Abstract
Therapeutic interventions based on metabolic inhibitor-based therapies are expected to be less prone to acquired resistance. However, there has not been any study assessing the possibility that the targeting of the tumor cell metabolism may result in unforeseeable resistance. We recently established a pre-clinical model of estrogen-dependent MCF-7 breast cancer cells that were chronically adapted to grow (> 10 months) in the presence of graded, millimolar concentrations of the anti-diabetic biguanide metformin, an AMPK agonist/mTOR inhibitor that has been evaluated in multiple in vitro and in vivo cancer studies and is now being tested in clinical trials. To assess what impact the phenomenon of resistance might have on the metformin-like “dirty” drugs that are able to simultaneously hit several metabolic pathways, we employed the ingenuity pathway analysis (IPA) software to functionally interpret the data from Agilent whole-human genome arrays in the context of biological processes, networks, and pathways. Our findings establish, for the first time, that a “global” targeting of metabolic reprogramming using metformin certainly imposes a great selective pressure for the emergence of new breast cancer cellular states. Intriguingly, acquired resistance to metformin appears to trigger a transcriptome reprogramming toward a metastatic stem-like profile, as many genes encoding the components of the degradome (KLK11, CTSF, FREM1, BACE-2, CASP, TMPRSS4, MMP16, HTRA1), cancer cell migration and invasion factors (TP63, WISP2, GAS3, DKK1, BCAR3, PABPC1, MUC1, SPARCL1, SEMA3B, SEMA6A), stem cell markers (DCLK1, FAK), and key pro-metastatic lipases (MAGL and Cpla2) were included in the signature. Because this convergent activation of pathways underlying tumor microenvironment interactions occurred in low-proliferative cancer cells exhibiting a notable downregulation of the G2/M DNA damage checkpoint regulators that maintain genome stability (CCNB1, CCNB2, CDC20, CDC25C, AURKA, AURKB, BUB1, CENP-A, CENP-M) and pro-autophagic features (i.e., TRAIL upregulation and BCL-2 downregulation), it appears that the unique mechanism of acquired resistance to metformin has opposing roles in growth and metastatic dissemination. While refractoriness to metformin limits breast cancer cell growth, likely due to aberrant mitotic/cytokinetic machinery and accelerated autophagy, it notably increases the potential of metastatic dissemination by amplifying the number of pro-migratory and stemness inputs via the activation of a significant number of proteases and EMT regulators. Future studies should elucidate whether our findings using supra-physiological concentrations of metformin mechanistically mimic the ultimate processes that could paradoxically occur in a polyploid, senescent-autophagic scenario triggered by the chronic metabolic stresses that occur during cancer development and after treatment with cancer drugs.
Keywords: breast cancer, metabolism, metformin, AMPK, metastasis, degradome
In the era of personalized medicine, all initially successful molecularly targeted therapies are limited by the invariable and often rapid occurrence of resistance in tumor cells. Although new cancer drugs have been developed to specifically and efficiently interfere with defined genetic aberrations, resistance commonly occurs through the acquisition of compensatory mechanisms that bypass the function of the cancer gene that is pharmacologically targeted.1 Interestingly, one of the available pathways that can bypass the driver status of the genetic target is a common feature across multiple types of cancer: deregulated cellular metabolism.2-6 The metabolic properties of cancer cells are remarkably different from those of normal cells, and mounting evidence supports the idea that metabolic reprogramming is linked not only to the efficacy of classical therapeutic approaches in cancer, such as radiotherapy, hormonotherapy, and chemotherapy but also to the efficacy of newly developed molecularly targeted drugs.7-19
While it might appear intuitive that deregulated cancer metabolism can activate pro-survival signaling and decrease drug-induced apoptosis to provide a general, unspecific protection against cell injuries induced by multiple types of cytotoxicities, it is worth noting that resistance to oncogene-mediated targeted therapy has been shown to require a shift toward the very same metabolic state that is controlled by growth factor signaling.20,21 In cancer cells sensitive to lapatinib, the small-molecule dual inhibitor of the oncogenes EGFR and HER2, receptor tyrosine kinase signaling is disrupted, and activity of its Ras, PI3K, and mTOR downstream effectors is abrogated; because oncogene-dependent metabolic rewiring is prevented, cancer cell death is observed. In drug-resistant cells, however, the resistance mechanism does not involve the expected reactivation of the Ras, PI3K or mTOR pathways, but rather involves the reactivation of multiple metabolic processes, including the unfolded protein response, autophagy, glycolysis, and gluconeogenesis, which ensures a metabolic rewiring that permits cancer cell proliferation even upon the removal of any activity from canonical growth factor signaling pathways.
The latter observations strongly confirm that mutations that activate oncogenes or inactivate tumor suppressors appear to “softwire” cancer genes to metabolism, because these cancer driver genes directly regulate metabolic enzymes.22-25 Importantly, because metabolic reprogramming is a central (re)wiring or convergence point of many, if not all, cancer-related signaling pathways, tumor cells might be unable to adapt to the molecular challenges imposed by multifaceted drugs that act on cell metabolism at multiple levels. Not surprisingly, the area of cancer metabolism research is undergoing an unstoppable renaissance, because therapeutic interventions based on metabolic inhibitor-based therapies should be less prone to acquired resistance, assuming that the changes in tumor metabolism are similar across multiple cancerous tissues and affect many cancer cell types, including cancer stem cells (CSCs). In this regard, there have been no studies assessing the possibility that targeting tumor cell metabolism may face yet-to-be discovered resistance.
Metabolic reprogramming may not be a “passenger” phenomenon, but rather an active driver of the transformed phenotype. For this reason, we recently speculated that currently proposed antitumor drugs that target various metabolic pathways would impose great selective pressure for the emergence of resistant cells. An ever-growing amount of in vitro studies have confirmed that the anti-diabetic drug metformin can exert anticancer activity by decreasing the activation of the mammalian target of rapamycin (mTOR), a unique sensor that coordinates nutrient availability and energy metabolism with cell responses to growth factors. In vivo studies have shown that metformin can negatively affect the growth of human tumors even in the presence of activating mutations in the PIK3CA oncogene, another evolutionary conserved regulator of cell metabolism that converges with and impinges on the mTOR pathway.10,26-37 To anticipate the potential mechanisms of acquired resistance to metformin during the course of treatment, we recently established metformin-resistant pooled cell populations from the MCF-7 breast carcinoma cell line. Thus, to assess what impact the resistance phenomenon might have on metformin-based therapies, genome-wide analyses using Agilent 44K Whole Human Genome Arrays were evaluated using a bioinformatics approach with the ingenuity pathway analysis (IPA) software. Here, we reveal for the first time that the genomic spaces related to chronic adaptation to the AMPK agonist/mTOR inhibitor metformin involve a degradome-related metastasis aggressiveness gene expression-like signature.
Results
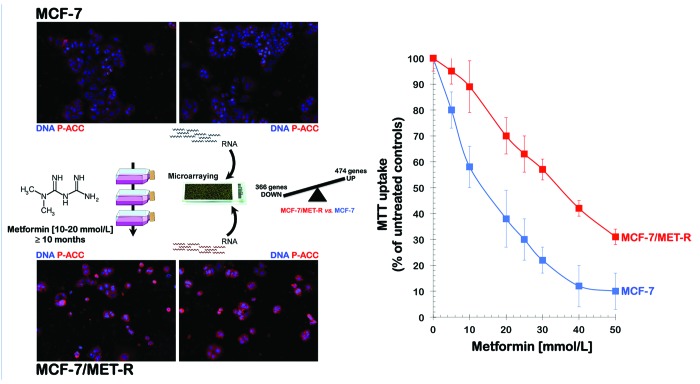
To anticipate the potential mechanisms of acquired resistance to metformin during the course of treatment, we established a pooled population of metformin-adapted cancer cells from metformin-naïve MCF-7 breast cancer cells. To simulate the clinic where patients receive metformin on a daily chronic basis, we developed a model of acquired adaptation to metformin by chronically exposing MCF-7 cells to graded concentrations of metformin for longer than 10 mo before starting any experimental procedure (Fig. 1, left panels). We have now isolated the metformin-refractory pooled populations of MCF-7/MET-R cells that are capable of growing in the presence of 30 to 40 mmol/L metformin, a range of metformin concentrations that are highly cytotoxic to the parental MCF-7 cells, as confirmed by 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide [MTT]-based metabolic assays (Fig. 1, right panel).
Figure 1. Discovery of a transcriptomic signature defining the acquisition of resistance to metformin. Left: A schematic depicting the experimental approach designed to establish metformin-adapted population of MCF-7 breast cancer cells. RNA was extracted from metformin-naïve MCF-7 parental cells and metformin-resistant MCF-7/MET-R cells and then hybridized to G4112F Agilent Human Whole Genome Microarrays. Gene expression was analyzed as described in the “Materials and Methods” section. For the complete gene data, see Tables S1 and S2. Figure shows also representative immunofluorescence images demonstrating a significant augmentation of phospho-acetyl-CoA carboxylase (P-ACC) expression, a marker of metformin-enhanced AMPK activity, as well as the reduced number and altered morphology of metformin-adapted MCF-7/MET-R cells compared with MCF-7 parental cells. Right: Figure shows dose-response MTT uptake curves confirming that MCF-7/MET-R cells exhibit increased cell viability in the presence of extremely high concentrations of metformin. Similar optical density values of MTT uptakes were obtained in untreated MCF-7 (approx. 0.8) and MCF-7/MET-R cells (approx. 0.7) after a 5-d culture period.
Characterization of a pathway-based transcriptomic signature in MCF-7 breast cancer cells with acquired resistance to metformin
To determine the gene expression effects related to metformin efficacy in breast cancer cells, we performed genome-wide analyses by comparing the global transcriptomic profiles of metformin-naïve MCF-7 cells to those obtained from a pooled population of metformin-adapted MCF7/MET-R cells. After RNA hybridization to an Agilent 44K (double density) Whole Human Genome Oligo Microarray, which contains 45 220 probes representing 41 000 unique human genes and transcripts, the normalized and filtered data from all experimental groups were simultaneously analyzed using the SAM algorithm. Using a 2.0-fold-change cut-off value relative to the transcriptome of metformin-naïve MCF-7 parental cells, genes that showed significant expression changes were identified. Only genes with well-annotated transcripts (i.e., not partial cds for hypothetical proteins, hypothetical insert cDNA clones, etc.) were selected, and genes that could not be identified were eliminated. We identified 840 genes (474 upregulated and 366 downregulated) that were differentially expressed in the MCF-7/MET-R cells. Tables S1 and S2 summarize the upregulated and downregulated gene transcripts, respectively, in the “metformin adaptation” transcriptomic signature.
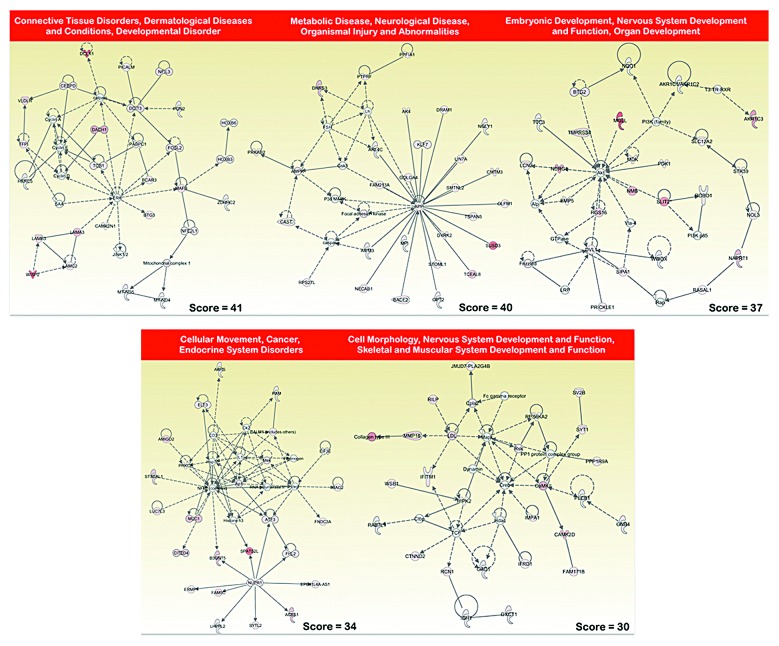
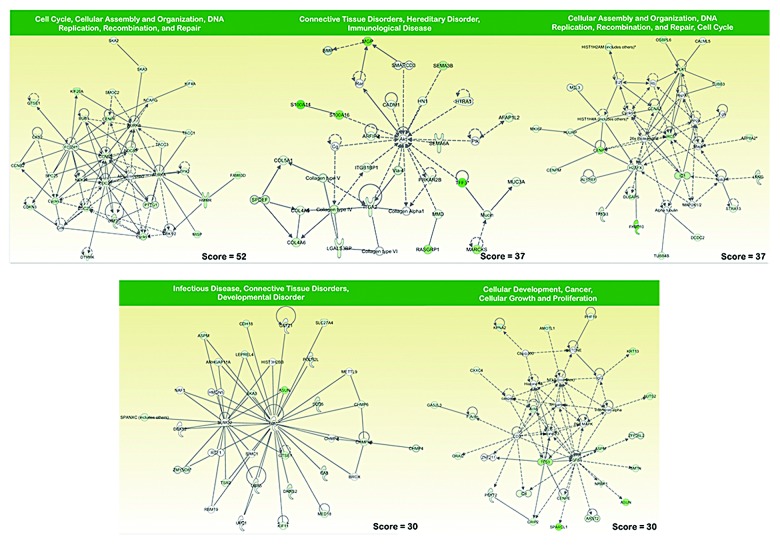
To identify functions that were significantly altered under the metabolic selective pressure (i.e., metformin treatment), we used an experimental approach that focused on gene pathways. Although several computational methods have been proposed for incorporating biological pathway information and gene sets into microarray data analysis, we decided to employ Ingenuity Pathway Analysis (IPA) using the Ingenuity® software. We utilized the “core analysis” function included in the software package to interpret the metformin resistance-related global transcriptomic profiles in the context of biological processes, networks, and pathways. The IPA software algorithmically generates networks of up- and downregulated functionally related annotated genes based on their connectivity and assigns a score that considers both the number of the focus genes in a network and the size of the network to approximate the relevance of each network in relation to the original list of focus genes. Figure 2 graphically illustrates the 5 gene network functions that were most significantly (score > 30) upregulated (red), and Figure 3 illustrates the 5 gene network functions that were most significantly (score > 30) downregulated (green) in the metformin resistance-related transcriptomic signature of MCF-7 breast cancer cells.
Figure 2. Network analysis of genes overexpressed in MCF-7/MET-R cells that have acquired resistance to metformin. A data set containing the differentially upregulated genes (called the focus molecules = 474) between metformin-refractory MCF-7/MET-R cells and metformin-sensitive MCF-7 parental cells was overlaid onto a global molecular network developed from information contained in the Ingenuity Pathway (IPA) Knowledge Base. Networks of these focus molecules were then algorithmically generated based on their connectivity. The figure shows upregulated networks with the 5 highest IPA scores (a composite measure that indicates the statistically significance of the interconnection between the molecules depicted in the network). The focus molecules are colored according to the gene expression (fold-change). The nodes are displayed using various shapes that represent the functional class of the gene product. Edges with dashed lines indicate indirect interactions, while continuous lines represent direct interactions.
Figure 3. Network analysis of genes under-expressed in MCF-7/MET-R cells that have acquired resistance to metformin. A data set containing the differentially downregulated genes (called the focus molecules = 366) between metformin-refractory MCF-7/MET-R cells and metformin-sensitive MCF-7 parental cells was overlaid onto a global molecular network developed from information contained in the Ingenuity Pathway (IPA) Knowledge Base. Networks of these focus molecules were then algorithmically generated based on their connectivity. The figure shows downregulated networks with the 5 highest IPA scores (a composite measure that indicates the statistically significance of the interconnection between the molecules depicted in the network). The focus molecules are colored according to the gene expression (fold-change). The nodes are displayed using various shapes that represent the functional class of the gene product. Edges with dashed lines indicate indirect interactions, while continuous lines represent direct interactions.
The top functions of the upregulated gene networks (Fig. 2) were related to: (1) Connective tissue disorders, dermatological diseases and conditions, developmental disorder (score = 41), including the cancer stem cell marker DCLK1, the enhancer of the cell motility and metastasis BCAR3 (breast cancer antiestrogen resistance 3) gene, and the PABPC1 (poly A binding protein 1) gene, a component of the ezrin-driven metastatic phenotype. Intriguingly, this gene network included the LAMA3, LAMB3, and LAMC2 genes, which encode 3 polypeptide chains, alpha3, β3, and gamma2, respectively, of laminin 5, which anchors epithelial cells to the underlying basement membrane and negatively regulates tumor invasion, and the tumor suppressor DACH1, whose expression is lost in some forms of metastastic cancer but is highly expressed in other metastastic carcinomas; (2) Metabolic disease, neurological disease, organismal injury, and abnormalities (score = 40), including genes coding for one of the indirect targets of metformin, AMPK, β2 non-catalytic subunit (PRKAB2), focal adhesion kinase (FAK), which is a prominent determinant in breast cancer initiation, progression, and metastasis via the maintenance of mammary cancer stem cells, beta-secretase 2 (BACE-2), which is a type I integral membrane glycoprotein and aspartic protease belonging to the peptidase A1 protein family, and the calpain inhibitor calpastatin (CASP), which plays a key, opposing role within the calpain/calpastatin system in initial tumor growth and subsequent metastastic dissemination. This gene network was identified around the amyloid precursor protein (APP), an androgen-induced gene associated with breast cancer cell proliferation; (3) Embryonic development, nervous system development and function, organ development (score = 37), a gene network that was identified around the Akt gene and included the gene coding for the transmembrane protease, serine 4 (TMPRSS4), which is a promoter of migration, invasion, and metastasis by facilitating EMT-like phenomena; the gene coding for the serine hydrolases monoacylglycerol lipase (MGLL), which is elevated in aggressive human cancer cells and plays a key role in cancer metastasis; and the genes coding for neuronal repellent Slit2 (SLIT2) and its transmembrane receptor ROBO, a key autonomous duo with oncogenic effects on tumor cells that may regulate tumorigenesis and metastasis through a mechanism related to contact inhibition. Intriguingly, this gene network included genes such as N-myc downstream regulated gene 1 (NDRG1), which has been shown to act as a metastatic suppressor in a number of human cancers; (4) Cellular movement, cancer, endocrine system disorders (score = 34), a gene network that was identified, at least in part, around NUPR1, a gene that has been found to aid the establishment of metastasis and to play a key role in the progression of several malignancies, including breast cancer, by inducing chemoresistance, protection from apoptosis, and genome instability. The other sub-network was identified around the NFkB complex and included genes such as those coding for the transmembrane mucin MUC1, whose overexpression is frequently associated with metastastic progression, the pro-metastatic gene AMIGO2, and the transcription factor ATF3, a molecule that functions as an integration point for cellular communication during changes in homeostasis and in the subsequent adaptation in response to those changes during breast cancer development and metastasis; (5) Cell morphology, nervous system development and function, skeletal and muscular system development and function (score = 30), a gene network that was identified around Creb and included genes such as those coding for the matrix metalloproteinase 16 (MMP16) and cytoplasmic phospholipase A2 (Cpla2), whose metabolites play critical roles in tumor metastasis via the promotion of angiogenesis and MMP expression. The top functions of the downregulated gene networks (Fig. 3) were related to: (1) Cell cycle, cellular assembly and organization, DNA replication, recombination, and repair (score = 52), a gene network that was identified around the gene coding for cyclin B1 (CCNB1), whose downregulation results in polyploidization during DNA damage-induced senescence, and this network included genes coding for cell cycle checkpoint proteins such as CCNB2, CDC25C, as well as spindle assembly checkpoint proteins such as CDC20, whose downregulation induces aberrant mitosis, the Aurora kinases AURKA and AURKB, which play important roles in chromosome alignment, segregation, and cytokinesis during mitosis, BUB1, whose inhibition results in genomic instability and anchorage-independent growth, and 2 out of the 3 human TACC (transforming acidic coiled-coil) genes (TACC1, TACC3) that participate in the oncogenic processes and whose downregulation alters the control of mRNA homeostasis in polarized cells; (2) Connective tissue disorders, hereditary disorder, immunological disease (score = 37), a gene network that was identified around Akt, included genes coding for semaphorins, which are a large family of secreted and membrane-bound molecules that have been found to regulate cell adhesion and cell motility, angiogenesis, immune function, and tumor progression, such as SEMA3B, a putative tumor suppressor gene, and SEMA6A, an angiogenesis inhibitor, the serine protease HTRA1, a tumor suppressor whose downregulation activates EMT-like phenomena and ATM DNA damage response pathways, and different types of collagen (IV, V, and VI); (3) Cellular assembly and organization, DNA replication, recombination, and repair, cell cycle (score = 37), a gene network that was identified around the gene coding for the anti-apoptotic protein BRIC5 (survivin), the downregulation of which may allow cancer cells to exit mitosis without achieving proper chromosome alignment, leading to the formation of polyploid nuclei, and this network included genes coding for centromere and nucleosomal proteins (CENP-M, CENP-A), and members of the histone cluster 1 (HIST1H4A, HIST1H2AM); (4) Infectious disease, connective tissue disorders, developmental disorder (score = 30), a gene network that was identified around the gene coding for ubiquitin (UBC), and this network included several genes coding for charged multivesicular body proteins (CHMP4, CHMP4B, CHMP6) and the mammalian asunder gene (ASUN), the downregulation of which leads to nucleus–centrosome uncoupling, abnormal spindles, and multinucleation; (5) Cellular development, cancer, cellular growth, and proliferation (score = 30), a gene network that was identified around the gene coding for Histone H3 and included the gene coding for p63 (TP63), an “epithelial organizer” that suppresses tumorigenesis and metastasis by directly impinging on EMT, stemness, senescence, cell death, and cell cycle arrest, the Secreted protein, acidic and rich in cysteine-like 1 (SPARCL1), whose downregulation increases the migratory, invasive, and metastastic properties of cancer cells, and CXXC4, whose decreased expression promotes a malignant phenotype by activating the Wnt stemness signaling pathway.
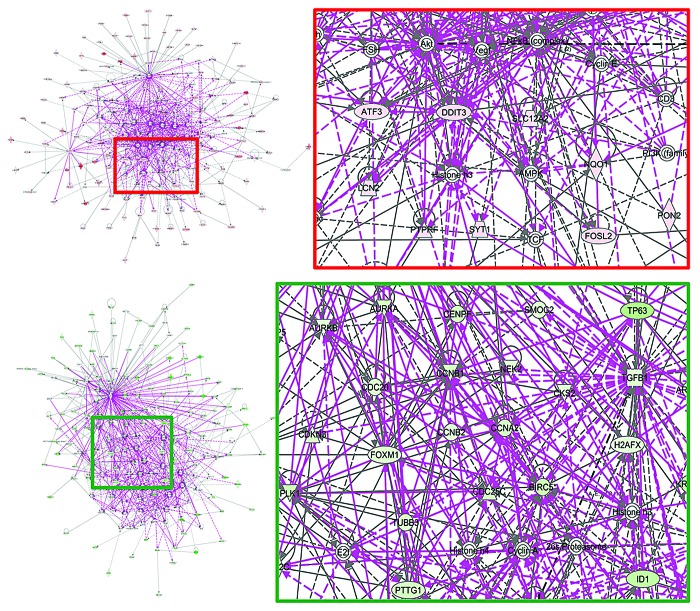
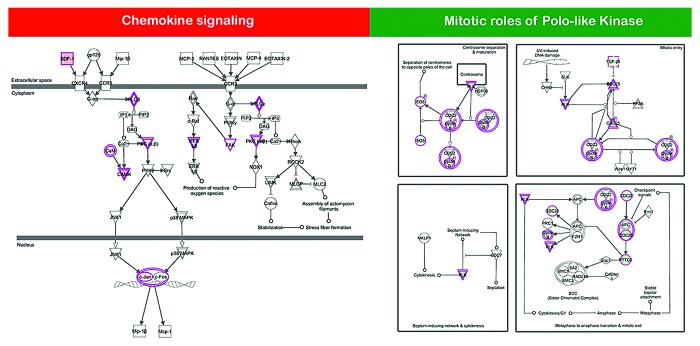
The ATF3 and DDIT3 genes, 2 autophagy-related members of cell stress responses related to mTOR inhibition, and the AMPK gene, which codes for one of the indirect targets of metformin, were central in a merged network combining the top 5 upregulated signaling networks with the highest IPA scores in the transcriptomic signature of metformin-adapted MCF-7/MET-R cells (Fig. 4, top panels). The CCNB1, CCNB2, CCNA2, and CDC25C genes, all of them coding for checkpoint proteins that regulate the cell cycle, were central in a merged network combining the top 5 downregulated signaling networks with the highest IPA scores in the transcriptomic signature of metformin-adapted MCF-7/MET-R cells (Fig. 4, bottom panels). When the IPA software was used to determine the canonical pathway analysis enrichment categories, “chemokine signaling”, “axonal guidance signaling”, and “VDR/RXR activation” were the most statistically significant maps that were modulated by the upregulated genes within the metformin-unresponsiveness transcriptomic signature (Table 1; Fig. 5, left panel). “Mitotic roles of polo-like kinases”, “Axonal guidance signaling”, “Cell cycle: G2/M DNA damage checkpoint regulation”, and “Remodeling of epithelial adherens junctions” were the most statistically significant maps that were modulated by the downregulated genes within the metformin-unresponsiveness transcriptomic signature (Table 1; Fig. 5, right panel).
Figure 4. Merged networks combining major signaling networks associated with the transcriptomic signature of MCF-7/MET-R cells that have acquired resistance to metformin.
Table 1. Top canonical pathways in the transcriptomic signature of metformin-adapted MCF-7/MET-R cells.
| Name | P value | Ratio |
|---|---|---|
| Chemokine signaling | 5,57E-05 | 9/74 (0,122) |
| Axonal guidance signaling | 3,02E-04 | 23/476 (0,048) |
| VDR/RXR activation | 5,7E-04 | 8/87 (0,092) |
| CXCR4 signaling | 1,66E-03 | 11/172 (0,064) |
| α-Adrenergic signaling | 2,08E-03 | 8/106 (0,075) |
| Mitotic roles of polo-like kinase | 1,7E-05 | 8/68 (0,118) |
| Axonal guidance signaling | 3,79E-04 | 19/476 (0,04) |
| Cell Cycle: G2/M DNA damage checkpoint regulation | 1,1E-03 | 5/48 (0,104) |
| Remodeling of epithelial adherens junctions | 7,28E-03 | 5/68 (0,074) |
| Role of osteoblasts, osteoclasts and chondrocytes in rheumatoid arthritis | 7,68E-03 | 10/244 (0,041) |
Figure 5. Canonical pathway analysis of differentially expressed genes in MCF-7/MET-R cell populations chronically adapted to grow in the presence of metformin. Using the “canonical pathways” feature of the IPA global functional analysis, we were able to identify metformin response-related pathways that were significantly impacted in metformin-refractory MCF-7/MET-R cells when compared with metformin-naïve MCF-7 parental cells. Figure shows 2 selected canonical pathways with the lowest P value differentially activated (left panel) or deactivated (right panel) in MCF-7 cells upon acquisition of resistance to metformin.
Discussion
Many genetic lesions important for cancer converge to promote proliferative metabolism in cancer cells, thus suggesting that “cancer metabolism” is a single entity that differs from “normal cell metabolism”. Targeting cancer metabolism for cancer therapy has been suggested as a simpler approach than targeting the mutated gene products to eliminate all cancerous cells simultaneously. Because the extent of metabolic reprogramming that occurs in cancer cells goes far beyond glycolytic behavior (the Warburg effect) and encompasses nearly all metabolic routes, including glutaminolysis, lipogenesis, fatty acid oxidation, gluconeogenesis, and the pentose phosphate pathway, and given the extremely high metabolic flexibility of cancer cells, exclusively targeting glycolysis or specific metabolic pathways in cancer might be just as complicated as targeting somatic mutations, if not more so.38,39
We began to recognize that cancer cells can escape death from metabolic inhibitors by turning off the glycolytic pathway and switching to aerobic respiration and high oxidative capacity phenotype.40 If glycolysis-addicted cancer cells can easily perform these metabolic tricks to hide among the non-proliferative oxidative phosphorylation-dependent normal cells until the treatment is over, then the possibility exists that the metabolic features of cancer cells will come back after the cessation of treatment with glycolysis inhibitors.41 An alternative approach may involve the use of “dirty” drugs, which are able to hit several metabolic pathways simultaneously. In this regard, there is considerable excitement and an increasing number of clinical trials testing the efficacy of the anti-diabetic biguanide metformin in cancer treatment, and these trials are based on epidemiological observations linking metformin use in diabetics to reduced cancer incidence and the multi-faceted ability of metformin to redundantly reprogram energy metabolism at both the organismal and cellular levels.10-37 Given the intrinsic metabolic flexibility of cancer cells, we recently envisioned that cancer cells could elude the metabolic stress-mediated signal transduction pathway targeted by metformin. To anticipate these obstacles, we explored the transcriptomic and signaling pathways activated upon the chronic metformin exposure of MCF-7 cells, a widely studied model for hormone-dependent human breast cancer. Our current findings establish, for the first time, that a “global” targeting of metabolic reprogramming using metformin certainly imposes great selective pressure for the emergence of resistant breast cancer cells. Intriguingly, acquired resistance to metformin in breast cancer cells appears to trigger a transcriptome reprogramming toward a degradome-related metastatic profile, as many genes encoding extracellular matrix secreted and cell membrane-associated proteases, all of which are commonly involved in cancer cell migration and invasion, were included in the signature. These findings suggest a convergent activation of pathways underlying tumor-microenvironment interactions when the cancer cells adapt to the metabolic challenges of drugs targeting various metabolic pathways, such as the biguanide metformin.
Metformin-refractory MCF-7/MET-R cells drastically increased (30-fold upregulation vs. metformin-naïve MCF-7 cells) the expression of KLK11 (Table 2), a gene encoding a cell-surface-expressed type II, trypsin-like transmembrane serine protease that was originally identified as one of the most highly upregulated genes in prostate cancer.42-46 Kallikrein-related peptidases (KLKs) are enzymes with extracellular hydrolysis activities, such as the activation and/or degradation of their substrates, including growth factors, extracellular matrix (ECM) proteins, other cancer-associated proteases, cell membrane-bound, and adhesion proteins. These serine proteases were among the first proteolytic enzymes to be studied extensively in the “degradome”, i.e., the complete set of proteases expressed at a given time within a cell, tissue, or organism.47-51 KLK11 protein expression has been shown to be highly expressed at sites of bone metastasis and in late-stage primary tumors, suggesting a role in tumor progression. Accordingly, in vivo studies demonstrated that overexpression of KLK11 led to tumor progression and metastasis. KLK11 expressed in ER-positive breast cancer cells, such as MCF-7, has been suggested to play a crucial role in breast cancer progression by increasing the bioavailability of insulin growth factors (IGFs) via the degradation of IGF binding protein-3 (IGFBP-3).52
Table 2. Top molecules in the transcriptomic signature of metformin-adapted MCF-7/MET-R cells.
| Molecules | Fold-change |
|---|---|
| KLK11 | ↑30,460 |
| CTSF | ↑24,119 |
| TNFSF10 | ↑17,960 |
| TNFAIP2 | ↑13,558 |
| TMTC1 | ↑13,390 |
| DCLK1 | ↑12,452 |
| MGLL | ↑12,360 |
| WISP2 | ↑10,812 |
| FRAS1 | ↑10,342 |
| CYP1B1 | ↑10,268 |
| CXorf61 | ↓23,062 |
| PMP22 | ↓14,517 |
| S100A14 | ↓14,192 |
| TFF3 | ↓13,100 |
| DKK1 | ↓12,359 |
| TFF1 | ↓10,680 |
| BCL2 | ↓10,660 |
| CYP26B1 | ↓9,515 |
| MGP | ↓9,131 |
| S100A16 | ↓8,254 |
Metformin-refractory MCF-7/MET-R cells exhibited a drastic increase (24-fold upregulation vs. metformin-naïve MCF-7 cells) in the expression of CTSF (Table 2), a gene coding for Cathepsin F, a member of the degradome cysteine proteases.53 Although only little data are available on Cathepsin F, several human cancer cell lines have increased expression of CTSF compared with its normal counterpart, suggesting that this enzyme could be involved in degradative processes during tumor progression.53-55 Cathepsins are a class of globular proteases that were initially described as intracellular peptide hydrolases, although several cathepsins also have extracellular functions. Most studies have confirmed that cathepsins are highly expressed in invasive tumors, and they mediate degradation of the ECM and collagen, increase the motility and invasion of cancer cells, mediate the dissemination of cancer cells, and induce the EMT and angiogenesis.56-59 Cathepsins can also activate other proteases, thereby indirectly affecting invasion by participating in proteolytic cascades; moreover, cathepsins can inactivate key cell adhesion factors involved in the maintenance of the epithelial phenotype by cleaving cell surface proteins, such as E-cadherin, thus abrogating its cell–cell adhesion function and promoting tumor cell invasion.
Metformin-refractory MCF-7/MET-R cells intriguingly had increased (18-fold upregulation vs. metformin-naïve MCF-7 cells) expression of tumor necrosis factor (ligand) superfamily, member 10 (TNFSF10/TRAIL) (Table 2).60-63 The protein encoded by this gene is a cytokine that belongs to the tumor necrosis factor (TNF) ligand family, which preferentially induces apoptosis in transformed and tumor cells, but does not appear to kill normal cells, even though it is expressed at a significant level in most normal tissues. However, in cells with a weak caspase-3 signaling cascade, the apoptotic effects of TNFSF10 require the caspase-8-mediated cleavage of the BH3-only BCL2/Bcl-2 family member BID to activate the intrinsic apoptosis pathway.64 Indeed, there are mechanisms that tightly control TNFSF10-induced apoptosis, which are utilized by cancer cells to counteract the cytotoxicity of TNFSF10. In this regard, it has been reported that TNFSF10 is able to induce autophagy in certain cancer cells, protecting them by blunting the cytotoxicity of TNFSF10 and possibly contributing to TNFSF10 resistance.65-68 The anti-apoptotic BCL2 family proteins, such as BCL2, bind beclin-1 (BECN1) to inhibit autophagy, and the dissociation of BCL2 family proteins from BECN1 promotes autophagy.69-71 Because MCF-7 human breast carcinoma cells do not express caspase-3, earlier studies have shown that TNFSF10 induces autophagy in MCF-7 cells, and autophagy is protective against the cytotoxicity of TNFSF10 in these cells,72 the prominent augmentation of TNFSF10 gene expression is accompanied by a severe inhibition of BCL2 (11-fold downregulation vs. metformin-naïve MCF-7 cells), which strongly suggests that the activation of protective autophagy plays a causative role in the acquisition of resistance to metformin. The metformin-refractory MCF-7/MET-R cells also activated (14-fold vs. metformin-naïve MCF-7 cells) the expression of TNFAIP2, which encodes tumor necrosis factor α (TNFα)-inducible protein 2. Similar to TNFα, which is an inflammatory cytokine that is present in the microenvironment of many tumors and is known to promote tumor progression, TNFAIP2 is a cell migration- and invasion-promoting protein, and its expression predicts shorter metastasis-free survival in cancer patients.73
It is worth noting that acquired resistance to metformin resulted in a drastic augmentation (12-fold upregulation in metformin-refractory MCF-7/MET-R cells vs. metformin-naïve MCF-7 cells) of DCLK1 gene expression (Table 2). Doublecortin-like kinase 1 was originally described as a marker that was able to distinguish between Dclk1-positive tumor stem cells and Dclk1-negative normal stem cells in the intestine.74 Later studies confirmed that Dclk1 regulates pluripotency and angiogenic factors via microRNA-dependent mechanisms, and its expression marks a morphologically distinct subpopulation of cells with stem cells properties in pancreatic cancer.75,76 In this scenario, it is tempting to suggest that chronic adaptation to metformin accelerates the retrogression from a differentiated cancer cell state to a more stem-like state endowed with enhanced migratory capacities (i.e., the “migrating cancer stem cells” concept originally proposed by Thomas Brabletz).77-80 It is also worth mentioning that the other protease gene that is notably upregulated upon acquisition of metformin resistance, in addition to KLK11 and CTSF, is the gene coding for the serine hydrolase enzyme monoacylglycerol lipase (MAGL) (Table 2). MAGL is overexpressed in aggressive types of tumor cells, where it regulates a fatty acid network enriched in oncogenic signaling lipids that promote migration, invasion, survival, and in vivo tumor growth.81-83 The overexpression of MAGL in nonaggressive cancer cells is sufficient to increase their pathogenicity by recapitulating this fatty acid network, thus revealing how cancer cells can co-opt a lipolytic enzyme to translate their lipogenic state into an array of protumorigenic signals. Indeed, MAGL’s unique role of providing lipolytic sources of free fatty acids (FFAs) for the synthesis of oncogenic signaling lipids that promote cancer aggressiveness, together with the fact that MAGL blockade impairs cell migration, invasiveness, and tumorigenicity by lowering the levels of FFAs and protumorigenic signaling lipids,84-86 strongly suggest that, in response to the expected chronic inactivation of several lipogenic enzymes and lipogenesis imposed by metformin, the metformin-refractory MCF-7/MET-R cells re-activate the very same lipogenic state that is commonly controlled by metformin’s targets (AMPK, acetyl-CoA carboxylase, mTOR) via MAGL. The serine proteinase degradome gene FREM1 (FRAS1-related extracellular matrix 1/signalase-like 1)87,88 and Wnt-induced signaling protein-2 (WISP2/CCN5), a gene coding for a metalloproteinase substrate implicated in the modification of the ECM, invasion, and angiogenesis that has been linked to a variety of human cancer types and may contribute to cancer metastasis,89,90 were also significantly upregulated in the metformin-refractory MCF-7/MET-R cells (Table 2).
Metformin-refractory MCF-7/MET-R cells drastically decreased (14-fold downregulation vs. metformin-naïve MCF-7 cells) the expression of PMP22/GAS3 (Table 2), a putative tumor suppressor gene. PMP22/gas3 overexpression was originally found to induce an apoptotic-like phenotype,91 and recent studies have revealed that the induction of GAS3 inhibits breast cancer by inhibiting the attachment and proliferation of the tumor cells, at least in part by blocking the interaction of β1 integrin with fibronectin.92 Indeed, the tumor-suppressive activity of GAS3 is related to the significantly increased metastasis-free survival of breast cancer patients. Another top molecule notably decreased upon acquisition of metformin resistance was the S100A14 (S100 calcium binding protein A14) gene (14-fold downregulation vs. metformin-naïve MCF-7 cells; Table 2). The levels of the protein encoded by the S100A14 gene have been found to be lower in cancerous tissues and are associated with higher metastatic potential and advanced clinical stage, suggesting this gene has a tumor suppressor function.93,94 The expression of the Trefoil factor 3 (TFF3) gene, which has been identified as a part of a gene expression signature of biologically aggressive basal-like and claudin-low breast carcinomas that are characterized by reduced expression levels or loss of epigenetic biomarker genes, such as E-cadherin and estrogen receptor, due to aberrant DNA hypermethylation,95 was found to be notably decreased (13-fold downregulation) in the metformin-refractory MCF-7/MET-R cells compared with the metformin-naïve MCF-7 cells (Table 2). The MCF-7/MET-R cells notably lost (12-fold downregulation vs. metformin-naïve MCF-7 cells) the expression of the Dickkopf1 (DKK1) gene (Table 2), which encodes a secreted inhibitor of the Wnt/β-catenin pathway and may have tumor suppressor functions.96-98 Exogenous expression of DKK1 in human malignant breast cancer cells with mesenchymal-like phenotype significantly reduces the expression of EMT-promoting factors, such as SLUG and TWIST;99 conversely, silencing DKK1 expression in non-tumorigenic epithelial breast cells leads to increased invasive capacity and decreased E-cadherin expression.100 Together, these findings strongly suggest that the negative effect of DKK1 on the EMT is part of the suppressive reprogramming that occurs when epithelial MCF-7 breast cancer cells adapt to the continuous presence of metformin.
Mounting evidence supports the idea that deregulated cellular metabolism is linked to drug resistance in cancer therapy.7-19,101 Although the demonstration of resistance to oncogene-mediated targeted therapy through the adaptation of cellular metabolism suggests that the rewiring of cellular metabolism plays a fundamental, convergent role for oncogenes and signal transduction in promoting tumorigenesis, little is known about how the cancer signaling networks are remodeled and which pathways are invoked to sustain survival in the presence of drugs targeting central key signaling metabolic hubs (e.g., AMPK, mTOR) that respond to an array of signaling metabolic inputs and regulate a range of downstream effector metabolic pathways. Together, our current findings suggest, for the first time, that chronic adaptation to high doses of the AMPK agonist/mTOR inhibitor metformin appears to causally involve 2 highly intertwined molecular phenomena underlying enhanced cancer aggressiveness. On the one hand, low-proliferative MCF-7/MET-R cells appear to circumvent mitotic catastrophe-induced cell death by becoming polyploid cells and increasing genome instability; on the other hand, genomically unstable MCF-7/MET-R cells appear to simultaneously acquire a metastatic profile, as many genes encoding extracellular matrix secreted and cell membrane-associated proteases involved in cancer cell migration and invasion were included in the signature. Because adhesion-dependent loss of genomic surveillance mechanisms can significantly increase genome instability, the possibility of a reciprocal relationship exists between the activation of the cellular degradome and increased genome instability as a previously unrecognized mechanism of resistance to multi-targeted metabolic drugs, such as metformin. Indeed, it is reasonable to suggest that the unique mechanism of acquired resistance to metformin has opposing roles in growth and metastatic dissemination, while refractoriness to metformin limits breast cancer cell growth, likely due to an aberrant mitotic/cytokinetic machinery and accelerated autophagy, it notably increases the potential of metastatic dissemination by amplifying the number of pro-migratory and stemness inputs via the activation of a significant number of proteases and EMT regulators. Future studies should unambiguously elucidate whether our findings using supra-physiological concentrations of metformin mechanistically recapitulate the processes through which the induction of a migratory-stemness cellular state paradoxically occurs in a polyploid, senescent–autophagic scenario102-105 that is triggered by the chronic metabolic stresses that commonly occur during cancer development and after treatment with cancer drugs.
Materials and Methods
Cell viability assays
The effect of metformin on cell viability was determined using a standard colorimetric 3,4,5-dimethylthiazol-2-yl-2,5-diphenyl-tetrazolium bromide (MTT) reduction assay. For each treatment, the percent cell viability was calculated using the following equation: (OD570 of the treated sample/OD570 of the untreated sample) ×100.
Agilent gene chip analyses
Total RNA isolated from metformin-naïve MCF-7 parental cells and one pooled population of metformin-refractory MCF-7 cells (i.e., MCF-7/MET-R cells) grown in the presence of metformin was isolated with TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. The RNA quantity and quality were determined using the RNA 6000 Nano Assay kit on an Agilent 2100 BioAnalyzer (Agilent Technologies) as recommended. Agilent Human Whole Genome Microarrays (G4112F) containing 45220 probes were then hybridized. Briefly, 500 ng of total RNA from each sample was amplified by Oligo-dT-T7 reverse transcription and labeled by in vitro transcription with T7 RNA polymerase in the presence of Cy5-CTP or Cy3-CTP using the Quick Amp Labeling Kit (Agilent) and purified using RNeasy columns (Qiagen). After fragmentation, 825 ng of labeled cRNA from each of the 2 samples was cohybridized in in situ hybridization buffer (Agilent) for 17 h at 65 °C and washed at room temperature (RT) for 1 min in Gene Expression Wash Buffer 1 (Agilent) and 1 min at 37 °C in Gene Expression Wash Buffer 2 (Agilent).
Statistical analysis of microarray data
The images were generated on a confocal microarray scanner (G2565BA, Agilent) at 5-μm resolution and quantified using GenePix 6.0 software (Molecular Dynamics). Spots with signal intensities that were twice that of the local background, not saturated, and not flagged by GenePix were considered reliable. Extracted intensities were background-corrected, and the log2 ratios were normalized in an intensity-dependent fashion by the global LOWESS method (intra-chip normalization). Normalized log2 ratios were scaled between arrays to allow comparisons between all data. The raw data were processed using MMARGE, a web implementation of Limma (a microarray analysis library developed within the Bioconductor Project in the R statistical environment). To identify genes that were differentially expressed, the multiclass SAM (significance analysis of microarrays) procedure was applied. Probes with Q values (FDR) below 5% and fold changes exceeding 2.0 in absolute value were initially selected as the relevant spots. The microarray probes were collapsed to genes by considering the median log2 ratio of the respective probes per gene.
Ingenuity analysis
Gene networks were constructed using Ingenuity Pathway Analysis (Ingenuity® Systems). Data sets containing identifiers of genes that were >2.0-fold up- or downregulated were uploaded into the application. These “focus genes” were overlaid onto a global molecular network developed from information contained in the Ingenuity Pathway Knowledge Base. Networks of these “focus genes” (nodes) were algorithmically generated based on the principle that highly connected gene networks are the most biologically meaningful networks. All edges were supported by at least one reference from the literature stored in the Ingenuity Pathway Knowledge Base (the IPA interaction database is manually curated by scientists and updated quarterly). Briefly, the user-input or “focus genes” list was compared with the “global molecular network” (GMN) database, consisting of thousands of genes and interactions. The focus genes were sorted based on highest to lowest connectivity within the GMN, and networks of approximately 35 genes were grown starting with the most connected focus gene. IPA assigns a P value for a network of size n and an input focus gene list of size f by calculating the probability of finding f or more focus genes in a randomly selected set of n genes from the GMN. The intensity of the node color indicated the degree of expression (green scale for downregulated nodes; red scale for upregulated nodes). The nodes were displayed using various shapes, each of which represents a functional class of the gene products. The score indicated the likelihood of the genes in a network being found together due to random chance.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was financially supported by the Ministerio de Ciencia e Innovación (SAF2012–38914), Plan Nacional de I+D+I, MICINN, Spain.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/27982
References
- 1.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 2013;13:714–26. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 2.Muñoz-Pinedo C, El Mjiyad N, Ricci JE. Cancer metabolism: current perspectives and future directions. Cell Death Dis. 2012;3:e248. doi: 10.1038/cddis.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butler EB, Zhao Y, Muñoz-Pinedo C, Lu J, Tan M. Stalling the engine of resistance: targeting cancer metabolism to overcome therapeutic resistance. Cancer Res. 2013;73:2709–17. doi: 10.1158/0008-5472.CAN-12-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galluzzi L, Kepp O, Vander Heiden MG, Kroemer G. Metabolic targets for cancer therapy. Nat Rev Drug Discov. 2013;12:829–46. doi: 10.1038/nrd4145. [DOI] [PubMed] [Google Scholar]
- 5.Sotgia F, Martinez-Outschoorn UE, Lisanti MP. Cancer metabolism: new validated targets for drug discovery. Oncotarget. 2013;4:1309–16. doi: 10.18632/oncotarget.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao Y, Butler EB, Tan M. Targeting cellular metabolism to improve cancer therapeutics. Cell Death Dis. 2013;4:e532. doi: 10.1038/cddis.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menendez JA, Joven J, Cufí S, Corominas-Faja B, Oliveras-Ferraros C, Cuyàs E, Martin-Castillo B, López-Bonet E, Alarcón T, Vazquez-Martin A. The Warburg effect version 2.0: metabolic reprogramming of cancer stem cells. Cell Cycle. 2013;12:1166–79. doi: 10.4161/cc.24479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cufí S, Vazquez-Martin A, Oliveras-Ferraros C, Corominas-Faja B, Cuyàs E, López-Bonet E, Martin-Castillo B, Joven J, Menendez JA. The anti-malarial chloroquine overcomes primary resistance and restores sensitivity to trastuzumab in HER2-positive breast cancer. Sci Rep. 2013;3:2469. doi: 10.1038/srep02469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cufí S, Vazquez-Martin A, Oliveras-Ferraros C, Corominas-Faja B, Urruticoechea A, Martin-Castillo B, Menendez JA. Autophagy-related gene 12 (ATG12) is a novel determinant of primary resistance to HER2-targeted therapies: utility of transcriptome analysis of the autophagy interactome to guide breast cancer treatment. Oncotarget. 2012;3:1600–14. doi: 10.18632/oncotarget.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cufi S, Corominas-Faja B, Vazquez-Martin A, Oliveras-Ferraros C, Dorca J, Bosch-Barrera J, Martin-Castillo B, Menendez JA. Metformin-induced preferential killing of breast cancer initiating CD44+CD24-/low cells is sufficient to overcome primary resistance to trastuzumab in HER2+ human breast cancer xenografts. Oncotarget. 2012;3:395–8. doi: 10.18632/oncotarget.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirsch HA, Iliopoulos D, Tsichlis PN, Struhl K. Metformin selectively targets cancer stem cells, and acts together with chemotherapy to block tumor growth and prolong remission. Cancer Res. 2009;69:7507–11. doi: 10.1158/0008-5472.CAN-09-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iliopoulos D, Hirsch HA, Struhl K. Metformin decreases the dose of chemotherapy for prolonging tumor remission in mouse xenografts involving multiple cancer cell types. Cancer Res. 2011;71:3196–201. doi: 10.1158/0008-5472.CAN-10-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skinner HD, Sandulache VC, Ow TJ, Meyn RE, Yordy JS, Beadle BM, Fitzgerald AL, Giri U, Ang KK, Myers JN. TP53 disruptive mutations lead to head and neck cancer treatment failure through inhibition of radiation-induced senescence. Clin Cancer Res. 2012;18:290–300. doi: 10.1158/1078-0432.CCR-11-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Storozhuk Y, Hopmans SN, Sanli T, Barron C, Tsiani E, Cutz JC, Pond G, Wright J, Singh G, Tsakiridis T. Metformin inhibits growth and enhances radiation response of non-small cell lung cancer (NSCLC) through ATM and AMPK. Br J Cancer. 2013;108:2021–32. doi: 10.1038/bjc.2013.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zannella VE, Dal Pra A, Muaddi H, McKee TD, Stapleton S, Sykes J, Glicksman R, Chaib S, Zamiara P, Milosevic M, et al. Reprogramming metabolism with metformin improves tumor oxygenation and radiotherapy response. Clin Cancer Res. 2013;19:6741–50. doi: 10.1158/1078-0432.CCR-13-1787. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 16.Lonardo E, Cioffi M, Sancho P, Sanchez-Ripoll Y, Trabulo SM, Dorado J, Balic A, Hidalgo M, Heeschen C. Metformin targets the metabolic achilles heel of human pancreatic cancer stem cells. PLoS One. 2013;8:e76518. doi: 10.1371/journal.pone.0076518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pollak M. Potential applications for biguanides in oncology. J Clin Invest. 2013;123:3693–700. doi: 10.1172/JCI67232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morgillo F, Sasso FC, Della Corte CM, Festino L, Manzo A, Martinelli E, Troiani T, Capuano A, Ciardiello F. Metformin in lung cancer: rationale for a combination therapy. Expert Opin Investig Drugs. 2013;22:1401–9. doi: 10.1517/13543784.2013.828691. [DOI] [PubMed] [Google Scholar]
- 19.Hirsch HA, Iliopoulos D, Struhl K. Metformin inhibits the inflammatory response associated with cellular transformation and cancer stem cell growth. Proc Natl Acad Sci U S A. 2013;110:972–7. doi: 10.1073/pnas.1221055110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komurov K, Tseng JT, Muller M, Seviour EG, Moss TJ, Yang L, Nagrath D, Ram PT. The glucose-deprivation network counteracts lapatinib-induced toxicity in resistant ErbB2-positive breast cancer cells. Mol Syst Biol. 2012;8:596. doi: 10.1038/msb.2012.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Locasale JW. Metabolic rewiring drives resistance to targeted cancer therapy. Mol Syst Biol. 2012;8:597. doi: 10.1038/msb.2012.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009;23:537–48. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin-Castillo B, Dorca J, Vazquez-Martin A, Oliveras-Ferraros C, Lopez-Bonet E, Garcia M, Del Barco S, Menendez JA. Incorporating the antidiabetic drug metformin in HER2-positive breast cancer treated with neo-adjuvant chemotherapy and trastuzumab: an ongoing clinical-translational research experience at the Catalan Institute of Oncology. Ann Oncol. 2010;21:187–9. doi: 10.1093/annonc/mdp494. [DOI] [PubMed] [Google Scholar]
- 27.Martin-Castillo B, Vazquez-Martin A, Oliveras-Ferraros C, Menendez JA. Metformin and cancer: doses, mechanisms and the dandelion and hormetic phenomena. Cell Cycle. 2010;9:1057–64. doi: 10.4161/cc.9.6.10994. [DOI] [PubMed] [Google Scholar]
- 28.Vazquez-Martin A, Oliveras-Ferraros C, Del Barco S, Martin-Castillo B, Menendez JA. The anti-diabetic drug metformin suppresses self-renewal and proliferation of trastuzumab-resistant tumor-initiating breast cancer stem cells. Breast Cancer Res Treat. 2011;126:355–64. doi: 10.1007/s10549-010-0924-x. [DOI] [PubMed] [Google Scholar]
- 29.Vazquez-Martin A, Oliveras-Ferraros C, Cufí S, Martin-Castillo B, Menendez JA. Metformin and energy metabolism in breast cancer: from insulin physiology to tumour-initiating stem cells. Curr Mol Med. 2010;10:674–91. doi: 10.2174/156652410792630625. [DOI] [PubMed] [Google Scholar]
- 30.Del Barco S, Vazquez-Martin A, Cufí S, Oliveras-Ferraros C, Bosch-Barrera J, Joven J, Martin-Castillo B, Menendez JA. Metformin: multi-faceted protection against cancer. Oncotarget. 2011;2:896–917. doi: 10.18632/oncotarget.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oliveras-Ferraros C, Cufí S, Vazquez-Martin A, Menendez OJ, Bosch-Barrera J, Martin-Castillo B, Joven J, Menendez JA. Metformin rescues cell surface major histocompatibility complex class I (MHC-I) deficiency caused by oncogenic transformation. Cell Cycle. 2012;11:865–70. doi: 10.4161/cc.11.5.19252. [DOI] [PubMed] [Google Scholar]
- 32.Cufí S, Vazquez-Martin A, Oliveras-Ferraros C, Quirantes R, Segura-Carretero A, Micol V, Joven J, Bosch-Barrera J, Del Barco S, Martin-Castillo B, et al. Metformin lowers the threshold for stress-induced senescence: a role for the microRNA-200 family and miR-205. Cell Cycle. 2012;11:1235–46. doi: 10.4161/cc.11.6.19665. [DOI] [PubMed] [Google Scholar]
- 33.Menendez JA, Oliveras-Ferraros C, Cufí S, Corominas-Faja B, Joven J, Martin-Castillo B, Vazquez-Martin A. Metformin is synthetically lethal with glucose withdrawal in cancer cells. Cell Cycle. 2012;11:2782–92. doi: 10.4161/cc.20948. [DOI] [PubMed] [Google Scholar]
- 34.Corominas-Faja B, Quirantes-Piné R, Oliveras-Ferraros C, Vazquez-Martin A, Cufí S, Martin-Castillo B, Micol V, Joven J, Segura-Carretero A, Menendez JA. Metabolomic fingerprint reveals that metformin impairs one-carbon metabolism in a manner similar to the antifolate class of chemotherapy drugs. Aging (Albany NY) 2012;4:480–98. doi: 10.18632/aging.100472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vazquez-Martin A, Cufi S, Lopez-Bonet E, Corominas-Faja B, Oliveras-Ferraros C, Martin-Castillo B, Menendez JA. Metformin limits the tumourigenicity of iPS cells without affecting their pluripotency. Sci Rep. 2012;2:964. doi: 10.1038/srep00964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Menendez JA, Joven J. One-carbon metabolism: an aging-cancer crossroad for the gerosuppressant metformin. Aging (Albany NY) 2012;4:894–8. doi: 10.18632/aging.100523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cufí S, Corominas-Faja B, Lopez-Bonet E, Bonavia R, Pernas S, López IÁ, Dorca J, Martínez S, López NB, Fernández SD, et al. Dietary restriction-resistant human tumors harboring the PIK3CA-activating mutation H1047R are sensitive to metformin. Oncotarget. 2013;4:1484–95. doi: 10.18632/oncotarget.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamaguchi R, Perkins G. Finding a Panacea among combination cancer therapies. Cancer Res. 2012;72:18–23. doi: 10.1158/0008-5472.CAN-11-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamaguchi R, Perkins G. Challenges in targeting cancer metabolism for cancer therapy. EMBO Rep. 2012;13:1034–5. doi: 10.1038/embor.2012.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson GL, Dinsdale D, Macfarlane M, Cain K. Switching from aerobic glycolysis to oxidative phosphorylation modulates the sensitivity of mantle cell lymphoma cells to TRAIL. Oncogene. 2012;31:4996–5006. doi: 10.1038/onc.2012.13. [DOI] [PubMed] [Google Scholar]
- 41.Ros S, Santos CR, Moco S, Baenke F, Kelly G, Howell M, Zamboni N, Schulze A. Functional metabolic screen identifies 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 as an important regulator of prostate cancer cell survival. Cancer Discov. 2012;2:328–43. doi: 10.1158/2159-8290.CD-11-0234. [DOI] [PubMed] [Google Scholar]
- 42.Diamandis EP, Okui A, Mitsui S, Luo LY, Soosaipillai A, Grass L, Nakamura T, Howarth DJ, Yamaguchi N. Human kallikrein 11: a new biomarker of prostate and ovarian carcinoma. Cancer Res. 2002;62:295–300. [PubMed] [Google Scholar]
- 43.Nakamura T, Scorilas A, Stephan C, Jung K, Soosaipillai AR, Diamandis EP. The usefulness of serum human kallikrein 11 for discriminating between prostate cancer and benign prostatic hyperplasia. Cancer Res. 2003;63:6543–6. [PubMed] [Google Scholar]
- 44.Paliouras M, Borgono C, Diamandis EP. Human tissue kallikreins: the cancer biomarker family. Cancer Lett. 2007;249:61–79. doi: 10.1016/j.canlet.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 45.Kontos CK, Scorilas A. Kallikrein-related peptidases (KLKs): a gene family of novel cancer biomarkers. Clin Chem Lab Med. 2012;50:1877–91. doi: 10.1515/cclm-2012-0247. [DOI] [PubMed] [Google Scholar]
- 46.Borgoño CA, Michael IP, Diamandis EP. Human tissue kallikreins: physiologic roles and applications in cancer. Mol Cancer Res. 2004;2:257–80. [PubMed] [Google Scholar]
- 47.Puente XS, Sánchez LM, Overall CM, López-Otín C. Human and mouse proteases: a comparative genomic approach. Nat Rev Genet. 2003;4:544–58. doi: 10.1038/nrg1111. [DOI] [PubMed] [Google Scholar]
- 48.Quesada V, Ordóñez GR, Sánchez LM, Puente XS, López-Otín C. The Degradome database: mammalian proteases and diseases of proteolysis. Nucleic Acids Res. 2009;37:D239–43. doi: 10.1093/nar/gkn570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ordóñez GR, Puente XS, Quesada V, López-Otín C. Proteolytic systems: constructing degradomes. Methods Mol Biol. 2009;539:33–47. doi: 10.1007/978-1-60327-003-8_2. [DOI] [PubMed] [Google Scholar]
- 50.Fraile JM, Ordóñez GR, Quirós PM, Astudillo A, Galván JA, Colomer D, López-Otín C, Freije JM, Puente XS. Identification of novel tumor suppressor proteases by degradome profiling of colorectal carcinomas. Oncotarget. 2013;4:1931–2. doi: 10.18632/oncotarget.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shahinian H, Loessner D, Biniossek ML, Kizhakkedathu JN, Clements JA, Magdolen V, Schilling O. Secretome and degradome profiling shows that Kallikrein-related peptidases 4, 5, 6, and 7 induce TGFβ-1 signaling in ovarian cancer cells. Mol Oncol. 2014;8:68–82. doi: 10.1016/j.molonc.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sano A, Sangai T, Maeda H, Nakamura M, Hasebe T, Ochiai A. Kallikrein 11 expressed in human breast cancer cells releases insulin-like growth factor through degradation of IGFBP-3. Int J Oncol. 2007;30:1493–8. [PubMed] [Google Scholar]
- 53.Santamaría I, Velasco G, Pendás AM, Paz A, López-Otín C. Molecular cloning and structural and functional characterization of human cathepsin F, a new cysteine proteinase of the papain family with a long propeptide domain. J Biol Chem. 1999;274:13800–9. doi: 10.1074/jbc.274.20.13800. [DOI] [PubMed] [Google Scholar]
- 54.Krueger S, Kellner U, Buehling F, Roessner A. Cathepsin L antisense oligonucleotides in a human osteosarcoma cell line: effects on the invasive phenotype. Cancer Gene Ther. 2001;8:522–8. doi: 10.1038/sj.cgt.7700341. [DOI] [PubMed] [Google Scholar]
- 55.Vazquez-Ortiz G, Pina-Sanchez P, Vazquez K, Duenas A, Taja L, Mendoza P, Garcia JA, Salcedo M. Overexpression of cathepsin F, matrix metalloproteinases 11 and 12 in cervical cancer. BMC Cancer. 2005;5:68. doi: 10.1186/1471-2407-5-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mohamed MM, Sloane BF. Cysteine cathepsins: multifunctional enzymes in cancer. Nat Rev Cancer. 2006;6:764–75. doi: 10.1038/nrc1949. [DOI] [PubMed] [Google Scholar]
- 57.López-Otín C, Matrisian LM. Emerging roles of proteases in tumour suppression. Nat Rev Cancer. 2007;7:800–8. doi: 10.1038/nrc2228. [DOI] [PubMed] [Google Scholar]
- 58.Palermo C, Joyce JA. Cysteine cathepsin proteases as pharmacological targets in cancer. Trends Pharmacol Sci. 2008;29:22–8. doi: 10.1016/j.tips.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 59.Obermajer N, Jevnikar Z, Doljak B, Kos J. Role of cysteine cathepsins in matrix degradation and cell signalling. Connect Tissue Res. 2008;49:193–6. doi: 10.1080/03008200802143158. [DOI] [PubMed] [Google Scholar]
- 60.Abdulghani J, El-Deiry WS. TRAIL receptor signaling and therapeutics. Expert Opin Ther Targets. 2010;14:1091–108. doi: 10.1517/14728222.2010.519701. [DOI] [PubMed] [Google Scholar]
- 61.Allen JE, El-Deiry WS. Regulation of the human TRAIL gene. Cancer Biol Ther. 2012;13:1143–51. doi: 10.4161/cbt.21354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Walczak H. Death receptor-ligand systems in cancer, cell death, and inflammation. Cold Spring Harb Perspect Biol. 2013;5:a008698. doi: 10.1101/cshperspect.a008698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dimberg LY, Anderson CK, Camidge R, Behbakht K, Thorburn A, Ford HL. On the TRAIL to successful cancer therapy? Predicting and counteracting resistance against TRAIL-based therapeutics. Oncogene. 2013;32:1341–50. doi: 10.1038/onc.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Crowder RN, El-Deiry WS. Caspase-8 regulation of TRAIL-mediated cell death. Exp Oncol. 2012;34:160–4. [PubMed] [Google Scholar]
- 65.Han J, Hou W, Goldstein LA, Lu C, Stolz DB, Yin XM, Rabinowich H. Involvement of protective autophagy in TRAIL resistance of apoptosis-defective tumor cells. J Biol Chem. 2008;283:19665–77. doi: 10.1074/jbc.M710169200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hou W, Han J, Lu C, Goldstein LA, Rabinowich H. Enhancement of tumor-TRAIL susceptibility by modulation of autophagy. Autophagy. 2008;4:940–3. doi: 10.4161/auto.6769. [DOI] [PubMed] [Google Scholar]
- 67.Herrero-Martín G, Høyer-Hansen M, García-García C, Fumarola C, Farkas T, López-Rivas A, Jäättelä M. TAK1 activates AMPK-dependent cytoprotective autophagy in TRAIL-treated epithelial cells. EMBO J. 2009;28:677–85. doi: 10.1038/emboj.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hou W, Han J, Lu C, Goldstein LA, Rabinowich H. Autophagic degradation of active caspase-8: a crosstalk mechanism between autophagy and apoptosis. Autophagy. 2010;6:891–900. doi: 10.4161/auto.6.7.13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Levine B, Sinha S, Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy. 2008;4:600–6. doi: 10.4161/auto.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Germain M, Slack RS. Dining in with BCL-2: new guests at the autophagy table. Clin Sci (Lond) 2010;118:173–81. doi: 10.1042/CS20090310. [DOI] [PubMed] [Google Scholar]
- 71.Zhou F, Yang Y, Xing D. Bcl-2 and Bcl-xL play important roles in the crosstalk between autophagy and apoptosis. FEBS J. 2011;278:403–13. doi: 10.1111/j.1742-4658.2010.07965.x. [DOI] [PubMed] [Google Scholar]
- 72.He W, Wang Q, Xu J, Xu X, Padilla MT, Ren G, Gou X, Lin Y. Attenuation of TNFSF10/TRAIL-induced apoptosis by an autophagic survival pathway involving TRAF2- and RIPK1/RIP1-mediated MAPK8/JNK activation. Autophagy. 2012;8:1811–21. doi: 10.4161/auto.22145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen LC, Chen CC, Liang Y, Tsang NM, Chang YS, Hsueh C. A novel role for TNFAIP2: its correlation with invasion and metastasis in nasopharyngeal carcinoma. Mod Pathol. 2011;24:175–84. doi: 10.1038/modpathol.2010.193. [DOI] [PubMed] [Google Scholar]
- 74.Nakanishi Y, Seno H, Fukuoka A, Ueo T, Yamaga Y, Maruno T, Nakanishi N, Kanda K, Komekado H, Kawada M, et al. Dclk1 distinguishes between tumor and normal stem cells in the intestine. Nat Genet. 2013;45:98–103. doi: 10.1038/ng.2481. [DOI] [PubMed] [Google Scholar]
- 75.Sureban SM, May R, Qu D, Weygant N, Chandrakesan P, Ali N, Lightfoot SA, Pantazis P, Rao CV, Postier RG, et al. DCLK1 regulates pluripotency and angiogenic factors via microRNA-dependent mechanisms in pancreatic cancer. PLoS One. 2013;8:e73940. doi: 10.1371/journal.pone.0073940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bailey JM, Alsina J, Rasheed ZA, McAllister FM, Fu YY, Plentz R, Zhang H, Pasricha PJ, Bardeesy N, Matsui W, et al. DCLK1 marks a morphologically distinct subpopulation of cells with stem cell properties in preinvasive pancreatic cancer. Gastroenterology. 2014;146:245–56. doi: 10.1053/j.gastro.2013.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Opinion: migrating cancer stem cells - an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5:744–9. doi: 10.1038/nrc1694. [DOI] [PubMed] [Google Scholar]
- 78.Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–95. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 79.Brabletz T. To differentiate or not--routes towards metastasis. Nat Rev Cancer. 2012;12:425–36. doi: 10.1038/nrc3265. [DOI] [PubMed] [Google Scholar]
- 80.Brabletz T. EMT and MET in metastasis: where are the cancer stem cells? Cancer Cell. 2012;22:699–701. doi: 10.1016/j.ccr.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 81.Nomura DK, Long JZ, Niessen S, Hoover HS, Ng SW, Cravatt BF. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell. 2010;140:49–61. doi: 10.1016/j.cell.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ye L, Zhang B, Seviour EG, Tao KX, Liu XH, Ling Y, Chen JY, Wang GB. Monoacylglycerol lipase (MAGL) knockdown inhibits tumor cells growth in colorectal cancer. Cancer Lett. 2011;307:6–17. doi: 10.1016/j.canlet.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 83.Nomura DK, Lombardi DP, Chang JW, Niessen S, Ward AM, Long JZ, Hoover HH, Cravatt BF. Monoacylglycerol lipase exerts dual control over endocannabinoid and fatty acid pathways to support prostate cancer. Chem Biol. 2011;18:846–56. doi: 10.1016/j.chembiol.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fowler CJ. Monoacylglycerol lipase - a target for drug development? Br J Pharmacol. 2012;166:1568–85. doi: 10.1111/j.1476-5381.2012.01950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mulvihill MM, Nomura DK. Therapeutic potential of monoacylglycerol lipase inhibitors. Life Sci. 2013;92:492–7. doi: 10.1016/j.lfs.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Louie SM, Roberts LS, Mulvihill MM, Luo K, Nomura DK. Cancer cells incorporate and remodel exogenous palmitate into structural and oncogenic signaling lipids. Biochim Biophys Acta. 2013;1831:1566–72. doi: 10.1016/j.bbalip.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Smyth I, Du X, Taylor MS, Justice MJ, Beutler B, Jackson IJ. The extracellular matrix gene Frem1 is essential for the normal adhesion of the embryonic epidermis. Proc Natl Acad Sci U S A. 2004;101:13560–5. doi: 10.1073/pnas.0402760101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pavlakis E, Chiotaki R, Chalepakis G. The role of Fras1/Frem proteins in the structure and function of basement membrane. Int J Biochem Cell Biol. 2011;43:487–95. doi: 10.1016/j.biocel.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 89.Ji J, Jia S, Ji K, Jiang WG. Wnt1 inducible signalling pathway protein-2 (WISP-2/CCN5): Roles and regulation in human cancers. Oncol Rep. 2013 doi: 10.3892/or.2013.2909. [DOI] [PubMed] [Google Scholar]
- 90.Russo JW, Castellot JJ. CCN5: biology and pathophysiology. J Cell Commun Signal. 2010;4:119–30. doi: 10.1007/s12079-010-0098-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fabbretti E, Edomi P, Brancolini C, Schneider C. Apoptotic phenotype induced by overexpression of wild-type gas3/PMP22: its relation to the demyelinating peripheral neuropathy CMT1A. Genes Dev. 1995;9:1846–56. doi: 10.1101/gad.9.15.1846. [DOI] [PubMed] [Google Scholar]
- 92.Li YJ, Liu G, Li Y, Vecchiarelli-Federico LM, Liu JC, Zacksenhaus E, Shan SW, Yang BB, Li Q, Dash R, et al. mda-7/IL-24 Expression inhibits breast cancer through upregulation of growth arrest-specific gene 3 (gas3) and disruption of β1 integrin function. Mol Cancer Res. 2013;11:593–603. doi: 10.1158/1541-7786.MCR-12-0496. [DOI] [PubMed] [Google Scholar]
- 93.Kim G, Chung JY, Jun SY, Eom DW, Bae YK, Jang KT, Kim J, Yu E, Hong SM. Loss of S100A14 expression is associated with the progression of adenocarcinomas of the small intestine. Pathobiology. 2013;80:95–101. doi: 10.1159/000342394. [DOI] [PubMed] [Google Scholar]
- 94.Sapkota D, Costea DE, Blø M, Bruland O, Lorens JB, Vasstrand EN, Ibrahim SO. S100A14 inhibits proliferation of oral carcinoma derived cells through G1-arrest. Oral Oncol. 2012;48:219–25. doi: 10.1016/j.oraloncology.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 95.Roll JD, Rivenbark AG, Sandhu R, Parker JS, Jones WD, Carey LA, Livasy CA, Coleman WB. Dysregulation of the epigenome in triple-negative breast cancers: basal-like and claudin-low breast cancers express aberrant DNA hypermethylation. Exp Mol Pathol. 2013;95:276–87. doi: 10.1016/j.yexmp.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 96.Menezes ME, Devine DJ, Shevde LA, Samant RS. Dickkopf1: a tumor suppressor or metastasis promoter? Int J Cancer. 2012;130:1477–83. doi: 10.1002/ijc.26449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Qi L, Sun B, Liu Z, Li H, Gao J, Leng X. Dickkopf-1 inhibits epithelial-mesenchymal transition of colon cancer cells and contributes to colon cancer suppression. Cancer Sci. 2012;103:828–35. doi: 10.1111/j.1349-7006.2012.02222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Barbolina MV, Liu Y, Gurler H, Kim M, Kajdacsy-Balla AA, Rooper L, Shepard J, Weiss M, Shea LD, Penzes P, et al. Matrix rigidity activates Wnt signaling through down-regulation of Dickkopf-1 protein. J Biol Chem. 2013;288:141–51. doi: 10.1074/jbc.M112.431411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.DiMeo TA, Anderson K, Phadke P, Fan C, Perou CM, Naber S, Kuperwasser C. A novel lung metastasis signature links Wnt signaling with cancer cell self-renewal and epithelial-mesenchymal transition in basal-like breast cancer. Cancer Res. 2009;69:5364–73. doi: 10.1158/0008-5472.CAN-08-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mitra A, Menezes ME, Shevde LA, Samant RS. DNAJB6 induces degradation of beta-catenin and causes partial reversal of mesenchymal phenotype. J Biol Chem. 2010;285:24686–94. doi: 10.1074/jbc.M109.094847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Marignac VM, Smith S, Toban N, Bazile M, Aloyz R. Resistance to Dasatinib in primary chronic lymphocytic leukemia lymphocytes involves AMPK-mediated energetic re-programming. Oncotarget. 2013 doi: 10.18632/oncotarget.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Erenpreisa J, Cragg MS. Cancer: a matter of life cycle? Cell Biol Int. 2007;31:1507–10. doi: 10.1016/j.cellbi.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 103.Salmina K, Jankevics E, Huna A, Perminov D, Radovica I, Klymenko T, Ivanov A, Jascenko E, Scherthan H, Cragg M, et al. Up-regulation of the embryonic self-renewal network through reversible polyploidy in irradiated p53-mutant tumour cells. Exp Cell Res. 2010;316:2099–112. doi: 10.1016/j.yexcr.2010.04.030. [DOI] [PubMed] [Google Scholar]
- 104.Erenpreisa J, Salmina K, Huna A, Kosmacek EA, Cragg MS, Ianzini F, Anisimov AP. Polyploid tumour cells elicit paradiploid progeny through depolyploidizing divisions and regulated autophagic degradation. Cell Biol Int. 2011;35:687–95. doi: 10.1042/CBI20100762. [DOI] [PubMed] [Google Scholar]
- 105.Erenpreisa J, Cragg MS. Three steps to the immortality of cancer cells: senescence, polyploidy and self-renewal. Cancer Cell Int. 2013;13:92. doi: 10.1186/1475-2867-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.