT network of the YSL. Map9-YFP, YFP, and map9 RNA (200 pg) were injected separately in 1-cell-stage embryos and imaged at 4.3 hpf. The MT network is labeled with an anti-α-tubulin antibody (red), and Map9-YFP is labeled with an anti-GFP antibody. Nuclei are stained with Hoechst 33258. (E) Map9-YFP RNA was injected in 1-cell stage embryos. Map9 colocalizes with α-tubulin on the mitotic spindles of YSL nuclei, (scale bars, 20 μm).
First, we examined the MT cytoskeleton in MOex5 morphants and MOmis-injected controls at ~4.3 hpf (~30% epiboly) by using an antibody against α-tubulin (red; Fig. 6B and C) and confocal microscopy. At this stage, control embryos displayed dense MT spindles/arrays that expanded toward the vegetal pole (Fig. 6B). In MOex5 morphants, MT spindles were often absent in YSL, and the yolk MT array was not formed properly. These defects are reminiscent of the phenotype (defective yolk cell MTs) of zebrafish embryos in which eomesa, a T-box transcription factor involved in dorsal–ventral patterning, epiboly, and endoderm specification, is mutated.34 Second, we showed that Map9 is associated to the MT network of the YSL (Fig. 6D) as well as to the MTs of the mitotic spindles of YSL nuclei (Fig. 6E). As a control, injection of map9ΔCter-YFP RNA lacking the MAP domain led to the loss of localization of Map9 on the mitotic spindle (Fig. 1D). Map9 localization on the MT network of the YSL and co-localization with α-tubulin suggest that these MT defects in the YSL are Map9-specific.
Then, we directly injected MOex5 or MOmis (control) together with dextran-rhodamine in the YSL35 at the beginning of its formation (~23/4-3 hpf, 512–1 k cell stage30) and followed the injected embryos up to the end of gastrulation (~9 hpf, 80% epiboly). Control embryos developed normally up to 96 hpf, whereas the few MOex5-injected embryos that survived up to 10 hpf (~20%) were abnormal and died before 24 hpf (not shown). MOex5 morphants apparently developed normally up to 30% epiboly (~4.7 hpf), without obvious differences with controls (Fig. 7A–F), but could not reach 80% epiboly (Fig. 7G–L). At this stage, epiboly was stopped, the blastoderm tended to separate from the yolk, and embryos died. This phenotype was mostly caused by defects of the YSL MT network that, as a consequence, could not drive epiboly (i.e., the extension of the blastoderm toward the vegetal pole). Previous studies reported similar phenotypes resulting from the premature constriction of the actin–myosin ring of the YSL.36 We tried to rescue this phenotype by co-injecting either map9-YFP or map9 mRNA in the YSL, but these experiments were unsuccessful (not shown). Injection of MOex5 in the yolk instead of the YSL did not have any effect (not shown). To ascertain that the observed effects are YSL-specific, we also injected MOex5 or MOmis together with a fluorescent dye in random groups of cells in 64-cell stage embryos (Fig. S2). Differently from what observed following injection in 1-cell stage embryos, in which all daughter cells are affected, injection of MOex5 in 64-cell stage embryos did not impair epiboly, gastrulation, and normal embryo development. However, while cells in which only the dye was injected contributed to the 24 hpf embryo, cells in which MOex5 and the dye were injected did not. This indicates that map9 knocking down in individual cells of 64-cell stage embryos only leads to the death of the injected cells. This is an excellent illustration of the vertebrate developmental plasticity, and proves that in the embryo, map9 activity is mainly cell-autonomous.
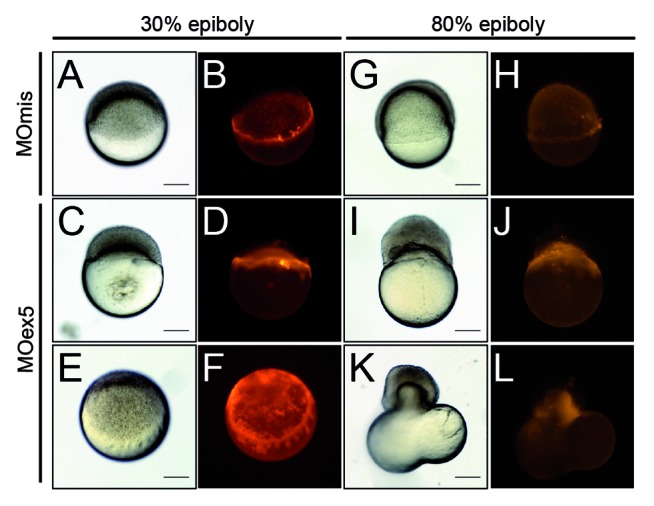
Figure 7. Inhibition of map9 expression in the YSL blocks epiboly. (A–L) At the beginning of YSL formation (23/4-3 hpf), 1 pmol of control MOmis or MOex5 and dextran-rhodamine (red) were co-injected in the YSL. Embryos were observed at 42/3 hpf (30% epiboly) and at ~8 hpf (80% epiboly). Scale bars, 200 μm.
Altogether these findings suggest that map9 is required for 2 of the main processes of zebrafish development, i.e., for individual cell divisions and for YSL to drive epiboly and gastrulation. These processes rely heavily on the YSL MT network.
Map9 knockdown perturbs different signaling pathways
Finally, to gain insight into the effects of map9 knockdown on gene expression, we measured by qRT-PCR, at mid-epiboly, the relative expression of a number of genes involved in the main pathways of early zebrafish development, knowing that changes in gene expression might be indirect effects of map9 inhibition by perturbating multiple regulation pathways.
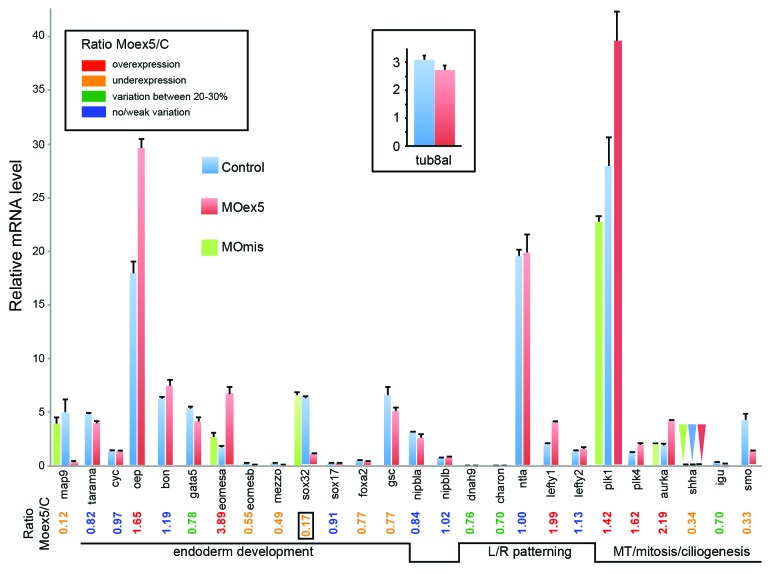
RNA was prepared from MOex5 and MOmis (controls) morphants at 7 hpf (50% epiboly, i.e., before the onset of embryo death at 9–10 hpf). As already described in Figure 2, map9 expression (Fig. 8) was strongly reduced (×0.12) in morphants, as a result of a splicing block and/or low stability of the mis-spliced zygotic mRNA, or of a feedback loop regulation. First, we examined the expression of genes that are involved in endoderm development.37 Oep and eomesa were overexpressed (×1.7 and ×3.9, respectively); eomesb, mezzo (og9x), and sox32 (or casanova, cas) were downregulated (×0.6, 0.5, <0.2, respectively), whereas gata 5 (faust) and foxa2 displayed variations comprised between 20 and 30%. Cyc (or ndr2 for nodal-related 2), tarama (acvr1b or alk4), ntla, bon (or mixer), and sox17 did not show any variation in comparison to control embryos (Fig. 8). Surprisingly, the expression of sox17 and foxa2 were not or were only slightly affected, despite the fact that sox32 is a direct transcriptional activator of these 2 genes.38 It is therefore possible that the residual sox32 activity is sufficient to activate sox17 and foxa2, or that at this stage sox17 and foxa2 expression is not sox32-dependent.
Figure 8. Depletion of map9 deregulates the expression of genes involved in different signaling pathways. Embryos were injected at the 1-cell stage with 1 pmol of MOex5 or MOmis (controls). RNA from 7 hpf map9 morphants and control embryos was used to quantify gene expression by RT-qPCR. Expression data were normalized to β-actin. Results are presented as the relative quantification of gene expression in MOex5 morphants and control embryos (C). For each gene, the morphant/control ratio is indicated below the graph, with the lowest value surrounded by a square. Inset, relative quantification of tuba8l, a maternal and miR-430 target gene whose expression during the early stages of development relies on miR-430 expression. Experiments were made in quadruplicate; n = 42 control embryos and 50 MOex5 morphants.
The expression of the mesendodermal marker and organizer-specific gene gsc39,40 was not affected. We also examined the expression of genes involved in LR patterning, which relies mainly on the function of the KV.41 Although the KV is formed later during segmentation (~112/3 hpf), the expression of dnah9 (or lrdr1 for left-right dynein-related1), which is responsible for the motility of monocilia in the KV and the inhibition of which hampers LR specification,41 was moderately downregulated (25–30%). Lefty2, another nodal-induced gene, which is expressed in the left LPM of the heart region, was stable, whereas lefty1, another gene involved in LR patterning was overexpressed (×1.99). Charon (Cerberus-related protein), which inhibits the expression of southpaw (spaw) in the right LPM,42 was downregulated (25–30%) in MOex5 morphants in comparison to control MOmis-injected embryos (Fig. 8). As Muto et al.43 recently described mitosis progression defects in nipbl-deficient (by MO) zebrafish embryos, we also checked the expression of nipbla and b and found that they were not affected by map9 knockdown.
We then measured the expression of a few genes involved in MT dynamics during mitosis and ciliogenesis. Shha and smo were downregulated (×0.3), plk1 and plk4 were upregulated (×1.4 and ×1.6, respectively), whereas igu showed a moderate decrease of about 30% (Fig. 8). As a supplementary control, the measure of the expression of 7 representative genes in MOmis-injected embryos revealed no significant variation with respect to non-injected embryos (Fig. 8).
In addition, since all gene expression levels were normalized to β-actin, we verified that the relative decrease of gene expression such as sox32 or smo was not the result of a lack of degradation of maternal β-actin transcripts in morphants. Comparison of the qPCR curves (Cp) between morphants and controls indicated that β-actin expression did not change between morphants and controls (not shown). Also, we checked that the apparent increase of gene expression was not due to the repression of miR-430 expression in morphants. Indeed, miR-430 is expressed at the onset of zygotic transcription and promotes clearance of maternal mRNAs. Thus, in mutants defective for miR-430, the mRNAs that are miR-430 targets are not degraded and appear overexpressed with respect to controls.44-46 By screening the Targetscan database (http://www.targetscan.org/fish_62/), we found that only plk4 is a potential miR-430 target, and we show here that it displays a significant increase in morphants (~×1.62). However, eomesa (×3.9) and aurka (×2.2) are not miR-430 targets, even considering weak sites (Targetscan). Furthermore, rescue experiments of mutants defective for miR-430 show that the expression of these genes is not modified, indicating that they are most likely not regulated by the expression of other miR-430-dependent genes (GEO profiles, GDS1771 from Giraldez lab, http://www.ncbi.nlm.nih.gov/geoprofiles/?term=GDS1771). To confirm these data, we measured the expression of tuba8l, a maternal gene strongly regulated by miR-430.46 Indeed in mutants defective for miR-430, tuba8l appears overexpressed by~700%.46 Our results indicate that tuba8l expression does not change significantly between controls and 7 hpf morphants (it is even slightly lower by ~13% in morphants) suggesting that miR-430 is not repressed in map9 morphants (inset, Fig. 8).
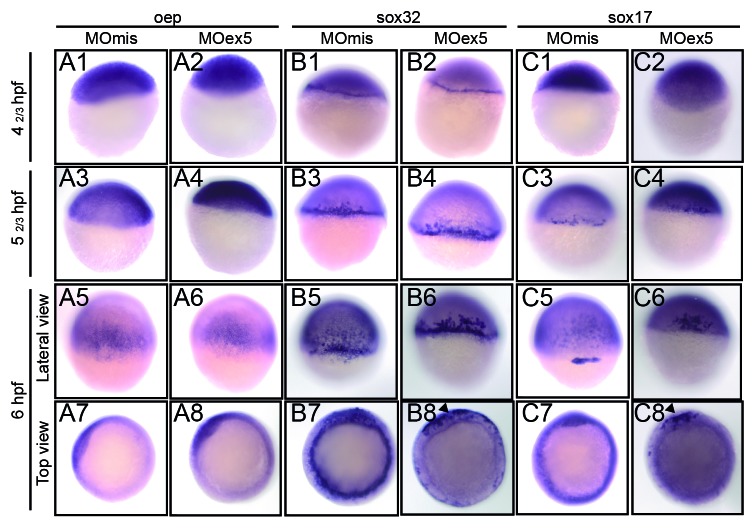
We then assessed by in situ hybridization the expression of oep, sox32, and sox17, the function of which is required for endoderm development. Oep, one of the first genes of the nodal cascade, was expressed early in the blastoderm and localized to the dorsal neural plate at 50% epiboly (shield stage, 6 hpf), as previously described,47 without obvious difference between MOex5 morphants and control embryos (Fig. 9). Sox32 and sox17 were strongly expressed in the marginal cells of the blastoderm, the YSL, and the presumptive endodermal cells from the earliest assessed stage (dome, 41/3 hpf) to the shield (6 hpf) stage in control embryos, as previously described.38 Conversely, in MOex5 morphants, their expression was restricted to only a portion of the marginal cells of the blastoderm (Fig. 9, B8 and C8).
Figure 9. In situ hybridization of selected genes of the nodal pathway that are involved in endoderm development. Embryos injected at the 1-cell stage with 1 pmol of control MOmis or MOex5, were fixed at the indicated times that correspond in MOmis-injected control embryos to the following developmental stages: dome (41/3 hpf), germ-ring (52/3 hpf), and shield (6 hpf). In situ hybridization was performed using antisense oep (nodal co-receptor), sox32 (downstream of oep) and sox17 (activated by sox32 and required for endoderm specification) RNA probes. In map9 morphants, expression of sox32 and sox17 was restricted to a portion of the blastoderm margin at the shield stage (arrowheads, B8, C8).
Discussion
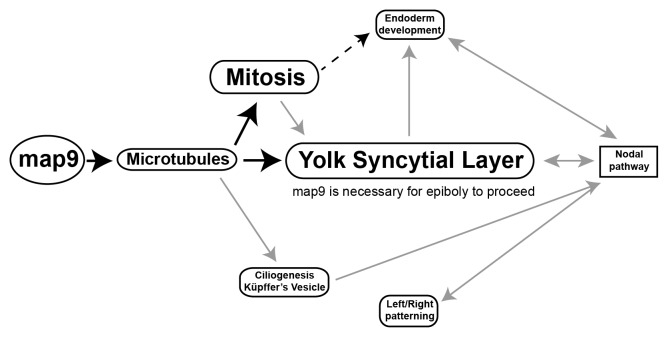
In this study we investigated the role of Map9 during zebrafish embryo development. A schematic representation (Fig. 10) of the pathways that are deregulated following morpholino-mediated map9 knockdown in zebrafish embryos recapitulates our findings and helps understanding how its inhibition can lead to profound developmental defects and/or embryo death.
Figure 10. Tentative model for the role of Map9 in zebrafish development. Map9 is required for MT dynamics. MTs are key components of the mitotic spindle (mitosis) and cilia. MTs and mitosis are essential for the formation of the YSL and epiboly. YSL is required for endoderm development and nodal signaling and the cilia of the KV are responsible for propagating nodal signals involved in left/right asymmetry. As indicated on the schematic, most of these pathways are interdependent. Black arrows indicate the possible role of Map9 during zebrafish development suggested by this and previous studies,9-11,19 and gray arrows summarize literature data.
Specifically, Map9 localizes to the mitotic spindle in embryonic cells, and morpholino-mediated knockdown of map9 leads to mitotic defects similar to those observed following inhibition of the mitotic kinase plk1,17 which, as also shown in mammalian cells (Eot-Houlier et al.11 and references therein), is essential for cell division in eukaryotes.48 We also showed that MAP9 is phosphorylated by PLK1,11 and that it is localized to the spindle pole by PLK1, thus contributing to spindle pole stability. Altogether, these observations suggest that deregulation of the Plk1–Map9 pathway in zebrafish embryos is associated with mitotic defects that can lead to apoptosis, developmental abnormalities, and, eventually, cell death. Interestingly, other studies in zebrafish linked apoptosis, YSL, cytoskeleton, and epiboly.36,49
We also show that morpholino-mediated knockdown of map9 not only affects the blastula cells, but also the extra-embryonic YSL. Injection of anti-map9 MOs in 1-cell stage embryos disrupts the MT network of YSL, and anti-map9 MO injection directly in the nascent YSL at a later developmental stage (~3 hpf) prevents normal epiboly progression, while Map9 deficiency in blastula cells leads to cell-autonomous death. Every YSL nucleus is surrounded by a MT array that directs its migration toward the vegetal pole. Interfering with the MT network by exposure to UV light or treatment with nocodazole or taxol (a MT stabilizing drug) leads to defective epiboly movements of both the YSL and the blastoderm.31 In map9 morphants, the yolk cell longitudinal MT array is defective with large areas without MT and epiboly is hampered, as previously described in eomesa mutants.34 The deregulated expression of the eomes genes in map9 morphants (overexpression of eomesa and downregulation of eomesb) could contribute to the observed phenotype. Similarly, the endodermal marker sox32, a fish-specific HMG-box transcription factor, is downregulated and its expression restricted to only a portion of the marginal cells of the blastoderm in map9 morphants, as previously reported in the eomesa mutants as a result of YSL defects.34 The restriction of sox32 and sox17 expression in endodermal cells could also contribute to the lethal phenotype of map9 morphants. Moreover, knockdown experiments have shown that Sox17 is necessary for the formation of the Kupffer vesicle and LR patterning in zebrafish.50
In addition, map9 knockdown might affect also the nodal signaling pathways that are induced in the YSL at the onset of gastrulation6 to specify the fates of mesodermal and endodermal progenitor cells along the animal–vegetal axis and to establish the LR body axis.51,52 In zebrafish, cyc and oep are part of the YSL nodal signaling pathways.53 Cyc is essential for organizer development and, consequently, also for mesoderm and endoderm formation. Multiple signaling pathways are involved earlier in dorsal forerunner cells (DFC) specification, and perturbation of DFC maturation and organization leads to failure in KV organogenesis and function at later stages.54 The propagation of posterior to anterior (PA) waves in zebrafish left–right development55 depends on the function of the KV cilia and on the hedgehog signaling pathway.56,57 In this pathway, the sonic hedgehog ligand binds to and activates the patched receptor that triggers the translocation of the G protein-coupled receptor smo to the tip of the cilia, where it allows accumulation of the transcription factor Gli2.57-59 The strong downregulation of shha, which is essential for embryo development,56,60 and of smo in map9 morphants, together with the overexpression of plk1 and plk4, which play important functions in ciliogenesis,61-63 suggest that map9 depletion may have consequences on the function of the primary cilia and on cilia-dependent signaling pathways as well.
Finally, map9 inhibition induced the deregulation of many genes involved early in endoderm development and LR patterning. However it is also possible that these deregulations are secondary effects of Map9 inhibition and/or epiboly arrest. Similar observations have been made in nipbl-deficient zebrafish43 and in Nipbl+/− mouse64 models of Cornelia de Lange syndrome, in which many genes are mis-expressed following nipbl inhibition.
Although the main phenotype of map9 morphants (i.e., gastrulation defects followed by early embryo death) is likely to result from MT-related defects (mitosis defects and YSL-based epiboly failure), it will be important to determine whether the observed gene deregulations are secondary effects and how they contribute to this phenotype.
Materials and Methods
Zebrafish
Experiments were done using the zebrafish golden mutant65 purchased through a local company (Antinea) and maintained according to standard protocols.66 Embryos were obtained by natural spawning of breeding pairs and grown at 28.5 °C in tank water. They were injected and staged according to standard procedures. Ages were expressed as hours post-fertilization (hpf).
Ethics statement
All experimental animal procedures were performed according to the guidelines of the French Council on Animal Care (Arrété du 27 Décembre 1994, ref RESR9401964A, Annexe I). Experiments were approved by the Ethics Committee for animal experimentation of the authors’ institution (Direction départementale de la protection des populations - Hérault).
Cloning and sequencing zebrafish map9
The full-length zebrafish map9 ORF (clone IMAGp998O0412053Q) was ordered from ImaGenes GmbH, sequenced, and the sequence deposited in the GenBank database under the accession number JQ768417. A fragment with the same sequence was amplified by RT-PCR using RNA from the golden zebrafish line used in the laboratory. The map9 ORF and a map9ΔCter cDNA (lacking the MAP domain corresponding to amino acids 451–678) were cloned in phase with YFP in the pCS2+ vector. mRNA was in vitro transcribed using the mMessage mMachine Kit (Ambion, Life Technologies Corporation).
Injection of morpholino oligomers (MOs)
Translation-blocking map9 MO (MO-ATG: 5′-CGTCCATCGT AGCTCCCAAA GAAAC-3′) and splice-blocking map9 MO targeting the exon 5 acceptor splice junction (MOex5: 5′-ACAACCTGTT TACATAAAAA GGTGT-3′) as well as a control MO similar to MOex5 but with 5 mismatches (MOmis, 5′-ACAACGTCTT TAGATAAATA CGTGT-3′) and the tp53 MO (5′-TTGATTTTGC CGACCTCCTC TCCAC-3′) were purchased from GeneTools. MO solutions (1 nL) were injected, with or without 1 ng of dextran-rhodamine, in the cytoplasm of 1-cell stage or 64-cell stage embryos or directly in the YSL. For overexpression assays, map9-YFP RNA was injected at the indicated concentrations in 1-cell stage embryos. For the localization of map9 on the mitotic spindles, 200 pg of map9-YFP RNA were injected in 1-cell stage embryos, and map9 was detected using a rabbit anti-GFP antibody (Biovalley, 1:1000) at 24 hpf. For rescue experiments, 1-cell stage embryos were injected with 1 pmol MOex5 or 200 pg map-YFP RNA, or both.
Apoptosis assay
For TUNEL assays, embryos were fixed in 4% paraformaldehyde overnight, dechorionated, washed in PBS/0.1% Triton X100 (PBT), incubated in equilibration buffer, and treated with terminal deoxynucleotidyl transferase in the presence of fluorescein-12-dUTP (DeadEnd Fluorometric TUNEL System, Promega). Embryos were washed in 2× SSC and PBT and incubated with Hoechst 33258 (1/1000 in PBS).
Whole mount immunofluorescence
Embryos at the indicated stages were fixed in 4% paraformaldehyde overnight, washed in 0.1% PBT, and permeabilized in 1% PBT at room temperature (RT) for 1 h 30 min. After washing in H2O and PBT, they were incubated in blocking buffer (1% BSA, 1% DMSO, 2% goat serum in PBS) at RT for 30 min and then with primary antibodies overnight at 4 °C. Secondary antibodies were added together with Hoechst 33258 (Sigma). Embryos were washed and mounted in FluorSave or PBS/50% glycerol.
The following primary antibodies were used for immunofluorescence: rabbit anti-phosphoSer10-histone H3 (Upstate, 1:200); mouse anti-α-tubulin (Sigma, 1:1000); and mouse anti-γ-tubulin (Abcam, 1:1000). Alexa 488 rabbit and Alexa 555 mouse fluor-conjugated secondary antibodies (Invitrogen) were diluted at 1:1000.
Whole mount in situ hybridization (ISH)
Embryos at the indicated stages were fixed in 4% paraformaldehyde overnight, dechorionated and dehydrated in PBT/methanol. After rehydration, ISH was performed in hybridization buffer (50% formamide, 5× SSC, 0.5 mg/ml tRNA, 0.05 mg/ml heparin, 10% Tween) using digoxigenin (DIG)-labeled antisense RNA probes synthesized with the DIG RNA Labeling Mix (Roche). Hybridized probes were detected by using alkaline phosphate-conjugated anti-DIG antibodies and visualized with NBT/BCIP (Roche).
The map9 and sox17 (cloned in the pCS2+ vector)67,68 antisense RNA probes were synthesized using T7 RNA polymerase (Promega); the oep (pCRII-TOPO) and sox32 (pGEM-T-Easy) probes using Sp6 RNA polymerase (Promega).
Microscopy observation
Embryos were observed using a Leica DM 2500 confocal microscope and the Leica Applications System, an Axiovert 200M Zeiss microscope and the AxioVision Imaging System, or an MVX10 Olympus microscope.
RT-PCR, qPCR
Total RNA from whole zebrafish embryos was isolated using the Nucleospin RNA II kit (Macherey-Nagel). First-strand cDNA was generated using the Superscript III reverse transcriptase (Invitrogen), and qPCR reactions were performed in quadruplicate in 96-well plates in a final volume of 10 μL, using the SYBR Green I Master reaction mix (Roche) on a 480-Light Cycler instrument (Roche). PCR amplifications were performed with an initial 5 min denaturation step at 95 °C followed by 42 cycles (95 °C for 20 s; 56–60 °C for 15 s; 72 °C for 15 s). The relative expression levels of each target gene was normalized to β-actin as control gene.69 For quantifying the 2 map9 mRNA species (wt and misspliced) in MOex5 morphants, primer pairs specific of wt (ZFovF and ZFdelR1) and misspliced (ZF18F and ZFjctR) mRNAs were used. Primer sequences are listed in Table S1.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors are grateful to Drs A Lander, A Muto, A Bruce, M Halpern, T Lepage, and M Poulain for providing the sox17, sox32, oep, and cyc cDNA clones to C Gonzalez, Drs M N’Guyen and K Kissa for advice and help in manipulating zebrafish embryos, to Dr Chris Jopling for help in map9 ISH experiments, and to Dr A Renucci for helpful discussions. We also would like to thank Montpellier Rio Imaging facility for microscopy support. We thank Drs E Andermarcher and R Kiernan for the careful reading of the manuscript and the English language corrections. This work was supported by the CNRS and grants from Association de la Recherche contre le Cancer (ARC N°4027), Ligue Nationale contre le Cancer (Comité Languedoc-Roussillon 2009), and the European Community’s Seventh Framework Programme [FP7-PEOPLE-2011-ITN] under grant agreement no. PITN-GA-2011–289209 for the Marie-Curie Initial Training Network FishForPharma. L.F. was a recipient of a MERT fellowship.
Glossary
Abbreviations:
- ASAP
aster-associated protein
- aurka
aurora kinase a
- bon
bonnie and clyde
- cyc
cyclops
- dnah9
dynein, axonemal, heavy polypeptide 9
- eomesa b
eomesodermins a, b
- foxa2
forkhead box A2
- gata5
gata-binding protein 5
- GFP
green fluorescent protein
- gsc
goosecoid
- hpf
hours post-fecondation
- igu
iguana
- KV
Küpffer's vesicle
- LR
left-right
- MAP
microtubule-associated protein
- ISH
in situ hybridization
- LPM
lateral plate mesoderm
- MO
morpholino oligonucleotide
- MT
microtubule
- nipbla b
nipped-b-like homologs a, b
- ntla
no tail a
- oep
one-eyed pinhead
- ORF
open reading frame
- PBS
phosphate-buffered saline
- plk1 4
polo-like kinases 1, 4
- shha
sonic hedgehog a
- smo
smoothened
- sox32 and sox17
SRY-box containing genes 32 and 17
- spaw
southpaw
- sqt
squint
- tp53
tumor protein 53
- TUNEL
terminal deoxynucleotidyl transferase-mediated fluorescein-dUTP nick-end labeling assay
- YFP
yellow fluorescent protein
- YSL
yolk syncytial layer
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/27944
References
- 1.Bonner MK, Poole DS, Xu T, Sarkeshik A, Yates JR, 3rd, Skop AR. Mitotic spindle proteomics in Chinese hamster ovary cells. PLoS One. 2011;6:e20489. doi: 10.1371/journal.pone.0020489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sauer G, Körner R, Hanisch A, Ries A, Nigg EA, Silljé HH. Proteome analysis of the human mitotic spindle. Mol Cell Proteomics. 2005;4:35–43. doi: 10.1074/mcp.M400158-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Karsenti E, Vernos I. The mitotic spindle: a self-made machine. Science. 2001;294:543–7. doi: 10.1126/science.1063488. [DOI] [PubMed] [Google Scholar]
- 4.Wittmann T, Hyman A, Desai A. The spindle: a dynamic assembly of microtubules and motors. Nat Cell Biol. 2001;3:E28–34. doi: 10.1038/35050669. [DOI] [PubMed] [Google Scholar]
- 5.Wadsworth P, Lee WL, Murata T, Baskin TI. Variations on theme: spindle assembly in diverse cells. Protoplasma. 2011;248:439–46. doi: 10.1007/s00709-010-0205-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carvalho L, Heisenberg CP. The yolk syncytial layer in early zebrafish development. Trends Cell Biol. 2010;20:586–92. doi: 10.1016/j.tcb.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 7.Carvalho L, Stühmer J, Bois JS, Kalaidzidis Y, Lecaudey V, Heisenberg CP. Control of convergent yolk syncytial layer nuclear movement in zebrafish. Development. 2009;136:1305–15. doi: 10.1242/dev.026922. [DOI] [PubMed] [Google Scholar]
- 8.Bonneau B, Popgeorgiev N, Prudent J, Gillet G. Cytoskeleton dynamics in early zebrafish development: A matter of phosphorylation? Bioarchitecture. 2011;1:216–20. doi: 10.4161/bioa.18116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saffin JM, Venoux M, Prigent C, Espeut J, Poulat F, Giorgi D, Abrieu A, Rouquier S. ASAP, a human microtubule-associated protein required for bipolar spindle assembly and cytokinesis. Proc Natl Acad Sci U S A. 2005;102:11302–7. doi: 10.1073/pnas.0500964102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venoux M, Basbous J, Berthenet C, Prigent C, Fernandez A, Lamb NJ, Rouquier S. ASAP is a novel substrate of the oncogenic mitotic kinase Aurora-A: phosphorylation on Ser625 is essential to spindle formation and mitosis. Hum Mol Genet. 2008;17:215–24. doi: 10.1093/hmg/ddm298. [DOI] [PubMed] [Google Scholar]
- 11.Eot-Houllier G, Venoux M, Vidal-Eychenié S, Hoang MT, Giorgi D, Rouquier S. Plk1 regulates both ASAP localization and its role in spindle pole integrity. J Biol Chem. 2010;285:29556–68. doi: 10.1074/jbc.M110.144220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basbous J, Knani D, Bonneaud N, Giorgi D, Brondello JM, Rouquier S. Induction of ASAP (MAP9) contributes to p53 stabilization in response to DNA damage. Cell Cycle. 2012;11:2380–90. doi: 10.4161/cc.20858. [DOI] [PubMed] [Google Scholar]
- 13.Otto EA, Hurd TW, Airik R, Chaki M, Zhou W, Stoetzel C, Patil SB, Levy S, Ghosh AK, Murga-Zamalloa CA, et al. Candidate exome capture identifies mutation of SDCCAG8 as the cause of a retinal-renal ciliopathy. Nat Genet. 2010;42:840–50. doi: 10.1038/ng.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Q, Tan G, Levenkova N, Li T, Pugh EN, Jr., Rux JJ, Speicher DW, Pierce EA. The proteome of the mouse photoreceptor sensory cilium complex. Mol Cell Proteomics. 2007;6:1299–317. doi: 10.1074/mcp.M700054-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–4. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 16.Gherman A, Davis EE, Katsanis N. The ciliary proteome database: an integrated community resource for the genetic and functional dissection of cilia. Nat Genet. 2006;38:961–2. doi: 10.1038/ng0906-961. [DOI] [PubMed] [Google Scholar]
- 17.Jeong K, Jeong JY, Lee HO, Choi E, Lee H. Inhibition of Plk1 induces mitotic infidelity and embryonic growth defects in developing zebrafish embryos. Dev Biol. 2010;345:34–48. doi: 10.1016/j.ydbio.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Amsterdam A, Nissen RM, Sun Z, Swindell EC, Farrington S, Hopkins N. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci U S A. 2004;101:12792–7. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Venoux M, Delmouly K, Milhavet O, Vidal-Eychenié S, Giorgi D, Rouquier S. Gene organization, evolution and expression of the microtubule-associated protein ASAP (MAP9) BMC Genomics. 2008;9:406. doi: 10.1186/1471-2164-9-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathavan S, Lee SG, Mak A, Miller LD, Murthy KR, Govindarajan KR, Tong Y, Wu YL, Lam SH, Yang H, et al. Transcriptome analysis of zebrafish embryogenesis using microarrays. PLoS Genet. 2005;1:260–76. doi: 10.1371/journal.pgen.0010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 22.Dosch R, Wagner DS, Mintzer KA, Runke G, Wiemelt AP, Mullins MC. Maternal control of vertebrate development before the midblastula transition: mutants from the zebrafish I. Dev Cell. 2004;6:771–80. doi: 10.1016/j.devcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 23.Thisse B, Thisse C. Fast Release Clones: A High Throughput Expression Analysis. ZFIN Direct Data Submission (http://zfinorg) 2004.
- 24.Adams M, Simms RJ, Abdelhamed Z, Dawe HR, Szymanska K, Logan CV, Wheway G, Pitt E, Gull K, Knowles MA, et al. A meckelin-filamin A interaction mediates ciliogenesis. Hum Mol Genet. 2012;21:1272–86. doi: 10.1093/hmg/ddr557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aspatwar A, Tolvanen ME, Jokitalo E, Parikka M, Ortutay C, Harjula SK, Rämet M, Vihinen M, Parkkila S. Abnormal cerebellar development and ataxia in CARP VIII morphant zebrafish. Hum Mol Genet. 2013;22:417–32. doi: 10.1093/hmg/dds438. [DOI] [PubMed] [Google Scholar]
- 26.Cole LK, Ross LS. Apoptosis in the developing zebrafish embryo. Dev Biol. 2001;240:123–42. doi: 10.1006/dbio.2001.0432. [DOI] [PubMed] [Google Scholar]
- 27.Jeon HY, Lee H. Depletion of Aurora-A in zebrafish causes growth retardation due to mitotic delay and p53-dependent cell death. FEBS J. 2013;280:1518–30. doi: 10.1111/febs.12153. [DOI] [PubMed] [Google Scholar]
- 28.Zhong H, Xin S, Zhao Y, Lu J, Li S, Gong J, Yang Z, Lin S. Genetic approach to evaluate specificity of small molecule drug candidates inhibiting PLK1 using zebrafish. Mol Biosyst. 2010;6:1463–8. doi: 10.1039/b919743e. [DOI] [PubMed] [Google Scholar]
- 29.Giet R, Glover DM. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J Cell Biol. 2001;152:669–82. doi: 10.1083/jcb.152.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimmel CB, Law RD. Cell lineage of zebrafish blastomeres. II. Formation of the yolk syncytial layer. Dev Biol. 1985;108:86–93. doi: 10.1016/0012-1606(85)90011-9. [DOI] [PubMed] [Google Scholar]
- 31.Solnica-Krezel L, Driever W. Microtubule arrays of the zebrafish yolk cell: organization and function during epiboly. Development. 1994;120:2443–55. doi: 10.1242/dev.120.9.2443. [DOI] [PubMed] [Google Scholar]
- 32.Lepage SE, Bruce AE. Zebrafish epiboly: mechanics and mechanisms. Int J Dev Biol. 2010;54:1213–28. doi: 10.1387/ijdb.093028sl. [DOI] [PubMed] [Google Scholar]
- 33.Solnica-Krezel L. Gastrulation in zebrafish -- all just about adhesion? Curr Opin Genet Dev. 2006;16:433–41. doi: 10.1016/j.gde.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 34.Du S, Draper BW, Mione M, Moens CB, Bruce A. Differential regulation of epiboly initiation and progression by zebrafish Eomesodermin A. Dev Biol. 2012;362:11–23. doi: 10.1016/j.ydbio.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arrington CB, Yost HJ. Extra-embryonic syndecan 2 regulates organ primordia migration and fibrillogenesis throughout the zebrafish embryo. Development. 2009;136:3143–52. doi: 10.1242/dev.031492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Popgeorgiev N, Bonneau B, Ferri KF, Prudent J, Thibaut J, Gillet G. The apoptotic regulator Nrz controls cytoskeletal dynamics via the regulation of Ca2+ trafficking in the zebrafish blastula. Dev Cell. 2011;20:663–76. doi: 10.1016/j.devcel.2011.03.016. [DOI] [PubMed] [Google Scholar]
- 37.Tam PP, Kanai-Azuma M, Kanai Y. Early endoderm development in vertebrates: lineage differentiation and morphogenetic function. Curr Opin Genet Dev. 2003;13:393–400. doi: 10.1016/S0959-437X(03)00085-6. [DOI] [PubMed] [Google Scholar]
- 38.Kikuchi Y, Agathon A, Alexander J, Thisse C, Waldron S, Yelon D, Thisse B, Stainier DY. casanova encodes a novel Sox-related protein necessary and sufficient for early endoderm formation in zebrafish. Genes Dev. 2001;15:1493–505. doi: 10.1101/gad.892301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen S, Kimelman D. The role of the yolk syncytial layer in germ layer patterning in zebrafish. Development. 2000;127:4681–9. doi: 10.1242/dev.127.21.4681. [DOI] [PubMed] [Google Scholar]
- 40.Long S, Ahmad N, Rebagliati M. The zebrafish nodal-related gene southpaw is required for visceral and diencephalic left-right asymmetry. Development. 2003;130:2303–16. doi: 10.1242/dev.00436. [DOI] [PubMed] [Google Scholar]
- 41.Essner JJ, Amack JD, Nyholm MK, Harris EB, Yost HJ. Kupffer’s vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development. 2005;132:1247–60. doi: 10.1242/dev.01663. [DOI] [PubMed] [Google Scholar]
- 42.Hashimoto H, Rebagliati M, Ahmad N, Muraoka O, Kurokawa T, Hibi M, Suzuki T. The Cerberus/Dan-family protein Charon is a negative regulator of Nodal signaling during left-right patterning in zebrafish. Development. 2004;131:1741–53. doi: 10.1242/dev.01070. [DOI] [PubMed] [Google Scholar]
- 43.Muto A, Calof AL, Lander AD, Schilling TF. Multifactorial origins of heart and gut defects in nipbl-deficient zebrafish, a model of Cornelia de Lange Syndrome. PLoS Biol. 2011;9:e1001181. doi: 10.1371/journal.pbio.1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–7. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–8. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 46.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–9. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 47.Thisse B, Pflumio S, Fürthauer M, Loppin B, Heyer V, Degrave A, Woehl R, Lux A, Steffan T, Charbonnier XQ, et al. Expression of the zebrafish genome during embryogenesis. ZFIN Direct Data Submission (http://zfinorg) 2001.
- 48.Petronczki M, Lénárt P, Peters JM. Polo on the Rise-from Mitotic Entry to Cytokinesis with Plk1. Dev Cell. 2008;14:646–59. doi: 10.1016/j.devcel.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 49.Arnaud E, Ferri KF, Thibaut J, Haftek-Terreau Z, Aouacheria A, Le Guellec D, Lorca T, Gillet G. The zebrafish bcl-2 homologue Nrz controls development during somitogenesis and gastrulation via apoptosis-dependent and -independent mechanisms. Cell Death Differ. 2006;13:1128–37. doi: 10.1038/sj.cdd.4401797. [DOI] [PubMed] [Google Scholar]
- 50.Aamar E, Dawid IB. Sox17 and chordin are required for formation of Kupffer’s vesicle and left-right asymmetry determination in zebrafish. Dev Dyn. 2010;239:2980–8. doi: 10.1002/dvdy.22431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hagos EG, Dougan ST. Time-dependent patterning of the mesoderm and endoderm by Nodal signals in zebrafish. BMC Dev Biol. 2007;7:22. doi: 10.1186/1471-213X-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fan X, Hagos EG, Xu B, Sias C, Kawakami K, Burdine RD, Dougan ST. Nodal signals mediate interactions between the extra-embryonic and embryonic tissues in zebrafish. Dev Biol. 2007;310:363–78. doi: 10.1016/j.ydbio.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feldman B, Gates MA, Egan ES, Dougan ST, Rennebeck G, Sirotkin HI, Schier AF, Talbot WS. Zebrafish organizer development and germ-layer formation require nodal-related signals. Nature. 1998;395:181–5. doi: 10.1038/26013. [DOI] [PubMed] [Google Scholar]
- 54.Matsui T, Bessho Y. Left-right asymmetry in zebrafish. Cell Mol Life Sci. 2012;69:3069–77. doi: 10.1007/s00018-012-0985-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang X, Yost HJ. Initiation and propagation of posterior to anterior (PA) waves in zebrafish left-right development. Dev Dyn. 2008;237:3640–7. doi: 10.1002/dvdy.21771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilson CW, Stainier DY. Vertebrate Hedgehog signaling: cilia rule. BMC Biol. 2010;8:102. doi: 10.1186/1741-7007-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–44. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.May SR, Ashique AM, Karlen M, Wang B, Shen Y, Zarbalis K, Reiter J, Ericson J, Peterson AS. Loss of the retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli. Dev Biol. 2005;287:378–89. doi: 10.1016/j.ydbio.2005.08.050. [DOI] [PubMed] [Google Scholar]
- 59.Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev Cell. 2008;15:801–12. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McMahon AP, Ingham PW, Tabin CJ. Developmental roles and clinical significance of hedgehog signaling. Curr Top Dev Biol. 2003;53:1–114. doi: 10.1016/S0070-2153(03)53002-2. [DOI] [PubMed] [Google Scholar]
- 61.Bettencourt-Dias M, Glover DM. Centrosome biogenesis and function: centrosomics brings new understanding. Nat Rev Mol Cell Biol. 2007;8:451–63. doi: 10.1038/nrm2180. [DOI] [PubMed] [Google Scholar]
- 62.Bettencourt-Dias M, Hildebrandt F, Pellman D, Woods G, Godinho SA. Centrosomes and cilia in human disease. Trends Genet. 2011;27:307–15. doi: 10.1016/j.tig.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carvalho-Santos Z, Machado P, Branco P, Tavares-Cadete F, Rodrigues-Martins A, Pereira-Leal JB, Bettencourt-Dias M. Stepwise evolution of the centriole-assembly pathway. J Cell Sci. 2010;123:1414–26. doi: 10.1242/jcs.064931. [DOI] [PubMed] [Google Scholar]
- 64.Kawauchi S, Calof AL, Santos R, Lopez-Burks ME, Young CM, Hoang MP, Chua A, Lao T, Lechner MS, Daniel JA, et al. Multiple organ system defects and transcriptional dysregulation in the Nipbl(+/-) mouse, a model of Cornelia de Lange Syndrome. PLoS Genet. 2009;5:e1000650. doi: 10.1371/journal.pgen.1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lamason RL, Mohideen MA, Mest JR, Wong AC, Norton HL, Aros MC, Jurynec MJ, Mao X, Humphreville VR, Humbert JE, et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 2005;310:1782–6. doi: 10.1126/science.1116238. [DOI] [PubMed] [Google Scholar]
- 66.Westerfield M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio). Eugene: University of Oregon Press, 2007. [Google Scholar]
- 67.Halpern ME, Hatta K, Amacher SL, Talbot WS, Yan YL, Thisse B, Thisse C, Postlethwait JH, Kimmel CB. Genetic interactions in zebrafish midline development. Dev Biol. 1997;187:154–70. doi: 10.1006/dbio.1997.8605. [DOI] [PubMed] [Google Scholar]
- 68.Poulain M, Lepage T. Mezzo, a paired-like homeobox protein is an immediate target of Nodal signalling and regulates endoderm specification in zebrafish. Development. 2002;129:4901–14. doi: 10.1242/dev.129.21.4901. [DOI] [PubMed] [Google Scholar]
- 69.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.