Abstract
In recent years, there has been a great expansion in the nature of new technologies for the study of all biologic subjects at the molecular and genomic level and these have been applied to the field of human nutrition. The latter has traditionally relied on a mix of epidemiologic studies to generate hypotheses, dietary intervention studies to test these hypotheses, and a variety of experimental approaches to understand the underlying explanatory mechanisms. Both the novel and traditional approaches have begun to carve out separate identities vís-a-vís their own journals, their own international societies, and their own national and international symposia. The present review draws on the advent of large national nutritional phenotype databases and related technological developments to argue the case that there needs to be far more integration of molecular and public health nutrition. This is required to address new joint approaches to such areas as the measurement of food intake, biomarker discovery, and the genetic determinants of nutrient-sensitive genotypes and other areas such as personalized nutrition and the use of new technologies with mass application, such as in dried blood spots to replace venipuncture or portable electronic devices to monitor food intake and phenotype. Future development requires the full integration of these 2 disciplines, which will provide a challenge to both funding agencies and to university training of nutritionists.
Introduction
The sequencing of the human genome and the rapid introduction of analytical technologies in genotyping, transcriptomics, proteomics, and metabolomics have transformed all aspects of research in biology, including human nutrition. During the same period, there has been a deepening of the recognition among policy makers of the importance of human nutrition in the prevention of noncommunicable chronic disease (1). Indeed, these 2 developments have led to a growing recognition of public health nutrition as a professional qualification, such as the Association of Nutritionists in the United Kingdom, which emerged from the professionalization program of the Nutrition Society (2). The UK health services will only employ public health nutritionists who have qualified for this professional registration, which is state recognized, much like national registers of dietitians. The discipline of public health nutrition now merits its own global conferences (3) and journal (4), and equally, the development of molecular nutrition has led to its own international society, the International Society for Nutrigenomics and Nutrigenetics (5), and related journals (6, 7). There is therefore a need to create an integrated approach to human nutrition in which the power of molecular nutrition is aligned to the grand societal challenges of diet-related disease. The present article sets out to explore some of these areas in which integration is opportune.
Current Status of Knowledge
The first such area is the measurement of food intake, which is at the heart of public health nutrition. Although many techniques exist to measure food and nutrient intake, all are burdened with a significant degree of energy underreporting. The field of epidemiology corrects for this based on a correlation between the intake of energy and the intakes of individual nutrients or body weight (8). However, such computational corrections cannot be readily applied to foods. In contrast to the intake of nutrients, individuals can choose to be consumers or nonconsumers of individual foods. Thus, the mean population intakes of foods can be determined by 1) the percentage of the population consuming the food, 2) the frequency of intake among consumers, and 3) intakes of the food at each eating occasion. The possibility that the field of metabolomics might be used to characterize populations or subgroups within populations on the basis of their urinary metabolomic profiles has thus received considerable attention (9, 10). Such subgrouping has been variably referred to as metabotypes (11) or nutri-clusters (12). Metabolomics has been used to identify specific patterns of food intake in acute intervention studies (13), short-term intervention studies (10), chronic intervention studies (14), and cross-sectional studies (12). In general, acute and short-term intervention studies have produced reasonably robust models. However, the longer term intervention studies and the cross-sectional studies lack precision. Although metabolomic profiling can yield quite accurate patterns for very specific food groups, there is scope for distortion of the metabolomic profiles when several food groups are included due to skewed high intakes of certain foods such as coffee or alcohol. It is, of course, not outside the scope of metabolomic methodologies to eliminate signals from such foods, but there needs to be consideration as to what foods to exclude and why they might be excluded. Coffee is, for example, not a nutrient-bearing beverage, but alcohol is a significant source of energy.
Collaborative work between those experts in food consumption studies and those with skills in metabolomic profiling will eventually lead to approaches to food intake studies that complement normal food intake studies. Cluster analysis and principal components analysis have been extensively used in human nutrition to characterize complex and diverse patterns of food and nutrient intake (15). Marrying that approach with urinary metabolomics will eventually lead to technologies that will allow individuals to be defined in terms of their habitual dietary patterns. However, at present, it is impossible to see how metabolomics can assist in the measurement of energy intake or of the intakes of carbohydrate or fat given that the metabolic end products of these nutrients are lost as heat and carbon dioxide.
A second area in which nutrigenomics and public health nutrition will meet is in the construction of large-scale nutritional phenotype databases (16, 17). Such databases go beyond the normal large-scale dietary surveys to embrace not only large cross-sectional studies of diet and health but also the study of groups of smaller size, which are more intensively studied. In these cases, traditional dietary data and blood biochemical data continue to be collected, but this is enhanced by the use of the various “omics” techniques (genotyping, transcriptomics, metabolomics, and proteomics). In addition, the intensive investigation of the smaller groups will include acute challenge tests such as glucose or lipid tolerance tests as well as detailed physiologic (e.g., resting energy metabolism, aerobic fitness) and body composition imaging (e.g., DXA, MRI) measures. Such large nutritional phenotype databases are targeted in the new approach to research funding in the European Union as part of the Joint Programming Initiative (JPI)6 and specifically the JPI targeted at food and health (18). The sharing of large databases that measure diet and hefdk;aalth is a key objective of this JPI, and these databases will include national dietary surveys, large collaborative dietary intervention studies, and small detailed studies involving some element of a dietary challenge.
An example of a nationally funded nutritional phenotype database is given in Table 1. This database was constructed by a consortium of Irish universities (19) over the period 2008–2013 and provides a powerful tool for the integration of public health nutrition and molecular nutrition. The project, also known under the acronym JINGO [Joint Irish Nutrigenomics Organization (20)] was funded by competitive national research funding bodies (Department of Agriculture, Food, and the Marine and the Health Research Board in the Republic of Ireland, which accounted for €10 million of investment, and the Department of Employment and Learning in Northern Ireland, with a contribution of €1.4 million). The JINGO project is governed by a consortium agreement, which covers issues such as standard operating procedures, intellectual property, publication and authorship rules, sample storage, sample transfer, and third-party access. The logic behind the project was to establish a large, comprehensive database of free-living adults across a spectrum of ages, to include a significant metabolically challenged cohort, all of which would embrace dietary and anthropometric data alongside physiologic and nutrigenomic data. It should be added that the existence of these databases has attracted further investment from the private sector involving private-public partnerships, which has enhanced, and will continue to enhance, the analytical depth and scope of the databases.
TABLE 1.
Details of the Irish National Nutrition Phenotype Database1
| National adult nutrition survey | Metabolically challenged cohort | Targeted elderly sample | |
| Study type | Cross-sectional | Acute intervention (OGTT, OLTT) | Longitudinal |
| Sample size, n | 1500 (free-living) | 214 (free-living) | 5200 (from outpatient/general practice clinics) |
| Age range, y | 18–90 | 18–60 | 60–102 |
| Dietary data | 4-d semi-weighed food diary | FFQ | Targeted questionnaire |
| Physical activity data | Accelerometry and questionnaire | Questionnaire | Questionnaire |
| Anthropometric data | Standard | Standard | Standard |
| Body composition data | Bioelectrical impedance | DXA, plethysmography | DXA |
| Physical function data | No | Resting metabolic rate, aerobic fitness, muscle function | No |
| Targeted blood analysis | Extensive fasting profile | Extensive fasting and postprandial | Targeted |
| Gene sequence data | Yes | Yes | Yes |
| Gene expression data | No | Yes | No |
| Metabolomics data | Urine | Plasma and urine | No |
OGTT, oral-glucose-tolerance test; OLTT, oral-lipid-tolerance test.
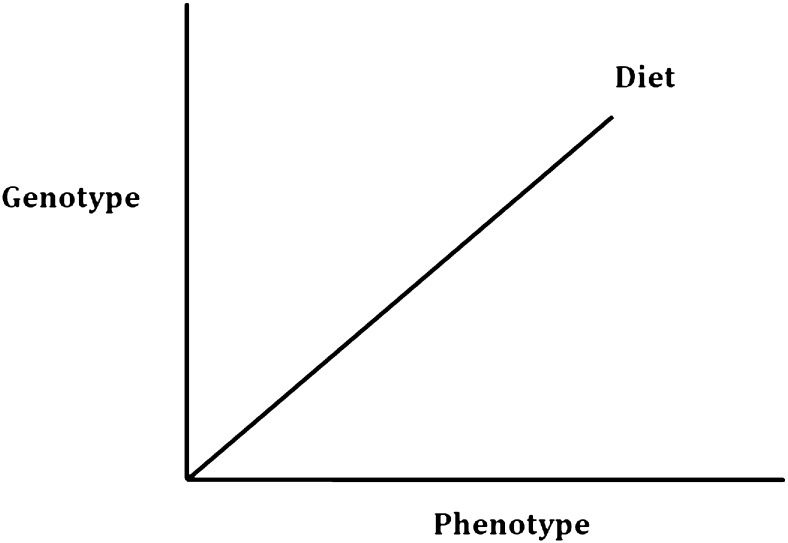
The template for the analysis of any 1 of the 3 cohorts is shown in Fig. 1, where the 3 dimensions integrate public health nutrition and molecular nutrition. The genotype dimension can be at the level of a single nucleotide polymorphism (SNP) or at a more detailed level involving multiple SNPs and a measure of gene expression. The dietary data can be confined to foods only, measures of nutrient intake, or biomarkers of nutrient intake. The phenotypic dimension can incorporate all of the measures outlined in Table 1. A typical example of the integration of these 3 dimensions will be discussed later in this article, in which studies involving a phenotype (blood pressure) are linked to dietary status (plasma riboflavin levels) and to a common genetic variant of a gene covering the synthesis of a key enzyme in one-carbon transfer.
FIGURE 1.
Generalized approach to the analysis of nutritional phenotype databases incorporating public health nutrition and molecular nutrition elements. A typical example as cited in the article might be the relation between blood pressure (phenotype) and riboflavin status (diet) as influenced by different single nucleotide polymorphisms of the gene methylenetetrahydrofolate reductase (MTHFR) (genotype).
The third area in which public health nutrition and molecular nutrition meet draws on detailed nutritional phenotype databases and involves biomarker discovery and phenotype characterization. One research tool that can provide valuable information is the nutritional challenge, in which an individual’s ability to cope with the consumption of a large nutrient load is tested. Both the dynamic metabolic response to, i.e., the signal generated, and recovery from the challenge provide critical information. This can be explained by using the concept of systems elasticity or resilience (21) in which a healthy person should generate only small signals and spring back to baseline levels rapidly, demonstrating good elasticity. In contrast a predisease state would result in a delayed response that is more sustained, with incomplete recovery to baseline, or a nonreturn to baseline in the case where disease is already present. Studying metabolic response and its pattern and components can thereby provide vital information on a person’s position within the spectrum of health to disease and their likelihood to further transition.
One such nutritional challenge test study is that which has been constructed within the Joint Irish Nutrigenomics Organisation “JINGO” Database initiative as outlined in Table 1. This study (NCT01172951) was developed specifically to test responses in a group of people deemed to be “healthy,” both by a series of fasting blood tests to rule out metabolic dysfunction and by the virtue that they did not require medication for any condition. In this study, which examined 65 adults with an average age of 32 y and a BMI of 25 kg/m2, was with respect to their postprandial responses to a glucose-tolerance test (22). Despite first being considered as a quite homogenous group of healthy adults, analysis based on the metabolic response to the large glucose load revealed 4 very distinct groups or “clusters” of individuals. The first cluster, with the most adversely perturbed glucose response also had the highest BMI, had the greatest adiposity, were the most unfit, and exhibited the poorest insulin sensitivity and biologic profile. Therefore, such analysis successfully captured different metabolic phenotypes and, most important, helped to identify those most at risk of developing a glucose-associated disease.
Other recent analyses within that challenge study have identified a relation between metabolic profile and aerobic fitness (23). In this study, maximal oxygen consumption (VO2max), a marker of aerobic fitness level, was measured after a cycle ergometer test. Participants were split into those of highest fitness (a VO2max value >57 for men, >43 mL ⋅ kg−1 ⋅ min−1 for women) and lowest fitness (<42 in men, <32 mL ⋅ kg−1 ⋅ min−1 in women) and their phenotypic characteristics compared. Unsurprisingly, those who were the fittest were less overweight. Further analysis also revealed a significantly increased fat oxidation rate in those participants when they were working at higher exercise intensities. This was accompanied by a more rapid return to normal breathing and heart rate levels after the test was completed, again demonstrative of a healthy or metabolically flexible phenotype. Targeted amino acid examination using principal components analysis further uncovered a clear separation based on fitness level, finding 20 metabolites to be significantly higher in those who were most unfit even after differences in gender, BMI, and age were taken into account. These are just 2 examples of the type of associations that a nutritional phenotype database allows to be discovered. The collation of databases into megadatabases of nutritional phenotype will truly test the power of such associations.
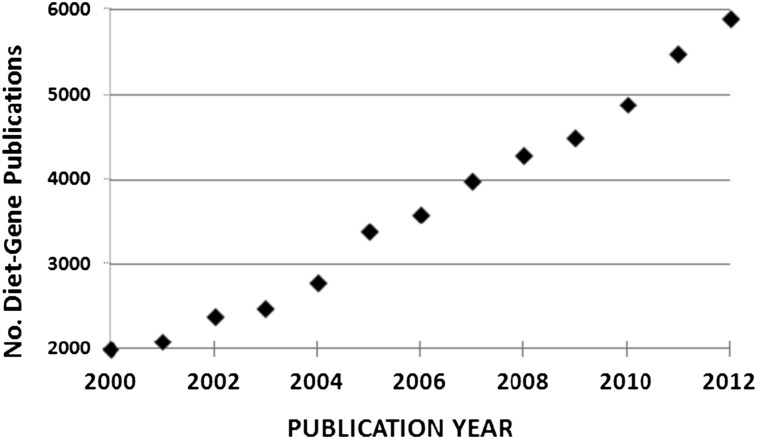
Another area in which public health nutrition and molecular nutrition need to come together is in making sense of the logarithmic growth in publications linking diet with genetic variation (Fig. 2). The majority of these are associations and as such do not prove cause and effect. The vast and expanding number of such studies must raise some concern as to their translation into public health nutrition or into understanding the manner in which a common genetic variant influences specific metabolic pathways. A strong correlation can exist between the presence of a given SNP and a metabolic response to a specific diet, but that can be confounded by the contribution of other SNPs that are not part of the target study. Even if very large genome-wide studies are used to simultaneously measure many SNPs for their role in some diet-related phenotype response, ethical considerations will require, in as far as possible, properly conducted dietary intervention studies to prove that the SNP or collection of SNPs are alone in determining the diet-related phenotype response. One of the very few examples that pursued this confirmatory path involves allelic variation of the gene for the folic acid enzyme methylenetetrahydrofolate reductase (MTHFR) and its role in the response of blood pressure (phenotype) to dietary riboflavin status as measured by biochemical status (diet determinant). Two related studies (24, 25) saw the recruitment of equal numbers of participants with the CC, TT or CT alleles of the gene for MTHFR. Half of each allelic group received an intervention consisting of a supplementation of 1.6 mg/d of riboflavin for 16 wk during which time blood pressure was monitored. The remainder received a placebo. Those participants with the C to T allele had a dramatic reduction in blood pressure with riboflavin intervention. The difference between the 2 studies is simply that the controls who received no treatment in the first study became the intervention group in the second study and vice versa. Each study confirmed this gene-nutrient interaction on blood pressure. The design and implementation of such confirmatory studies must draw on the skills of both the public health nutrition community and the molecular nutrition community.
FIGURE 2.
The growth in the number of publications identified under the term diet-gene in the PubMed database from the year 2000 to 2012.
A final area that will integrate the disciplines of both molecular and public health nutrition is the field of personalized nutrition (PN). The number of novel technologies that can be embraced to characterize health and to assess nutritional needs is rapidly advancing, not just in the field of the omics technologies but equally in the area of digitalized dietary analysis. The engagement of such innovations in information technology for the capture of dietary intake data and also electronic engineering via companion diagnostics is central in the advancement of PN. PN can be accomplished at varying degrees of complexity, from the basic level in which dietary advice is simply tailored according to an individual’s nutrient intake data, all the way to more sophisticated levels of PN, which are based on various magnitudes of phenotypic and genetic data (26). Many Web-based software tools are available to consumers for online nutritional assessment, including FFQs and 24-h recall methods (27). Such computerized dietary assessment tools can reduce the labor of more traditional methods in which data are entered manually by a nutritionist, generating automated feedback in a programmed system. The use of smart phone technology is also emerging as a new and convenient route for remote dietary intake data collection (28). However, the reliability of these methods is still a work in progress, and techniques to assess dietary intake and provide automated feedback and motivational text messages are currently being tested and developed as part of the Connecting Health and Technology Project, which began in 2012 (29).
Remote methods for the collection of phenotypic data are also emerging, with several self-assessment devices now on the market, ranging from blood glucose monitors to portable blood pressure monitors, physical activity monitors, sleep quality monitors, and many more (26). The scientific literature to merit the use of many commercial devices in the research setting is currently sparse, yet the potential for their use in PN is promising. The use of scientifically validated phenotyping devices could transform the range of health variables that can be assessed remotely by uploading data to secure Web-based systems or by sending biologically stable samples, such as buccal swabs, fecal samples, or dried blood spots, by mail to a clinician or researcher. The use of dried blood spot analysis has been used successfully for the analysis of a wide range of biochemical variables including lipids, hormones, cytokines, and vitamins (30–32).
A European Union Seventh Framework Program–integrated project, Food4Me (33), began in 2011 and set out to explore the scientific, business, and consumer aspects of PN and to determine whether dietary advice based on nutrient, phenotypic, and genetic information could deliver consumer benefits. As part of this endeavor, the project includes a proof-of-principle study in which >1280 participants from across 7 centers in Ireland, the United Kingdom, The Netherlands, Spain, Germany, Poland, and Greece have enrolled to receive online personalized dietary advice. The study is fully Internet-delivered and uses national postal services for the transfer of dried blood spot and buccal cell samples from the participants to their study center. The participants (men and women aged >18 y) who signed up enrolled via the study Web site (33), where they completed a series of screening questionnaires before being randomly assigned for 6 mo to 1 of 4 groups: level 0, nonpersonalized dietary advice (general healthy-eating guidelines); level 1, personalized advice (feedback based on individual nutrient intake data); level 2, personalized advice (feedback based on individual nutrient intake data and phenotypic data); and level 3, personalized advice (feedback based on individual nutrient intake data, phenotypic data, and genetic data). Nutrient intake data are determined by using a multilingual online FFQ. The phenotype data consist largely of simple anthropometric, blood biochemistry, and physical activity information. The genetic data include a selection of nutrient-sensitive genes, such as fat mass and obesity associated gene (FTO), MTHFR, transcription factor 7-like 2 (TCF7L2), fatty acid desaturase 1 (FADS1), apolipoprotein E (APOE), and their links to body weight and exercise, folate intake and cardiovascular health, weight loss from low-fat diets, omega-3 FAs, and saturated fat intake, respectively.
Half of the participants are assigned to receive more intensive feedback, whereby they are assessed more frequently, are provided with additional coaching, and have access to an online forum. The study is due for completion in 2014 and hopes to determine whether PN is more effective than conventional healthy-eating guidelines, whether more detailed feedback promotes increased compliance to dietary advice, and whether the Internet is a successful delivery method for PN.
The design and interpretation of such a study requires a wide range of expertise, which is reflected in the multidisciplinary team of Food4Me. The project extends beyond the biologic sciences to include experts in consumer science, business development, information technology, ethics, and legal studies for a comprehensive assessment of the state of the art in PN. Such multidisciplinary approaches pave the way for the future in nutrition research, particularly in the coming together of both molecular and public health nutrition.
In conclusion, the present review has examined the opportunities and challenges to the integration of public health nutrition and molecular nutrition. Arising from this, 2 areas need to be addressed. The first is to ensure that educational strategies in human nutrition ensure some level of competence and understanding in each of these fields. The second requires funding agencies to ensure that the full translation of research funding programs is optimized by the integration of molecular and public health nutrition.
Acknowledgments
M.J.G. prepared the manuscript; and B.A.M., M.F.R., and M.C.W. contributed to writing the manuscript. All authors read and approved the final manuscript.
Footnotes
Abbreviations used: JINGO, Joint Irish Nutrigenomics Organisation; JPI, Joint Programming Initiative; MTHFR, methylenetetrahydrofolate reductase; PN, personalized nutrition; SNP, single nucleotide polymorphism.
Literature Cited
- 1.Alwan A, Armstrong T, Bettcher D, Branca F, Chisholm D, Ezzati M, Garfield R, MacLean D, Mathers C, Mendis S, et al. Global status report on non-communicable diseases 2010. Geneva: World Health Organization; 2011 [Google Scholar]
- 2.Association for Nutrition. [Homepage on the Internet] [cited 2013 Dec 2]. Available from: http://www.associationfornutrition.org.
- 3. Nutrition 2014. Spain: III World Congress of Public Health Nutrition [cited 2013 Dec 2]. Available from: http://www.nutrition2014.org.
- 4. Cambridge University Press. Public Health Nutrition [serial on the Internet] [cited 2013 Dec 2]. Available from: http://journals.cambridge.org/action/displayJournal?jid=PHN.
- 5. International Society of Nutrigenetics/Nutrigenomics. [Homepage on the Internet] [cited 2013 Dec 2]. Available from: http://www.isnn.info/
- 6. Springer Link. Genes & Nutrition [serial on the Internet] [cited 2013 Dec 2]. Available from: http://link.springer.com/journal/12263.
- 7. Wiley Online Library. Molecular Nutrition & Food Research [serial on the Internet] [cited 2013 Dec 2]. Available from: http://onlinelibrary.wiley.com/journal/10.1002/(ISSN)1613-4133.
- 8.Rhee JJ, Cho E, Willett WC. Energy adjustment of nutrient intakes is preferable to adjustment using body weight and physical activity in epidemiological analyses. Public Health Nutr. 2013;23:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibney MJ, Walsh M, Brennan L, Roche HM, German B, van Ommen B. Metabolomics in human nutrition: opportunities and challenges. Am J Clin Nutr. 2005;82:497–503 [DOI] [PubMed] [Google Scholar]
- 10.Walsh MC, Brennan L, Pujos-Guillot E, Sébédio JL, Scalbert A, Fagan A, Higgins DG, Gibney MJ. Influence of acute phytochemical intake on human urinary metabolomic profiles. Am J Clin Nutr. 2007;86:1687–93 [DOI] [PubMed] [Google Scholar]
- 11.Bolca S, Van de Wiele T, Possemiers S. Gut metabotypes govern health effects of dietary polyphenols. Curr Opin Biotechnol. 2013;24(2):220–5 [DOI] [PubMed] [Google Scholar]
- 12.O'Sullivan A, Gibney MJ, Brennan L. Dietary intake patterns are reflected in metabolomic profiles: potential role in dietary assessment studies. Am J Clin Nutr. 2011;93(2):314–21 [DOI] [PubMed] [Google Scholar]
- 13.Lloyd AJ, Favé G, Beckmann M, Lin W, Tailliart K, Xie L, Mathers JC, Draper J. Use of mass spectrometry fingerprinting to identify urinary metabolites after consumption of specific foods. Am J Clin Nutr. 2011;94(4):981–91 [DOI] [PubMed] [Google Scholar]
- 14.Rasmussen LG, Winning H, Savorani F, Ritz C, Engelsen SB, Astrup A, Larsen TM, Dragsted LO. Assessment of dietary exposure related to dietary GI and fibre intake in a nutritional metabolomic study of human urine. Genes Nutr. 2012;7(2):281–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hearty ÁP, Gibney MJ. Dietary patterns in Irish adolescents: a comparison of cluster and principal component analyses. Public Health Nutr. 2013;16:848–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Ommen B, Bouwman J, Dragsted LO, Drevon CA, Elliott R, de Groot P, Kaput J, Mathers JC, Müller M, Pepping F, et al. Challenges of molecular nutrition research the nutritional phenotype database to store, share and evaluate nutritional systems biology studies. Genes Nutr. 2010;5:189–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Ommen B, Keijer J, Kleemann R, Elliott R, Drevon CA, McArdle H, Gibney M, Müller M. The challenges for molecular nutrition research 2: quantification of the nutritional phenotype. Genes Nutr. 2008;3:51–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Joint Programming Initiative. A healthy diet for healthy life [cited 2013 Dec 2]. Available from: http://www.healthydietforhealthylife.eu/
- 19.Irish Universities Nutrition Alliance. [Homepage on the Internet] [cited 2013 Dec 2]. Available from: http://www.iuna.net/
- 20. Joint Irish Nutrigenomics Organisation. [Homepage on the Internet] [cited 2013 Dec 2]. Available from: http://www.ucd.ie/jingo/
- 21.Wopereis S, Wolvers D, van Erk M, Gribnau M, Kremer B, van Dorsten FA, Boelsma E, Garczarek U, Cnubben N, Frenken L, et al. Assessment of inflammatory resilience in healthy subjects using dietary lipid and glucose challenges. BMC Med Genomics. 2013;6(1):44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris C, O'Grada C, Ryan M, Roche HM, Gibney MJ, Gibney ER, Brennan L. Identification of differential responses to an oral glucose tolerance test in healthy adults. PLoS One. 2013;22;8(8):e72890:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morris C, Grada CO, Ryan M, Roche HM, De Vito G, Gibney MJ, Gibney ER, Brennan L. The relationship between aerobic fitness level and metabolic profiles in healthy adults. Mol Nutr Food Res. 2013;57:1246–54 [DOI] [PubMed] [Google Scholar]
- 24.Horigan G, McNulty H, Ward M, Strain JJ, Purvis J, Scott JM. Blood pressure in treated hypertensive individuals with the MTHFR 677TT genotype is responsive to intervention with riboflavin: findings of a targeted randomized trial. J Hypertens. 2010;28:478–86 [DOI] [PubMed] [Google Scholar]
- 25.Wilson CP, McNulty H, Ward M, Strain JJ, Trouton TG, Hoeft BA, Weber P, Roos FF, Horigan G, McAnena L, et al. Blood pressure in treated hypertensive individuals with the MTHFR 677TT genotype is responsive to intervention with riboflavin: findings of a targeted randomized trial. Hypertension. 2013;61:1302–8 [DOI] [PubMed] [Google Scholar]
- 26.Gibney MJ, Walsh MC. The future direction of personalised nutrition: my diet, my phenotype, my genes. Proc Nutr Soc. 2013;72:219–25 [DOI] [PubMed] [Google Scholar]
- 27.Stumbo PJ, Weiss R, Newman JW, Pennington JA, Tucker KL, Wiesenfeld PL, Illner AK, Klurfield DM, Kaput J. Web enabled and improved software tools and data are needed to measure nutrient intake and physical activity for personalized health research. J Nutr. 2010;140:2104–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daugherty BL, Schap TE, Ettienne-Gittens R, Zhu FM, Bosch M, Delp EJ, Ebert DS, Kerr DA, Boushey CJ. Novel technologies for assessing dietary intake: evaluating the usability of a mobile telephone record among adults and adolescents. J Med Internet Res. 2012;14:e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kerr DA, Pollard CM, Howat P, Delp EJ, Pickering M, Kerr KR, Dhaliwal SS, Pratt IS, Wright J, Boushey CJ. Connecting Health and Technology (CHAT): protocol of a randomized controlled trial to improve nutrition behaviours using mobile devices and tailored text messaging in young adults. BMC Public Health. 2012;12:477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin RM, Patel R, Oken E, Thompson J, Zinovik A, Kramer MS, Vilchuck K, Bogdanovich N, Sergeichick N, Foo Y, et al. Filter paper blood spot enzyme linked immunoassay for insulin and application in the evaluation of determinants of child insulin resistance. PLoS ONE. 2012;7:e46752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mihalopoulos NL, Philips TM, Slater H. Validity and reliability of perinatal biomarkers of adiposity after storage as dried blood spots on paper. Am J Hum Biol. 2011;23:717–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lakshmy R, Mathur P, Gupta R. Measurement of cholesterol and triglycerides from a dried blood spot in an Indian Council of Medical Research–World Health Organisation multicentric study on risk factors for non-communicable diseases in India. J Clin Lipidol. 2012;6:33–41 [DOI] [PubMed] [Google Scholar]
- 33.Food4me.org. An integrated analysis of opportunities and challenges for personalized nutrition [cited 2013 Dec 2]. Available from: http://www.food4me.org/