Abstract
Blood plays an important role in homeostatic regulation with each of its cellular components having important therapeutic and diagnostic uses. Therefore, separation and sorting of blood cells has been of a great interest to clinicians and researchers. However, while conventional methods of processing blood have been successful in generating relatively pure fractions, they are time consuming, labor intensive, and are not optimal for processing small volume blood samples. In recent years, microfluidics has garnered great interest from clinicians and researchers as a powerful technology for separating blood into different cell fractions. As microfluidics involves fluid manipulation at the microscale level, it has the potential for achieving high-resolution separation and sorting of blood cells down to a single-cell level, with an added benefit of integrating physical and biological methods for blood cell separation and analysis on the same single chip platform. This paper will first review the conventional methods of processing and sorting blood cells, followed by a discussion on how microfluidics is emerging as an efficient tool to rapidly change the field of blood cell sorting for blood-based therapeutic and diagnostic applications.
Keywords: microfluidics, lab-on-a-chip, blood, cell separation, sample preparation
1. Introduction
Blood is inarguably one of the most important biospecimens and resources used in medicine and research. While the majority of blood is composed of water, it is at the same time multifaceted, comprised of a wide variety of molecules and cells. Examples of these molecules include carbohydrates, lipids, proteins, minerals and gases. Typical blood cells include red blood cells (RBCs), white blood cells (WBCs) and platelets.[1] Given the diversity of blood, it is not surprising that blood plays an important role in homeostatic regulation while providing a good source for markers useful in clinical diagnosis.[2] For example, platelet concentration (normally ranging between 1.5 × 105 – 4.5 × 105 cells μL−1) is often assayed as an indicator of impaired clotting and a marker for diseases such as leukemia or anemia.[3] WBCs (with its concentration normally ranging between 4 × 103 – 11 × 103 cells μL−1) play a critical role in mounting immune responses to foreign pathogens to help protect and fight against infections. Functional phenotyping of WBCs (or immunophenotyping) to examine their capability to release pro- and anti-inflammatory cytokines is a powerful approach for diagnosis of immune diseases (such as acquired immune deficiency syndrome, or AIDS, and tuberculosis, or TB) and assessments of patient immune responses to immunomodulatory therapies.[4]
There also exist rare cells within the circulatory system that are of significant clinical importance. For example, in cancers with a solid tumor origin, circulating tumor cells (CTCs) that originate from the tumor can be found circulating in the blood. Isolation of rare CTCs (0.3 – 100 cells mL−1) in blood for downstream molecular and cellular analysis has been of a great interest for both improving cancer prognosis and understanding the cancer metastatic process.[5] Other examples of rare cells found in the circulation include cancer-associated fibroblasts which, like CTCs, may provide important prognostic information for cancer patients.[6] Hematopoietic stem cells (HSCs) are typically found in bone marrow but can be induced to enter blood circulation after stimulation with granulocyte colony-stimulating factor (G-CSF), a hormone commonly used in cancer treatments and blood transfusion.[7] Isolation of rare fetal cells such as fetal nucleated red blood cells (FNRBCs) from maternal circulation or cord blood presents a non-invasive method of obtaining fetal DNA for prenatal genetic screening.[8]
Separation and sorting of different populations or subpopulations of blood cells from unprocessed or minimally processed blood specimens is of a prime interest to both clinical and biomedical applications and holds a central role in diagnosis and prognosis of physiologic and pathologic conditions such as infectious diseases, cancers and inflammatory responses. Conventional approaches of blood cell separation are on the macroscale level, and the methods are largely limited by factors such as required blood sample volume, component purity, cell quality, processing time and operation efficiency. These technical challenges are further aggravated by some stringent applications, such as: (i) removing RBCs from whole blood, (ii) avoiding spontaneous platelet-triggered agglutination, (iii) capturing rare cells such as CTCs from patient blood[9] and (iv) targeting particular WBC subpopulations at various status.
Conventional blood cell sorting methods include the use of antibodies and their specificity to protein markers on blood cells of interest. Antibody-based approaches possess the advantage of high specificity and sensitivity but are limited by the quality and high cost of antibodies. Label-free separation of cellular components of blood by their physical properties such as cell density and size has also been widely used; however, high-resolution separation of blood cells is difficult with centrifugation or size-based filtration approaches using fibrous membranes or track-etched polycarbonate filters. In addition, conventional macroscale blood cell isolation methods require a large volume of blood and involve many manual interventions prone to introducing artifacts, and they also commonly require skilled technicians and well-equipped, expensive laboratories.
Novel microfluidics and lab-on-a-chip (LOC) technology developments have been gaining in importance in recent years as efficient and powerful approaches for high-throughput blood cell separation, owing to their precise control of fluid behavior and the ability to scale down the required sample volume and achieve continuous non-invasive molecular and functional analysis of blood cells down to the single-cell level. In addition, exploring unique fluidic transport phenomena in confining microfluidic environments and integrating both physical and biochemical methods and analytical assays in a single-chip format provide comprehensive capabilities of integrated microfluidic approaches for blood cell sorting and analysis over conventional macroscale methods.
Over the last decade, there have been many novel developments in designing highly integrated and functional microfluidic devices and systems that incorporate different approaches for the separation and sorting of blood cells as well as rare cells in blood such as CTCs. Some of these microfluidic cell separation and sorting techniques have been discussed in several recent informative and insightful reviews.[10] Particularly, the blood-on-a-chip review[10g] by Toner and Irimia has discussed thoroughly the complexity and associated challenges of handling and processing blood using emerging microfluidics technologies. However, there are some novel microfluidic cell sorting approaches reported very recently, such as those utilizing microfiltration membranes,[11] hydrodynamic and inertia focusing,[12] and patterned adhesive protein arrays,[13] that have been proven effective for sorting cells from unprocessed or minimally processed blood specimens. In this review, we aim to provide readers with a comprehensive introduction for these recently developed approaches and their underlying working principles. Another major goal of this review is to provide a perspective on the future trend of this exciting field of sorting blood cells using novel microfluidic technologies.
Specifically, in this review we will focus on discussing recent microfluidic innovations for separation of different types or subpopulations of blood cells as well as rare cells (such as CTCs) from unprocessed or minimally processed blood specimens. To do so, we will first briefly describe commonly used macroscale approaches of processing blood into its different cellular components. Their advantages and limitations will serve as an introduction as well as motivation for our review of the promising microfluidic innovations that are playing a transformative role in integrating and advancing blood cell separation and sorting in a monolithic format. Examples of specific single-purpose microfluidic devices for blood cell sorting and separation will be described and categorized based on their operational mechanisms, together with a perspective on the exciting new trend of developing highly integrated functional microfluidic devices and systems for high-throughput, high-resolution, high-content analysis of blood specimens for different clinical and biomedical applications.
2. Macroscale Blood Cell Separation and Sorting
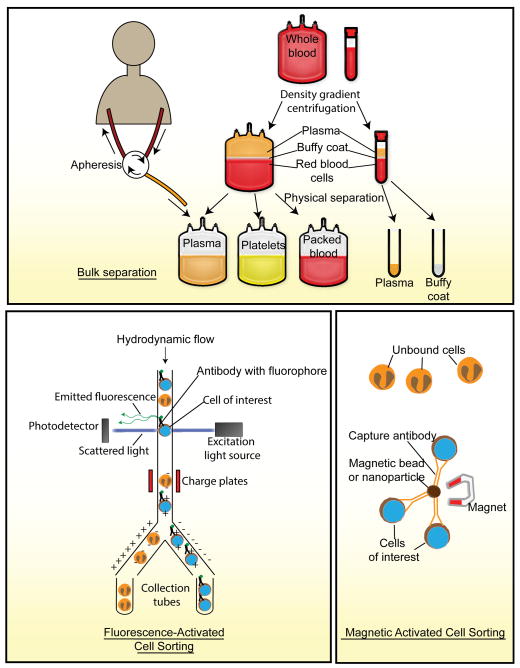
The proper handling and separation of blood into its cellular components is a multi-step process and is crucial for the success of downstream analytical procedures. Readers interested in detailed discussions on conventional macroscale methods of handling and separation of blood are referred to some excellent references elsewhere.[14] Here we will present a brief overview of these conventional methods followed by a description of the methods of analyzing blood and its components (Figure 1).
Figure 1.
Schematic representation of conventional blood processing and analytic methods.
2.1. Centrifugation
One main use of blood in medicine is for blood transfusion, in which components of the blood, such as RBCs, WBCs and platelets, are isolated from healthy donor blood before transferred into patients. Blood is first collected from donors in its uncoagulated form in transfusion bags containing additives to prevent coagulation.[15] The blood is then fractionated through centrifugation into three layers: RBCs, buffy coat (which contains WBCs) and plasma. These layers can be easily harvested by aspiration and used for downstream cell isolation.[14, 16] Apheresis is another method of obtaining blood components from a donor. In apheresis, an intravenous line is introduced into the donor, and blood is allowed to flow into an apheresis machine, in which the blood is fractionated using centrifugation, and the desired blood fraction is collected and any unused component is transfused back into the donor.[17] For clinical testing and research applications, the volume of blood required is smaller than that needed for transfusion. In such applications, a vacuum tube that contains additives to enhance or prevent blood coagulation can be used to harvest blood from a venipuncture site before the blood sample is fractionated through centrifugation into different components. While easy to conduct, the use of centrifugation does not allow for high-resolution separation of blood cells and is usually used as a method of bulk isolation prior to downstream specific isolation protocols.
2.2. Chemical Lysis or Density Gradient Centrifugation
Pure fractions of RBCs or plasma can be harvested after centrifugation by aspiration and often do not require further processing before downstream analyses. However, the buffy coat, a collection of many different blood cell types, exists as a thin layer of cells between the interface of RBCs and plasma, and cross contamination of the buffy coat with RBCs may occur. One method of removing RBC contaminants includes the use of a lysis buffer containing ammonium chloride.[18] Alternatively, this contamination can be reduced through the incorporation of a density gradient into the centrifugation step by using a polysaccharide, typically Ficoll.
2.3. Antibody-Based Approaches
Common antibody-based methods for isolation and sorting of blood cells include flow-based protocols such as fluorescence-activated cell sorting (FACS) or solid-state immobilization and capture of cells using magnetic-activated cell sorting (MACS). Specialized systems for isolating CTCs have been developed based on magnetic and imaging techniques (CellSearch™).[5b, 19] The following sections will describe some of the commonly used antibody-based assays employed for separation and sorting of blood cells.
2.3.1. Fluorescence Activated Cell Sorting (FACS)
FACS is one of the most commonly used methods to date for separation of live cell fractions. As its name suggests, fluorescence is used as the mechanism to sort a specific population of cells from a pool by operating on the same principles as flow cytometry with a few differences. Firstly, a fluorophore-conjugated antibody specific to a surface marker found on the cells of interest is added to the pool of cells, such as the buffy coat. The pool of cells is applied through the FACS machine, which ultimately generates droplets of single cells. If a droplet containing the appropriately labeled cell passes through the path of the laser, the emitted fluorophore is detected by a photodetector, and an electrical pulse is delivered to the droplet that changes its path into a collection tube (Figure 1).[20] FACS is an extremely specific and sensitive method for harvesting cells; however, a major drawback of FACS is its high purchase and maintenance cost. Furthermore, like flow cytometry, FACS cannot reliably handle small numbers of cells.
2.3.2. Magnetic-Activated Cell Sorter (MACS)
A cheaper alternative to FACS for cell sorting is the use of MACS. Unlike FACS which makes use of antibodies labeled with fluorophores, MACS employs magnetic beads conjugated with antibodies that are added to the pool of cells and bind to the cells expressing the specific protein marker. The pool of cells is flushed through a flow column that is placed within a magnetic field, which attracts magnetic beads and thus retains the cells of interest within the column (Figure 1).[21] While MACS is cheaper and readily available, the protocol is lengthier than FACS and involves multiple wash steps that can cause significant loss of the cells of interest.
2.3.3. CellSearch™
The CellSearch™ System is the only device approved by the US Food and Drug Administration (FDA) for isolation of CTCs from the blood of cancer patients. This system makes use of iron nanoparticles conjugated with an antibody that recognizes a specific marker on the CTC, such as the epithelial cell adhesion molecule (EpCAM), to capture CTCs. CTCs and non-CTCs are further differentiated by immunostaining and imaging.[22] Some useful references on isolation of CTCs from patient blood and their molecular and cellular analysis have been published elsewhere.[23] While the CellSearch™ System is highly efficient for isolating rare CTCs from blood specimens, it is hampered by the fact that surface expression of EpCAM on CTCs may be more heterogeneous than initially anticipated (for example, due to the epithelial-mesenchymal transition, EMT) or even absent altogether in some tumor types (such as melanoma).[24] Furthermore, the clinical importance of EpCAM is highly debatable,[25] making the use of EpCAM as a target for capturing CTCs controversial.[26]
3. Blood Cell Sorting Using Innovative Microfluidic Tools
Table 1 summarizes representative microfluidic technologies based on different physical and biochemical principles for separation and sorting of blood cells. In this section, we will focus on discussing microfluidic separation and purification of blood cells from blood samples using: (i) size- and deformability-based physical approaches; (ii) novel microscale fluid dynamics including hydrodynamic and hemodynamic phenomena; (iii) affinity- or topography-based schemes; and (iv) magnetophoresis, acoustophoresis and electrical methods. There are other novel microfluidic cell manipulation methods based on aqueous two-phase systems (ATPS),[27] optical methods,[28] sedimentation,[29] and mimicking phenomena of the microvasculature.[30] These methods are less common, and their utility for clinical blood samples still needs to be fully validated in the future; thus these methods are not covered in this review (interested readers are referred to recent reviews[10f, 10g, 31] published elsewhere for these topics).
Table 1.
Representative microfluidic devices for blood cell sorting based on various principles.
| Principles | Pre-treatment | Dilution ratioa) | Cell concentration [million/mL] | Target components | Throughput [mL/min] | Recovery efficiencyb) [%] | Purityc) [%] | Ref. |
|---|---|---|---|---|---|---|---|---|
| Physical filtration | ||||||||
| Linear filter array | RBCs washed with isotonic saline. | - | - | RBCs | 0.08 to 0.63 | - | - | [32] |
| Slanted cross-flow filter array | FNRBCs were isolated from women | - | 0.003 to 0.03 | FNRBCs | 0.1 to 0.45 | 74 | 1.7 | [33] |
| Diffusive cross-flow filter array | With freeze dried heparin anticoagulant | None | - | RBCs | 0.005 to 0.012 | 50 | 94 | [34] |
| Gradient filter array | - | 1: 1 to 10 | - | Neuroblastoma cells | - | - | - | [35] |
| Crescent-shaped pillar array | - | 1: 2 | 0.0001 | Cancer cell lines | - | 20 to 90 | 75 to 98 | [37] |
| Parylene membrane | - | None | 5,000 | Cancer cell lines | >0.2 | 90 | - | [40a] |
| PDMS membrane | RBC lysis | - | 0.5 to 6 | WBCs | 0.2 to 20 | 70 | >97 | [11] |
| Hydrodynamic mechanisms | ||||||||
| Serpentine inertial focusing | - | 1: 19 to 199 | - | RBCs | 1.5 | - | - | [47] |
| High aspect-ratio inertial focusing | - | 1: 19 | 250 | Cancer cell lines | 0.4 | >80 | - | [48] |
| Curvilinear inertial focusing | - | 1: 1 | 5,000 | CTCs | 0.05 | >85 | 10 | [49] |
| Bilateral hydrodynamic focusing | - | None | 5,000 | WBCs | 0.02 | 100 | - | [54a] |
| Single-sided hydrodynamic focusing | - | None | 5,000 | WBCs | 0.0003 | 97.2 | 96.9 | [12a] |
| Hydrophoresis | - | 1: 9 | 7,000 | Platelet | 0.02 | 76.8 | 82.8 | [55a] |
| Deterministic pillar array | - | None | 5,000 | RBCs & WBCs | 0.001 | - | - | [57] |
| Hemodynamic phenomena | ||||||||
| Fahraeus-Lindqvist effect | Centrifugation for various hematocrits | - | - | Serum | - | - | - | [63] |
| Zweifach-Fung effect | - | - | - | Plasma | 0.00017 | - | 100 | [66] |
| Cell-free & plasma skimming effect | Centrifugation for various hematocrits | - | - | Plasma | 0.00017 to 0.017 | - | - | [64] |
| Cell-free effect | - | None | 5,000 | WBCs | - | 67 | - | [65] |
| Surface affinity and topography | ||||||||
| Antibody-coated pillar array | Stored on a rocking platform | None | 5,000 | CTCs | 0.017 | 65 | 50 | [69] |
| Antibody-coated nanopillar | - | None | 5,000 | CTCs | 0.008 to 0.056 | >95 | - | [71] |
| Printed antibody array | RBC lysis | - | - | T-cells | 0.003 | - | >95 | [80] |
| Structured adhesive surface | - | - | 240 | Cancer cell lines | 0.83 to 1.8 | >76 | >95 | [74] |
| Nanotopography | RBC lysis | - | 100 | Cancer cell lines | 0.01 to 0.03 | >80 | 10 to 85 | [13b] |
| Magnetophoresis | ||||||||
| Bead-cell trapped by cavity | RBC lysis & bead conjugation | - | - | CTCs | 0.02 | >87 | - | [84a] |
| Bead based positive & negative selections | Bead conjugation | - | 8,000 | CTCs | 0.14 | >77 | >0.1 | [12b] |
| Self-assembled magnetic bead array | Mononuclear cell isolation by Ficoll | - | - | B-cells | - | >94 | >78 | [86] |
| Inherent magnetic property of cells | - | 1: 10 | - | WBCs | - | 87.7 | - | [87] |
| Temperature sensitive polymer | - | None | - | CD4+ T cells | 0.045 | - | 90 to 94 | [13c] |
| Electrospun nanofiber | - | None | 0.0001 | CMCs | 0.008 to 0.017 | 68 to 87 | - | [82] |
| Electrical methods | ||||||||
| Electrowetting on dielectric (EWOD) | Mononuclear cell isolation by Ficoll-Hypaque | - | 0.1 | CD8+ T-cells | - | >90 | - | [91] |
| Dielectrophoretic (DEP) | Dissolving lead ions | - | 20 | Normal RBCs | - | - | - | [93] |
| Acoustophoresis | ||||||||
| 1D acoustic alignment | RBC lysis | 1: 9 | 0.25 | Cancer cell lines | 0.07 | >72 | >79 | [97] |
| 2D acoustic concentration | - | 4999 | 0.025 to 0.1 | RBCs | 0.15 to 0.2 | >94 | - | [100] |
Ratio of volumes for samples diluted in diluent before chip loading;
Ratio of amounts of target components before and after chip processes;
Ratio of amounts of target components compared to all collected components.
3.1. Physical Filtration
Different types of blood cells have very different physical properties, such as density, size, shape and even deformability. Thus, microfluidic sieves and filters have been developed to sort blood cells according to their distinct physical characteristics. One of the earliest microfluidic devices to manipulate blood dates back to 1994.[32] It had a regular one- dimensional (1D) array of filters with a gap size of 5 μm to isolate and enrich RBCs (Figure 2A). As a suspension of RBCs in isotonic saline flowed across the 1D filter array integrated into a microfluidic channel, most RBCs were trapped and only some deformable RBCs were able to pass through the filter with minimal RBC destruction.[32]
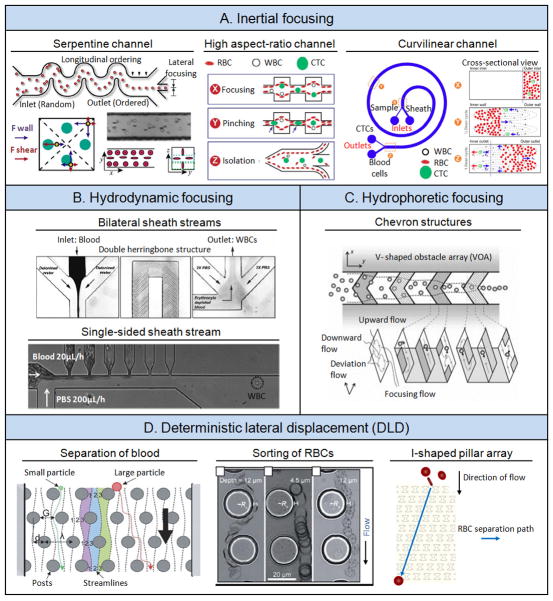
Figure 2.
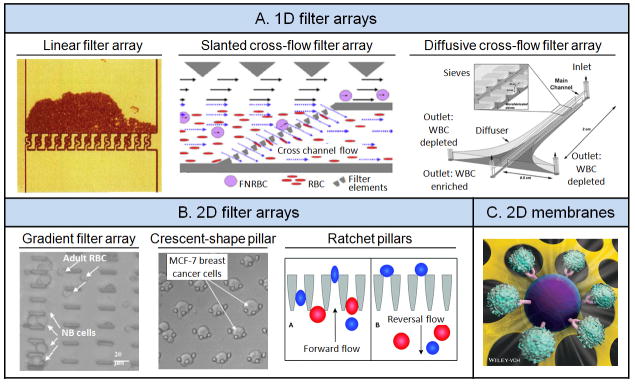
Physical filtration. (A) One-dimensional (1D) filter arrays. Left: Linear filter array. Reproduced with permission.[32] Copyright 1994, American Association for Clinical Chemistry. Middle: Slanted cross-flow filter array. Reproduced with permission.[33] Copyright 2010, Elsevier. Right: Diffusive cross-flow filter array. Reproduced with permission.[34] Copyright 2006, Royal Society of Chemistry. (B) Two-dimensional (2D) filter arrays. Left: Gradient array with successively narrower filters along the flow direction. Reproduced with permission.[35] Copyright 2004, The Institute of Electrical and Electronics Engineers. Middle: Crescent-shaped pillar arrays. Reproduced with permission.[37] Copyright 2009, Springer. Right: Ratchet pillars with reversal flow. Reproduced with permission.[38] Copyright 2012, Royal Society of Chemistry. (C) 2D microfiltration membranes made of PDMS. Reproduced with permission.[11] Copyright 2013, Wiley-VCH.
A major challenge in using sieves for cell sorting and separation is clogging and fouling. An effective solution to overcome this issue is to arrange filter structures in a cross- flow fashion to allow unfiltered remnants to travel with the primary stream, thus preventing accumulation or aggregation of un-sorted cells in situ at the sieving structures. Such a cross-flow microfluidic cell sorting device was successfully developed to isolate FNRBCs from adult anucleate RBCs (AARBCs) for non-invasive prenatal diagnosis based on their size difference, Figure 2A.[33] More specifically, this cross-flow microfluidic cell sorting device contained a slanted filter array that divided an upstream flow into two parallel channels, with one channel harvesting the filtrate that had permeated through the filter array while the other permitting continuous movement of unfiltered remnants toward another outlet. When a mixture of FNRBCs and AARBCs was passing through this device, FNRBCs tended to stay in the main stream with AARBCs passing through the filter array. This cross-flow microfluidic cell sorting device was reported to achieve a FNRBC recovery rate of 74% and an AARBC depletion rate of 46%.[33]
Cross-flow microfluidic filters have also been developed for leukapheresis, a common clinical blood processing procedure for depletion of WBCs from whole blood. Leukapheresis is commonly used for treating hematological malignancies and autoimmune diseases. A notable example for microfluidic leukapheresis was the diffusive cross-flow filtration scheme that could modulate flow resistance of each sieve element (Figure 2A).[34] The filtration section consisted of a pair of sieve arrays lining along the main channel. Using a diverging configuration of the sieve array, flow rates along the sieve array were uniform, thereby reducing accidental WBC entry through the sieves. Using an optimized diverging configuration of the sieve array, the diffusive cross-flow filtration was reported to achieve isolation of 50% RBCs while depleting more than 97% WBCs.[34]
Several research groups have developed two-dimensional (2D) filter arrays to increase separation speed and sample throughput. An example of such 2D filter arrays was the construction of a cell sorting device that had four segments of successively narrower filters (15, 10, 5 and 2.5 μm spacing) along the flow axis, Figure 2B.[35] As diluted blood spiked with neuroblastoma cells was introduced into the device, different cells were trapped in different segments of the filter array according to their physical sizes. Neuroblastoma cells were retained in filters with a gap size of 10 μm, whereas other blood cells passed through the entire device without being trapped. Isolation and enrichment of neuroblastoma cells from patient blood using such 2D filter arrays can provide a valuable cell source for cancer diagnosis, since neuroblastoma is the most common pediatric extracranial tumor.
Although microfluidic filter arrays[35–36] were able to isolate and enrich blood cells based on cell size, a successful implementation of such devices depends on a precise control of filter geometries. In addition, cell clogging and fouling of filter structures are common in such devices owing to their intrinsic design and operational features. A notable 2D filter array developed recently for isolation of CTCs from blood samples can overcome the pitfall of cell clogging. In this 2D filter array, each filter was constructed using 3 identical pillars arranged into a crescent shape (Figure 2B).[37] Once a filter or trap was filled with a CTC, other blood cells would escape the filled trap due to an increased flow resistance that detours incoming cells. Cancer cells were generally larger than blood cells, and as such this 2D filter array (with the gap between the pillars of 5 μm) only trapped cancer cells but not blood cells. The isolation efficiency was reported to be > 80% for breast and colon cancer cell lines spiked in blood samples under a low pressure operation.
Although a precise control of the microfluidic filter geometry is necessary for achieving optimal cell sorting performance, improved operations of these microfluidic filters can also result in increased cell separation and sorting efficiency. Such an attempt was successfully demonstrated using a structural ratchet mechanism created using funnel-shaped microscale constrictions under an oscillatory flow, Figure 2B.[38] When a cell suspension containing mouse lymphoma cells (MLCs) and human peripheral blood mononuclear cells (PBMCs) was injected through a 2D filter array consisted of funnel constrictions, PBMCs that were smaller and more deformable could easily flow across the constrictions in forward flow while larger and less deformable MLCs were excluded. When a reversal flow was applied, MLCs that were trapped at the constrictions would be released to unclog the funnel constrictions. PBMCs however were not able to pass back through the constrictions since the cells had to overcome a smaller opening on the leeward side of the funnel constrictions. By repeating the forward and reversal flow, MLCs and PBMCs were successfully separated with enhanced separation efficiencies and purities.
Exploiting and integrating microfabricated filtration membranes into microfluidic devices have recently emerged as an efficient approach for separation and sorting of blood cells. Recently our research laboratory developed a new surface micromachining technique to achieve wafer-scale high-fidelity lithographic patterning on polydimethylsiloxane (PDMS) for large microfiltration membranes (up to 3 cm × 3 cm, with the membrane thickness of about 10 μm) with high porosity (up to 30%).[11, 39] Since the PDMS surface micromachining technique was compatible with soft lithography and other silicon-based microfabrication methods, the PDMS microfiltration membranes could be easily integrated with other PDMS support structures to provide a superior mechanical strength. This is advantageous for microfiltration membranes with a large surface area and high porosity as well as for integration with other PDMS-based microfluidic molecular/cellular analytical modules. Microfluidic devices integrated with PDMS microfiltration membranes had successfully achieved on-chip isolation, enrichment, and functional analysis of PBMCs as well as subpopulations of WBCs such as CD14+ monocytes from lysed and whole blood specimens with high flow rate (up to 20 mL min−1) and excellent cell purity (> 97%).[11, 39] Another material that has been successfully utilized for microfabrication of filtration membranes was Parylene-C, a biocompatible polymer that possesses good mechanical strength and flexibility.[40] The Parylene microfiltration membranes (with the membrane surface area up to 36 mm2 and porosity about 7% - 15%) could contain well-defined pores of different geometries (circular, oval-shaped, and rectangular pores) with critical dimensions down to a few microns. The Parylene microfiltration membranes were successfully applied for capturing CTCs from 1 mL of whole blood in less than 5 min, achieving 90% capture efficiency, 90% cell viability, and 200-fold sample enrichment.[40] It is worth noting that other microfiltration membranes made by nickel electroplating[41] or through direct uses of commercial filtration papers[42] or plastic sheets[43] have also been successfully incorporated into microfluidic blood cell sorting devices.
The filtrate purity achieved by physical filtration of blood cells is inevitably compromised by stowaway cells. Blood cells are fairly deformable and can easily pass through a slit or constriction smaller than the cell size. Furthermore, there is a significant size overlap among various blood cell subpopulations as well as CTCs. In order to enhance separation resolution and thus purity for size-based blood cell separation, our research laboratory and others have recently developed a strategy to combine microfiltration membranes with antibody-conjugated microbeads for isolation and enrichment of subpopulations of WBCs as well CTCs. Microbeads conjugated with antibodies could bind target WBCs[11, 39] or CTCs[44] to increase the apparent size difference between target cells and other blood cells, thus enhancing separation resolution and purity from subsequent cell filtration process using microfiltration membranes, Figure 2C. Another recent study further utilized instability of fluid flow (such as the Taylor–Gortler instability phenomenon) to enhance mixing of antibody-coated microbeads and blood cells in microfluidic channels to improve the bead-cell conjugation process before downstream cell filtration process to isolate and enrich CTCs.[45]
3.2. Hydrodynamic Mechanisms and Hemodynamic Phenomena
The human circulatory system is made up of a complex network of blood vessels interacting as microfluidic systems for transporting blood. The rich and complex interactions among blood cells, blood plasma and confining blood vessels at the microscale level have allowed establishments of unique blood flow characteristics. Blood related hydrodynamic and hemodynamic phenomena have been the focus of attempts by researchers to design microfluidic networks that mimic in vivo flow conditions to enhance sorting and separation of different blood components.
Inertial focusing, for example, has attracted much attention to the microfluidic community in recent years as a novel strategy to control and steer particles in microfluidic channels.[46] As its name suggests, inertial focusing makes use of inertial forces generated as a result of fluidic flow within a confining microchannel. Shear-gradient lift and wall-induced lift generate a net lift force that drives particles toward equilibrium positions within the microchannel cross-section, turning an initial homogeneous microparticle stream into a highly focused microparticle stream within a short distance, Figure 3A.[47] The inertial force is mainly regulated by two parameters: the Dean number and the ratio of particle diameter to the hydraulic diameter of the microchannel. Manipulating these two parameters has allowed researchers to conveniently combine inertial focusing with other microfluidic methodologies to enhance blood cell separation and sorting. For example, inertial focusing was successfully applied for manipulations of position and alignment of RBCs within microchannels.[47] Inertial focusing was also incorporated into a microfluidic device for isolation and enrichment of CTCs from diluted blood samples (Figure 3A).[48] The design of this CTC sorting device uniquely contained a high aspect ratio rectangular microchannel structured with a contraction-expansion array. In the cell-focusing region, under the influence of shear modulated inertial lift force, all cells equilibrated efficiently along the channel sidewalls. Flowing through the rare cell pinching region, the center of mass of larger CTCs was aligned along the channel center while the smaller hematologic cells remained focused along the channel sidewalls. Bifurcating outlets allowed for collection of larger CTCs at the center outlet while other hematologic cells were collected from the side outlets. Efficiency of CTC recovery was further enhanced in this CTC sorting device by optimizing factors such as hematocrit, microchannel geometry and the Reynolds number.[48]
Figure 3.
Hydrodynamic mechanisms. (A) Inertial focusing. Left: In serpentine channel. Reproduced with permission.[47] Copyright 2007, The National Academy of Sciences, U.S.A. Middle: In high aspect-ratio channel. Reproduced with permission.[48] Copyright 2011, Royal Society of Chemistry. Right: In curvilinear channel. Reproduced with permission.[49] Copyright 2013, Nature Publishing Group. (B) Hydrodynamic focusing. Top: Bilateral sheath streams. Reproduced with permission.[54a] Copyright 2006, American Chemical Society. Bottom: A single-sided sheath stream. Reproduced with permission.[12a] Copyright 2010, John Wiley & Sons. (C) Hydrophoretic focusing. Reproduced with permission.[55a] Copyright 2011, Royal Society of Chemistry. (D) Deterministic lateral displacement (DLD). Left: Separation of blood. Reproduced with permission.[57] Copyright 2006, The National Academy of Sciences, U.S.A. Middle: Sorting of RBCs. Reproduced with permission.[58] Copyright 2012, Royal Society of Chemistry. Right: I-shaped pillar array. Reproduced with permission.[12c] Copyright 2013, Nature Publishing Group.
In a separate study, inertial focusing was used in tandem with the Dean drag force to isolate CTCs from diluted blood.[49] This was achieved by incorporating a curvilinear channel into the microfluidic device design (Figure 3A). As particles moved through the microchannel, the channel curvature resulted in an additional lift (the Dean drag force) owing to a centrifugal acceleration of fluid flow.[49] Depending on particle size, a net force between the Dean drag, shear-gradient lift and wall-induced lift determined the final particle position. CTCs were generally larger than hematologic cells and thus flowed closer to the inner wall, whereas hematologic cells flowed near the outer wall, resulting in efficient separation of CTCs from hematologic cells. Additional strategies have been implemented recently in conjunction with inertial focusing to further enhance blood cell sorting efficiency, such as using curvilinear microchannels with a trapezoid cross section,[50] phase partitioning in a hydrodynamic focusing setting[51] as well as multistage processing.[52]
Unlike inertial focusing that occurs in a single flow stream, hydrodynamic focusing is a technique capable of achieving narrow flow streams through sheath flows. Hydrodynamic focusing has been used in broad applications such as biological patterning and biochemical synthesis.[53] In a recent study, hydrodynamic focusing was successfully applied in a microfluidic lysis device for depletion of RBCs and enrichment of WBCs from blood for downstream genomic and phenotypic analysis, Figure 3B.[54] In this device, an input blood stream was flanked by two deionized water streams, resulting in a narrow blood stream interfacing with deionized water with a high surface-to-volume ratio. Following the inertial focusing section for lysing RBCs was a long serpentine channel with herringbone structures that facilitated rapid passive mixing to homogenize blood and lysis buffer. Although blood was processed in the device for only a few seconds, efficient RBC depletion was accomplished owing to enhanced blood-lysis buffer interaction resulted from hydrodynamic focusing. Moreover, short exposure time to lysis buffer could lead to a minimal adverse impact on WBCs.[54]
In another study, single-sided hydrodynamic focusing using lysis buffer as a sheath stream was applied in a microfluidic device to deplete RBCs and isolate and enrich WBCs from whole blood in a continuous-flow fashion, Figure 3B. The microfluidic device featured a sheath stream flowing perpendicularly with respective to the sample stream, from one side wall to the opposite wall having six small bifurcation outlet channels. As whole blood was thinned and pushed by the sheath flow, cellular components of blood with smaller size and greater deformability such as RBCs and platelets as well as blood plasma were shunted into the six bifurcation channels, leaving only WBCs in the main channel. By optimizing bifurcation channel dimensions and flow rates, WBC recovery rate of up to 97% and purity of 96.9% were achieved.[12a]
Hydrophoresis is another hydrodynamic mechanism used successfully in microfluidic devices for sorting blood cells. Hydrophoresis makes use of rotational flow for separating particles based on size.[55] A typical hydrophoretic device comprises of slanted obstacles that generate a rotational flow across the channel cross-section as fluid moves along the device (Figure 3C). This rotational flow exerts a force on particles within the flow stream, resulting in their separation based on size. Such devices have been successfully applied for separating platelets and RBCs from WBCs in blood specimens.[55] Another recent study applied hydrophoresis versus sedimentation onto a channel to separate WBCs from RBCs across slanted obstacles.[56]
Deterministic lateral displacement (DLD) is another novel hydrodynamic mechanism applied successfully in the microfluidic environment for sorting blood cells. In DLD, a micropost array is arranged with each row of posts slightly offset laterally with respect to the previous row above it. Larger particles (or cells) will display a greater deterministic lateral displacement as it interacts with the post array (Figure 3D). Thus, when a blood sample passes through the deterministic post array, cellular components such as RBCs, WBCs and platelets will be separated from one another.[57] The hydrodynamic interactions between the post array and traveling blood cells in DLD are resulted from complex interactions between the size, shape and deformability of blood cells as well as the post array geometry. Since its inception, DLD has been applied successfully for sorting and enrichments of RBCs (Figure 3D),[58] separation of platelets and healthy RBCs from WBCs and nucleated RBCs,[59] RBCs and WBCs from parasite trypanosomes,[60] and rare CTCs from blood cells.[61] Another notable DLD-based microfluidic device reported recently contained an I-shaped pillar array Figure 3D, in order to introduce rotational movements within the I-shaped pillars for RBCs to further enhance its separation from other blood cells.[12c]
Blood, as a complex medium, possesses many unique properties, and such hemodynamic behaviors have been used in microfluidics to enhance sorting of blood components. For example, the Fahraeus-Lindqvist, or sigma effect, describes a decrease in blood viscosity when blood is forced to travel through a tube of diameter less than 300 μm. This effect causes RBCs to aggregate at the leading edge of a meniscus, leading to capillary penetration failure when blood suspension flows in a small tube.[62] The Fahraeus-Lindqvist effect was utilized to construct a point-of-care (POC) device, Figure 4, to rapidly separate plasma from RBCs at various hematocrits for the pathological analysis of serum markers in resource-scarce countries.[63] Other hemodynamic behaviors include (i) the cell-free or liquid-skimming effect, which describes the phenomenon where RBCs tend to concentrate at the blood vessel center while WBCs remain near the vessel wall, and (ii) the plasma skimming or network Fahraeus effect, which describes the phenomenon that the concentration of RBCs in daughter vessels can be lower than that of the mother vessel. Microfluidic devices have been successfully developed to incorporate these hemodynamic mechanisms with the purpose of collecting plasma[64] and cellular components,[65] Figure 4. Another unique hemodynamic behavior is the Zweifach-Fung effect, also known as the bifurcation law, which describes the tendency of RBCs to travel into a vessel with a higher flow rate as it encounters a bifurcating region. This effect was successfully applied to separate plasma when blood flowed through a microchannel with multiple high-flow-resistance branches (Figure 4).[66]
Figure 4.
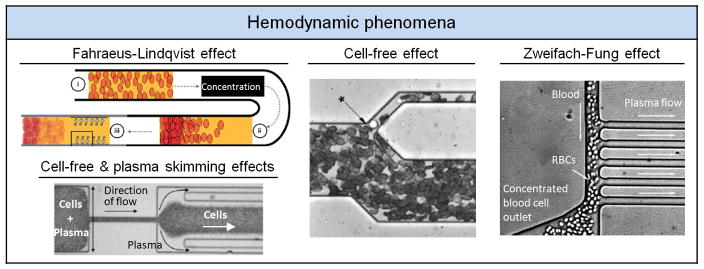
Hemodynamic phenomena. Top left: Fahraeus-Lindqvist effect. Reproduced with permission.[63] Copyright 2011, Royal Society of Chemistry. Bottom left: Cell-free effect and plasma skimming effect. Reproduced with permission.[64] Copyright 2006, IOS Press. Middle: Cell-free effect. Reproduced with permission.[65] Copyright 2005, American Chemical Society. Right: Zweifach-Fung effect. Reproduced with permission.[66] Copyright 2006, Royal Society of Chemistry.
Microfluidic sorting of blood cells using physical filtration and hydrodynamic and hemodynamic mechanisms may unavoidably introduce stresses to cells, and such unintended stimulations may alter molecular expressions and even cellular phenotypes of blood cells.[67] A recent relevant study compared activation of polymorphonuclear leukocytes (PMNs) when undergoing centrifugation in conventional centrifuges and curvilinear microchannels. The percentage of activated PMNs in both systems was found low compared to treatments such as RBC lysis.[50] In conclusion, it is important to conduct comparative assays to evaluate the effect of physical manipulations (such as physical filtration and shear flow) of cells in microfluidic environment on blood cell behaviors. These control assays will also be valuable for determining the optimized microfluidic device geometries and operational parameters.
3.3. Surface Affinity and Topography
Physical properties of blood cells such as cell size and deformability are the major characteristics that have been exploited for microfluidic cell sorting. However, cells in the biological and medical fields are typically identified by a pool of surface biomarkers. Given that using antibodies against specific biomarkers remains the most popular method to label and isolate cells, merging antibody-based approaches with microfluidics is a powerful approach for capture and identification of blood cells.
Antibody based microfluidic cell sorting has proven useful in isolating neutrophils [68] and CTCs[69] from patient blood for clinical diagnosis. In one notable antibody-based cell sorting device, patient blood was flowed across an array of pillars conjugated with antibodies against EpCAM (Figure 5A).[69] As the CTC population in cancer patient blood is generally very low, millions of pillars coated with capture antibodies could increase cell collision rates and thus capture efficiency of CTCs. Given the microfluidic laminar flow environment, it is probable that some CTCs would not collide with a pillar surface. To circumvent this limitation, another recent microfluidic CTC capture chip incorporated herringbone structures to generate passive chaotic mixing to enhance collision interactions between CTCs and the channel walls that were pre-coated with capture antibodies.[70] In a separate study, silicon nanopillar structures generated by chemical etching were integrated into microfluidic herringbone structures to enhance the capture efficiency of CTCs (Figure 5A).[71] Apart from chaotic mixing induced by herringbone structures, the silicon nanopillars offered a significantly increased anti-EpCAM coated CTC capture area to promote cell-surface interactions, critical for efficient CTC capture. Other nanomaterials and biomolecules, such as halloysite nanotubes[72] and aptamers,[73] respectively, were also recently incorporated successfully into microfluidic blood cell sorting applications.
Figure 5.
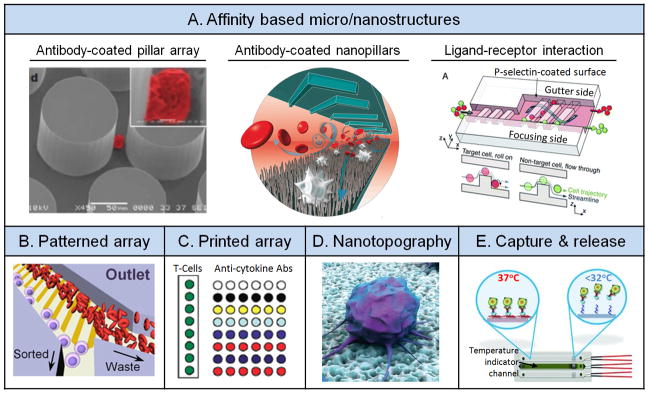
Surface affinity and topography. (A) Affinity based micro/nanostructures. Left: Antibody-coated pillar array. Reproduced with permission.[69] Copyright 2007, Nature Publishing Group. Middle: antibody-coated nanopillars. Reproduced with permission.[71] Copyright 2011, Wiley-VCH. Right: Ligand-receptor interaction. Reproduced with permission.[74] Copyright 2012, Royal Society of Chemistry. (B) Array by surface patterning. Reproduced with permission.[13a] Copyright 2013, Nature Publishing Group. (C) Array by robotic printing. Reproduced with permission.[80] Copyright 2008, Royal Society of Chemistry. (D) Nanotopography. Reproduced with permission.[13b] Copyright 2013, American Chemical Society. (E) Capture and release of cells. Reproduced with permission.[13c] Copyright 2012, Wiley-VCH.
In another recent study, P-selectin ligands were coated onto successive slanted ridges in a microfluidic channel to isolate WBCs from blood samples for clinical diagnostics (Figure 5A).[74] When leukemia cell lines with different P-selectin affinities flowed over the ridges, a lateral driving force induced by hydrophoresis enabled differential displacements along the ridge direction, thus enabling separation of leukemia cells in the device. Another more recent microfluidic cell sorting device utilized a plain feature-free surface functionalized with P-selectin ligands in a slanted strip pattern to generate lateral forces, in order to separate WBCs from RBCs in blood samples (Figure 5B).[13a] Similarly, such spatial patterning of adhesive ligands was applied successfully for separation of WBCs and CTCs from blood samples[75] as well as to create quadruplex capture of multiple WBC types.[76]
Glycosylation, the addition of sugar moieties to macromolecules such as proteins or lipids occurs frequently in nature and affects cell adhesion as well as protein structure and function.[77] In diseases such as cancer, aberrant glycosylation or the dysregulation of lectins (proteins that bind sugars) can occur, and both have been of great interest to researchers as potential targets for personalized cancer therapy.[78] It is conceivable that such a phenomenon can be utilized within a microfluidic system as an alternative to antibody-based approaches for cell capture and sorting. One such study involved functionalizing a multivalent surface with galactose and using its affinity for binding its partner galectin-3 (overexpressed on the surface of metastatic cells) to capture cancer cells.[79] The use of carbohydrates rather than antibodies also possesses the potential of allowing for the continuous monitoring of cancer cell mutations of particular antigenic structures.[79b] In addition, these sugar-functionalized surfaces allow the study of carbohydrate-protein interactions,[79b] adhesion and metastasis[79a] as well as glycomics.
As the microarray printer becomes more accessible, it has been increasingly used in microfluidics to print antibody arrays to aid in capture of desired blood cells. One such device employed the use of a robotic contact microarrayer to print a 150 – 250 μm antibody spot array on a polyethylene glycol (PEG)-diacrylate coated glass (Figure 5C).[80] The printed antibody array was able to capture specific T-cells from RBC-depleted human whole blood for multiplexed detection of cytokines secreted from T-cells. Such multi-parametric analyses of T-cell functions are valuable for diagnosis and monitoring drug response of infectious diseases such as AIDS and TB.
Our research laboratory has recently utilized the differential adhesion preferences of cancer cells to nanorough glass surfaces as compared to intrinsically non-adherent blood cells to achieve efficient CTC capture without using capture antibodies.[13b] To this end, we developed a simple yet precise controlled method to generate random nanoroughness on glass surfaces using reactive ion etching (RIE). These nanoroughened glass surfaces were shown to efficiently capture different kinds of cancer cells derived from different tissues (i.e., MCF-7, MDA-MB-231, HeLa, PCS, and SUM-149) spiked in blood samples, Figure 5D.[13b]
Affinity-based techniques for sorting blood cells can be very specific and thus cell purity resulted from such methods are superior to those achieved by other microfluidic cell sorting methods. However, it remains a challenge to release captured cells from an antibody coated surface without using enzymes or shear stress. To allow releasing target cells after their capture on antibody coated surfaces, a temperature sensitive polymer poly(N-isopropylacrylamide) (PNIPAAm) was explored recently. PNIPAAm has a unique property about its temperature sensitive interaction with proteins: above a critical temperature, PNIPAAm binds to proteins strongly whereas below the critical temperature, absorbed proteins are desorbed (Figure 5E).[81] Using PNIPAAm grafted silicon nanopillar structures, capture and stimulated release of CTCs were successfully achieved.[13c] Another creative method to circumvent the issue of releasing cells captured on a functionalized substrate was to use the laser microdissection (LMD) technique that is commonly used for isolating specific cells of interest from clinical tissue samples. In this work, a circulating melanoma cell (CMC) capture substrate was constructed by electrospinning nanofibers onto a LMD slide.[82] After CMCs were captured onto the antibody-coated nanofibers, individual single CMC could be retrieved using LMD to locally cut the LMD slide before downstream single-cell whole-genome amplification and sequence analysis. Other cell releasing strategies were also investigated with different levels of success. For example, using photocleavable 3-amino-3-(2-nitrophenyl)propionic acid (ANP) linker to bind antibodies to solid surfaces, selective capture and release of target cells via photochemical cleavage was demonstrated.[83]
From a biological point of view, all affinity-based methods for capture and enrichment of blood cells will introduce some specific interactions between antibodies or ligands and cell surface antigens. While some surface antigens may function simply as structural constituents of cell membranes, others may be involved in important signaling pathways that regulate molecular and cellular functions. Hence, there are concerns whether these affinity-based methods will alter the physiology of cells after cell sorting and isolation processes. Fortunately, the nature has evolved several intelligent ways to maintain cellular homeostasis and stable immune responses. One of them is co-stimulatory systems which require more than one extracellular signal to activate an intracellular pathway. A classic example is T-cell co-stimulation. Nonetheless, it is imperative to conduct control studies to evaluate the effect of such affinity-based methods on molecular and cellular functions that are pertinent to downstream molecular and cellular analysis. A recent example for such control studies demonstrated minimal changes of cell viability and culture potential of CD4+ cells isolated directly from blood specimens using microfluidic affinity-based methods.[81]
3.4. Magnetophoresis
The use of magnetophoresis is a common practice for cell isolation in biology. As the application of magnetic fields by a magnet is rapid and effective specifically to paramagnetic microparticles, there is almost no interference to other properties of cells and media. Importantly, it is convenient to conjugate antibodies of different affinities to magnetic microparticles to specifically target a variety of cell populations. This empowers researchers a complete spectrum of choices to isolate diverse types of blood cells as well as rare cells in human blood.
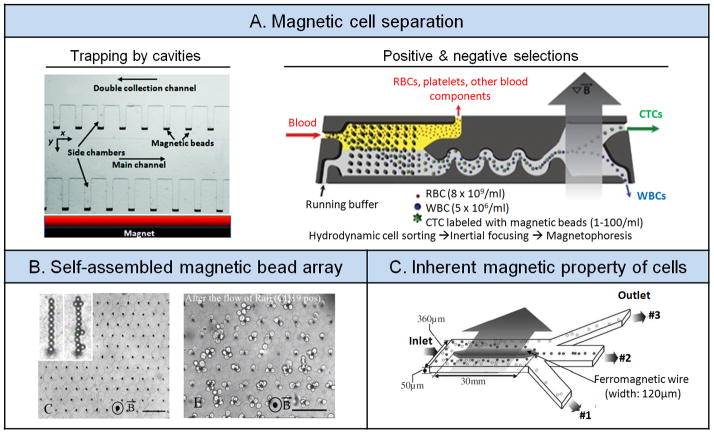
The simplest and most reliable way to isolate cells of interest using magnetophoresis is using antibody-coated magnetic beads to immobilize cells under a magnetic field.[84] One such application involved an array of microcavities lining along a main microfluidic channel to capture rare cells such as CTCs from blood specimens (Figure 6A).[84a] First, CTCs in RBC-depleted blood were labeled with magnetic beads before flowed through the microfluidic channel. Owing to the microfluidic laminar flow environment, there was little mass exchange between the microcavities and the main channel, and blood cells without magnetic bead labeling would exit through an outlet. Magnetically labeled CTCs, however, were drawn in and trapped inside the microcavities when a magnet was placed close to the main microfluidic channel. These CTCs could be retrieved conveniently for downstream molecular and cellular characterizations by moving the magnet to the other side of the microfluidic channel.
Figure 6.
Magnetophoresis. (A) Magnetic cell separation. Left: Trapping by cavities. Reproduced with permission.[84a] Copyright 2012, Royal Society of Chemistry. Right: Positive and negative selections. Reproduced with permission.[12b] Copyright 2013, American Association for the Advancement of Science. (B) Self-assembled magnetic bead array. Reproduced with permission.[86] Copyright 2010, The National Academy of Sciences, U.S.A. (C) Inherent paramagnetic properties of cells. Reproduced with permission.[87] Copyright 2006, Institution of Engineering and Technology.
Sophisticated microfluidic cell sorting systems integrating multiple cell separation modules have been reported for selective capture of rare cells such as CTCs from patient blood specimens. In a recent example, an integrated CTC capture system was demonstrated that was consisted of three different functional modules sequentially, a DLD module for depleting RBCs from blood samples, an inertial focusing module for alignment of blood cells, and a magnetophoretic module responsible for separating cells labeled with magnetic beads (Figure 6A).[12b] As discussed previously, surface expression of EpCAM on CTCs may be heterogeneous. Thus, isolation of CTCs based on their EpCAM expression may lose subpopulations of CTCs that may not express EpCAM. To differentiate and capture both EpCAM+ and EpCAM- CTC populations, in this work blood from human cancer patients was labeled separately with magnetic beads coated with either anti-EpCAM antibodies targeting EpCAM+ CTCs or antibodies against CD45 and CD15, markers of WBCs. After initial depletion of RBCs using the DLD module, remaining CTCs and WBCs were routed to the inertial focusing and magnetophoretic modules. EpCAM+ CTCs were positively enriched under the magnetic field whereas EpCAM- CTCs were enriched in a negative manner by depleting undesired CD45+ or CD15+ WBCs. Thus, this integrated CTC capture system was capable of capturing of both EpCAM+ and EpCAM- CTCs in the single microfluidic platform, which is not possible using only positive selection methods.
Although an external magnet can conveniently generate magnetic fields, embedding magnets directly into microfluidic devices can provide greater local magnetic forces. A recent work demonstrated a self-assembled magnetic microfluidic device, which contained a microfluidic channel built directly above a self-assembled NdFeB magnet. Highly efficient immunomagnetic separation of CTCs from blood samples was achieved in this device using negative immunomagnetic selection.[85] Another magnetic cell sorting microdevice (termed Ephesia system) was developed for maglignant B-cell immobilization and analysis (Figure 6B). This Ephesia system contained a column of functionalized superparamagnetic beads self-assembled onto an array of magnetic traps generated by microcontact printing. Using magnetic microbeads coated with antibodies specific to B-cells, B-cells from different clinical samples (blood, pleural effusion, and fine needle aspirates) were efficiently isolated with 98% of undesired T-cells depleted.[86] Moreover, the Ephesia immunophenotyping analysis using samples from various subjects (healthy donors and patients with chronic lymphocytic leukemia, mantle lymphoma and follicular lymphoma) was in good agreement with results from flow cytometry, supporting its utility as a rapid and low-cost clinical diagnostic system.
Because the presence of ions such as iron in blood confers magnetic properties to cells, magnetophoresis can directly differentiate blood cells without using chemical additives or magnetic microbeads. Although the efficiency of magnetophoresis is hampered by weak magnetic flux gradients on cells, this hurdle can be overcome by taking advantage of miniaturization and electroplating a ferromagnetic wire inside a microchip (Figure 6C).[87] As blood flowed through the wire magnetized by a permanent magnet, deoxyhemoglobin RBCs being paramagnetic were drawn closer to the wire whereas WBCs being diamagnetic were forced away from the wire. With an external magnetic flux of 0.2 T, this magnetic microfluidic cell sorting chip could isolate and enrich ~90% RBCs and WBCs into different collection channels.[87] Such strategy based on microfluidic magnetophoresis to examine paramagnetic signatures of blood cells was also recently utilized for disease diagnosis to differentiate and separate healthy RBCs from early- and late-stage malaria-infected RBCs.[88]
3.5. Electrical Methods and Acoustophoresis
In addition to magnetism, electrical fields can also be employed for microfluidic sorting of blood components with certain unique advantages over other principles. Firstly, electrical actuations and sensing are typically sensitive, rapid, convenient and robust. These characteristics can enable rapid, precise and well-controlled manipulations and measurements. Secondly, electrical methods can be easily integrated with other modules including electromagnetism, optoelectronics, electrochemical and electromechanical methods. Such compatibility and integrality of electrical methods allow multi-module strategies that can improve throughput, performance and functionality of microfluidic sorting of blood cells.
There are a variety of electrical methods for manipulation and sorting of blood components. Examples include electrohydrodynamics (EHD) for sorting and enrichment of RBCs from blood,[89] electroosmotic flow (EOF) for manipulation of WBCs,[90] electrowetting-on-dielectric (EWOD) for capture and enrichment of specific WBC subpopulations (Figure 7A),[91] electrophoresis (EP) for RBC lysis, separation, detection and focusing,[92] dielectrophoresis (DEP) for depleting RBCs polluted by lead ions (Figure 7A)[93] as well as plasmodium falciparum-infected RBCs[94] and separating WBCs and cancer cells,[95] and isoelectricity (IE) for high-throughput B-cell separation and characterization.[96]
Figure 7.
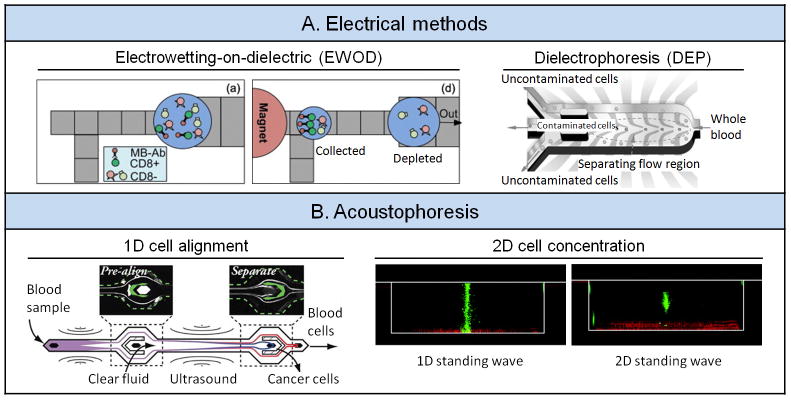
Electrical methods and acoustophoresis. (A) Left: Electrowetting-on-dielectric (EWOD). Reproduced with permission.[91] Copyright 2010, American Institute of Physics. Right: Dielectrophoresis (DEP). Reproduced with permission.[93] Copyright 2012, Wiley-VCH. (B) Left: 1D cell alignment. Reproduced with permission.[97] Copyright 2012, American Chemical Society. Right: 2D cell concentration. Reproduced with permission.[100] Copyright 2012, Royal Society of Chemistry.
The use of electrical methods in microfluidics, however, has often been associated with concerns about electrolysis that can generate hydrogen and oxygen gases and other undesired chemicals harmful to mammalian cells. In addition, elevated temperature due to Joule heating or high frequency in electrical methods can be difficult to control in a microfluidic environment, generating undesirable side effects when exceeding acceptable physiological ranges. Furthermore, electrical charges and ions are unevenly distributed within a cell. Thus, when electrical methods are applied to manipulate a cell, electrophysiological properties and molecular and cellular phenotypes of the cell may also be altered.
Acoustophoresis is another contact-free, label-free method that can focus particles laterally within microchannels. The generation of an ultrasound radiation force in an acoustically soft medium confined within acoustically rigid microchannel walls requires two criteria: (i) channel dimensions in concert with ultrasound frequencies, and (ii) a channel dimension that matches an integral multiple of half-wavelength in suspending fluid.[97] Under ultrasound fields, particles can move to either the pressure node or anti node of the ultrasound field in the microfluidic channel. As the magnitude of the acoustic force depends on particle size, density and compressibility, acoustophoresis has been utilized to isolate blood components such as peripheral blood progenitor cells (PBPCs) for transplantation,[98] blood plasma for biomarker detection,[99] and CTCs for cancer diagnostics (Figure 7B).[97] Moreover, 200-fold enrichment of RBCs was achieved under 2D acoustic standing wave in a microfluidic setting (Figure 7B).[100] It is worth noting that a recent study systematically examined the potential adverse effect of microfluidic acoustophoresis on phenotypic changes of microglia cells, human prostate cancer cells, human thrombocytes and WBCs, including their viability and proliferation and inflammatory responses. This study suggested that ultrasonic actuation with the operating voltage less than 10 Vpp would have negligible adverse impacts on many important cellular functions.[101]
4. Concluding Remarks
Conventional methods of sorting and separation of blood cells can be classified into two main categories and can be used alone or together. The first category falls under making use of the differences in physical properties between different blood cells for separation while the second category makes use of biological differences, such as surface protein markers to help discriminate between different cell types. While highly effective, there are limitations to such macroscale approaches, such as long processing time, high cost and the availability of good antibodies, and the amount of blood sample required. Owing to precise control over the cell microenvironment and the ability to scale down the operation to very small volumes of blood, recent advances in microfluidics have been gaining in importance as efficient and powerful approaches for high-throughput blood cell sorting and separation as well as non-invasive molecular and functional analysis down to a single-cell resolution. The field of blood cell preparation using microfluidics keeps evolving, and we foresee that new trends will lie in the following categories: (i) integration of multiple cell separation modules for advanced sorting of target blood cells, (ii) integration of upstream microfluidic blood cell sorting with downstream molecular, cellular and functional analysis on the single-chip platform, and (iii) complex, highly integrated microfluidic devices and systems with novel functionalities for high-throughput, high-content blood analysis. However, we need to reckon that current microfluidic cell sorting technologies are still facing significant challenges that need to be fully addressed in the near future, in order to fulfill their true potential and realize their impact on blood-based therapeutic and diagnostic applications. These challenges include the laborious fabrication process required for generating defect-free, intricate 3D microfluidic networks and their associated complex control and operation, and interfaces between delicate, highly integrated microfluidic systems with conventional macroscopic instruments. Nonetheless, given the potential and significant advantages of well-controlled microfluidic environment and the rich mechanisms that are available for manipulation of blood cells, we envision that microfluidic blood sorting systems may one day become a mainstay in the clinical and research laboratories.
Acknowledgments
We acknowledge valuable comments and suggestions on the manuscript from group members of the Integrated Biosystems and Biomechanics Laboratory. We thank Angela Hu, Mei Ki Cheung and Krystal Huijiao Guan for their assistance with the manuscript preparation. Work in Dr. Fu’s lab is supported by the National Science Foundation (CMMI 1129611, CBET 1149401, ECCS 1231826, CBET 1263889), the National Institute of Health (1R21HL114011), the American Heart Association (12SDG12180025), and the Department of Mechanical Engineering at the University of Michigan, Ann Arbor. Finally, we extend our apologies to colleagues in the field whose work we were unable to discuss or cite formally because of space constraints and imposed reference limitations.
Biographies

Dr. Zeta Tak For Yu has been a post-doctoral fellow of Mechanical Engineering at the University of Michigan, Ann Arbor since 2012. Dr. Yu received his Ph. D. degree from the University of California, Los Angeles (UCLA) and master and bachelor degrees from the Hong Kong University of Science and Technology (HKUST) in 2009, 2003 and 2001 respectively. Dr. Yu’s research focuses on applying microfluidics or lab-on-a-chip technology to perform cellular phenotyping and proteomics in immunology, cancer biology and stem cell biology.

Koh Meng Aw Yong received his B.Sc. (Hons) in Microbiology from the National University of Singapore in 2003 and his Ph.D in Pathobiology from The Johns Hopkins University in 2012. While most of his training was in cancer biology, Koh Meng is interested in using engineering tools to study how substrate mechanics influences cancer cell behavior as well as the development of microfluidic devices to improve cancer detection.

Dr. Jianping Fu has been an assistant professor of Mechanical and Biomedical Engineering at the University of Michigan, Ann Arbor since 2009. Dr. Fu received his Ph. D. degree from the Massachusetts Institute of Technology in 2007. He was an American Heart Association Postdoctoral Fellow at the University of Pennsylvania from 2007 to 2009. Dr. Fu’s research focuses on mechanobiology, stem cell biology, and applying microfabrication technology to illuminate biological systems at both the molecular and cellular levels. Dr. Fu is the recipient of the American Heart Association Scientist Development Grant (2012) and the National Science Foundation CAREER Award (2012).
References
- 1.Bruce Alberts AJ, Lewis Julian, Raff Martin, Roberts Keith, Walter Peter. Molecular Biology of the Cell. 4. New York: Garland Science; 2002. Table 22-1, Blood Cells. [Google Scholar]
- 2.a) Emery P, Dorner T. Ann Rheum Dis. 2011;70(12):2063–70. doi: 10.1136/ard.2010.148015. [DOI] [PubMed] [Google Scholar]; b) Iskandar HN, Ciorba MA. Transl Res. 2012;159(4):313–25. doi: 10.1016/j.trsl.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Rhea JM, Molinaro RJ. MLO Med Lab Obs. 2011;43(3):10–2. 16, 18. quiz 20, 22. [PubMed] [Google Scholar]; d) Szodoray P, Szabo Z, Kapitany A, Gyetvai A, Lakos G, Szanto S, Szucs G, Szekanecz Z. Autoimmun Rev. 2010;9(3):140–3. doi: 10.1016/j.autrev.2009.04.006. [DOI] [PubMed] [Google Scholar]; e) Vasan RS. Circulation. 2006;113(19):2335–62. doi: 10.1161/CIRCULATIONAHA.104.482570. [DOI] [PubMed] [Google Scholar]
- 3.a) Ganong WF. Review of medical physiology. Lange Medical Books/McGraw-Hill; New York: 2003. [Google Scholar]; b) Turgeon ML. Clinical Hematology: Theory and Procedures. Lippincott Williams & Wilkins; 2004. [Google Scholar]
- 4.a) Zhou Q, Kwa T, Liu Y, Revzin A. Expert Rev Anti Infect Ther. 2012;10(10):1079–81. doi: 10.1586/eri.12.102. [DOI] [PubMed] [Google Scholar]; b) Chen W, Huang NT, Li X, Yu ZT, Kurabayashi K, Fu J. Front Oncol. 2013;3:98. doi: 10.3389/fonc.2013.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Hayes DF. N Engl J Med. 2004;351(8):781–91. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]; b) Liu MC, Shields PG, Warren RD, Cohen P, Wilkinson M, Ottaviano YL, Rao SB, Eng-Wong J, Seillier-Moiseiwitsch F, Noone AM, Isaacs C. J Clin Oncol. 2009;27(31):5153–9. doi: 10.1200/JCO.2008.20.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Miller MC, Doyle GV, Terstappen LW. J Oncol. 2010;2010:617421. doi: 10.1155/2010/617421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones ML, Siddiqui J, Pienta KJ, Getzenberg RH. Prostate. 2013;73(2):176–81. doi: 10.1002/pros.22553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Greenbaum AM, Link DC. Leukemia. 2011;25(2):211–7. doi: 10.1038/leu.2010.248. [DOI] [PubMed] [Google Scholar]; b) Thomas J, Liu F, Link DC. Curr Opin Hematol. 2002;9(3):183–9. doi: 10.1097/00062752-200205000-00002. [DOI] [PubMed] [Google Scholar]
- 8.a) Bianchi DW. Br J Haematol. 1999;105(3):574–83. doi: 10.1046/j.1365-2141.1999.01383.x. [DOI] [PubMed] [Google Scholar]; b) Guetta E, Simchen MJ, Mammon-Daviko K, Gordon D, Aviram-Goldring A, Rauchbach N, Barkai G. Stem Cells Dev. 2004;13(1):93–9. doi: 10.1089/154732804773099290. [DOI] [PubMed] [Google Scholar]; c) Wachtel SS, Shulman LP, Sammons D. Clin Genet. 2001;59(2):74–9. doi: 10.1034/j.1399-0004.2001.590202.x. [DOI] [PubMed] [Google Scholar]
- 9.Dharmasiri U, Witek MA, Adams AA, Soper SA. Annu Rev Anal Chem. 2010;3:409–31. doi: 10.1146/annurev.anchem.111808.073610. [DOI] [PubMed] [Google Scholar]
- 10.a) Autebert J, Coudert B, Bidard FC, Pierga JY, Descroix S, Malaquin L, Viovy JL. Methods. 2012;57(3):297–307. doi: 10.1016/j.ymeth.2012.07.002. [DOI] [PubMed] [Google Scholar]; b) Bhagat AAS, Bow H, Hou HW, Tan SJ, Han J, Lim CT. Med Biol Eng Comput. 2010;48(10):999–1014. doi: 10.1007/s11517-010-0611-4. [DOI] [PubMed] [Google Scholar]; c) Cima I, Yee CW, Iliescu FS, Phyo WM, Lim KH, Iliescu C, Tan MH. Biomicrofluidics. 2013;7:1. doi: 10.1063/1.4780062. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Dong Y, Skelley AM, Merdek KD, Sprott KM, Jiang CS, Pierceall WE, Lin J, Stocum M, Carney WP, Smirnov DA. J Mol Diagn. 2013;15(2):149–157. doi: 10.1016/j.jmoldx.2012.09.004. [DOI] [PubMed] [Google Scholar]; e) Hou HW, Bhagat AAS, Lee WC, Huang S, Han J, Lim CT. Micromachines-Basel. 2011;2(3):319–343. [Google Scholar]; f) Lenshof A, Laurell T. Chem Soc Rev. 2010;39(3):1203–1217. doi: 10.1039/b915999c. [DOI] [PubMed] [Google Scholar]; g) Toner M, Irimia D. Annu Rev Biomed Eng. 2005;7:77–103. doi: 10.1146/annurev.bioeng.7.011205.135108. [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Yu L, Ng SR, Xu Y, Dong H, Wang YJ, Li CM. Lab Chip. 2013;13(16):3163–3182. doi: 10.1039/c3lc00052d. [DOI] [PubMed] [Google Scholar]
- 11.Chen W, Huang NT, Oh B, Lam RH, Fan R, Cornell TT, Shanley TP, Kurabayashi K, Fu J. Adv Healthc Mater. 2013 doi: 10.1002/adhm.201200378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a) Kim M, Mo Jung S, Lee KH, Jun Kang Y, Yang S. Artif Organs. 2010;34(11):996–1002. doi: 10.1111/j.1525-1594.2010.01114.x. [DOI] [PubMed] [Google Scholar]; b) Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E, Yu M, Chen PI, Morgan B, Trautwein J, Kimura A, Sengupta S, Stott SL, Karabacak NM, Barber TA, Walsh JR, Smith K, Spuhler PS, Sullivan JP, Lee RJ, Ting DT, Luo X, Shaw AT, Bardia A, Sequist LV, Louis DN, Maheswaran S, Kapur R, Haber DA, Toner M. Sci Transl Med. 2013;5(179):179ra47. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zeming KK, Ranjan S, Zhang Y. Nat Commun. 2013;4:1625. doi: 10.1038/ncomms2653. [DOI] [PubMed] [Google Scholar]
- 13.a) Bose S, Singh R, Hanewich-Hollatz M, Shen C, Lee CH, Dorfman DM, Karp JM, Karnik R. Sci Rep. 2013;3:2329. doi: 10.1038/srep02329. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Chen W, Weng S, Zhang F, Allen S, Li X, Bao L, Lam RH, Macoska JA, Merajver SD, Fu J. ACS Nano. 2013;7(1):566–75. doi: 10.1021/nn304719q. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Gurkan UA, Tasoglu S, Akkaynak D, Avci O, Unluisler S, Canikyan S, Maccallum N, Demirci U. Adv Healthc Mater. 2012;1(5):661–8. doi: 10.1002/adhm.201200009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardwick J. ISBT Science Series. 2008;3(2):148–176. [Google Scholar]
- 15.Kreuger A, Akerblom O, Hogman CF. Vox Sang. 1975;29(2):81–9. doi: 10.1111/j.1423-0410.1975.tb00483.x. [DOI] [PubMed] [Google Scholar]
- 16.a) Lozada JL, Caplanis N, Proussaefs P, Willardsen J, Kammeyer G. J Oral Implantol. 2001;27(1):38–42. doi: 10.1563/1548-1336(2001)027<0038:PPAISG>2.3.CO;2. [DOI] [PubMed] [Google Scholar]; b) Pasqualetti D, Ghirardini A, Cristina Arista M, Vaglio S, Fakeri A, Waldman AA, Girelli G. Transfus Apher Sci. 2004;30(1):23–8. doi: 10.1016/j.transci.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 17.Rossi EC, Simon TL, Wiley l. Rossi’s principles of transfusion medicine. Wiley-Blackwell; Chichester, UK; Hoboken, NJ: 2009. online. [Google Scholar]
- 18.Chernyshev AV, Tarasov PA, Semianov KA, Nekrasov VM, Hoekstra AG, Maltsev VP. J Theor Biol. 2008;251(1):93–107. doi: 10.1016/j.jtbi.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 19.de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, Doyle GV, Terstappen LW, Pienta KJ, Raghavan D. Clin Cancer Res. 2008;14(19):6302–9. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 20.Bonner WA, Hulett HR, Sweet RG, Herzenberg LA. Rev Sci Instrum. 1972;43(3):404–9. doi: 10.1063/1.1685647. [DOI] [PubMed] [Google Scholar]
- 21.Miltenyi S, Muller W, Weichel W, Radbruch A. Cytometry. 1990;11(2):231–8. doi: 10.1002/cyto.990110203. [DOI] [PubMed] [Google Scholar]
- 22.a) Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, Tibbe AG, Uhr JW, Terstappen LW. Clin Cancer Res. 2004;10(20):6897–904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]; b) Liberti PA, Rao CG, Terstappen LWMM. J Magn Magn Mater. 2001;225(1–2):301–307. [Google Scholar]
- 23.a) Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, Inserra E, Diederichs S, Iafrate AJ, Bell DW, Digumarthy S, Muzikansky A, Irimia D, Settleman J, Tompkins RG, Lynch TJ, Toner M, Haber DA. N Engl J Med. 2008;359(4):366–77. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Miyamoto DT, Lee RJ, Stott SL, Ting DT, Wittner BS, Ulman M, Smas ME, Lord JB, Brannigan BW, Trautwein J, Bander NH, Wu CL, Sequist LV, Smith MR, Ramaswamy S, Toner M, Maheswaran S, Haber DA. Cancer Discov. 2012;2(11):995–1003. doi: 10.1158/2159-8290.CD-12-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, Concannon KF, Donaldson MC, Sequist LV, Brachtel E, Sgroi D, Baselga J, Ramaswamy S, Toner M, Haber DA, Maheswaran S. Science. 2013;339(6119):580–4. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Yu M, Ting DT, Stott SL, Wittner BS, Ozsolak F, Paul S, Ciciliano JC, Smas ME, Winokur D, Gilman AJ, Ulman MJ, Xega K, Contino G, Alagesan B, Brannigan BW, Milos PM, Ryan DP, Sequist LV, Bardeesy N, Ramaswamy S, Toner M, Maheswaran S, Haber DA. Nature. 2012;487(7408):510–3. doi: 10.1038/nature11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.a) Frederick BA, Helfrich BA, Coldren CD, Zheng D, Chan D, Bunn PA, Jr, Raben D. Mol Cancer Ther. 2007;6(6):1683–91. doi: 10.1158/1535-7163.MCT-07-0138. [DOI] [PubMed] [Google Scholar]; b) Santisteban M, Reiman JM, Asiedu MK, Behrens MD, Nassar A, Kalli KR, Haluska P, Ingle JN, Hartmann LC, Manjili MH, Radisky DC, Ferrone S, Knutson KL. Cancer Res. 2009;69(7):2887–95. doi: 10.1158/0008-5472.CAN-08-3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Gun BT, Melchers LJ, Ruiters MH, de Leij LF, McLaughlin PM, Rots MG. Carcinogenesis. 2010;31(11):1913–21. doi: 10.1093/carcin/bgq187. [DOI] [PubMed] [Google Scholar]
- 26.a) Bohmer RM, Stroh HP, Johnson KL, LeShane ES, Bianchi DW. Fetal Diagn Ther. 2002;17(2):83–9. doi: 10.1159/000048014. [DOI] [PubMed] [Google Scholar]; b) Challen GA, Boles N, Lin KK, Goodell MA. Cytometry A. 2009;75(1):14–24. doi: 10.1002/cyto.a.20674. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Choolani M, O’Donoghue K, Talbert D, Kumar S, Roberts I, Letsky E, Bennett PR, Fisk NM. Mol Hum Reprod. 2003;9(4):227–35. doi: 10.1093/molehr/gag027. [DOI] [PubMed] [Google Scholar]; d) Mavrou A, Kouvidi E, Antsaklis A, Souka A, Kitsiou Tzeli S, Kolialexi A. Prenat Diagn. 2007;27(2):150–3. doi: 10.1002/pd.1640. [DOI] [PubMed] [Google Scholar]; e) Prieto B, Candenas M, Venta R, Ladenson JH, Alvarez FV. Clin Chem Lab Med. 2002;40(7):667–72. doi: 10.1515/CCLM.2002.114. [DOI] [PubMed] [Google Scholar]
- 27.a) SooHoo J, Walker G. Conf Proc IEEE Eng Med Biol Soc. 2007;2007:6319–22. doi: 10.1109/IEMBS.2007.4353800. [DOI] [PubMed] [Google Scholar]; b) Soohoo JR, Walker GM. Biomed Microdevices. 2009;11(2):323–9. doi: 10.1007/s10544-008-9238-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.a) Helleso OG, Lovhaugen P, Subramanian AZ, Wilkinson JS, Ahluwalia BS. Lab Chip. 2012;12(18):3436–40. doi: 10.1039/c2lc40375g. [DOI] [PubMed] [Google Scholar]; b) Kasukurti A, Potcoava M, Desai SA, Eggleton C, Marr DWM. Opt Express. 2011;19(11):10377–10386. doi: 10.1364/OE.19.010377. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Svoboda K, Block SM. Annu Rev Biophys Biomol Struct. 1994;23:247–285. doi: 10.1146/annurev.bb.23.060194.001335. [DOI] [PubMed] [Google Scholar]
- 29.Zhang XB, Wu ZQ, Wang K, Zhu J, Xu JJ, Xia XH, Chen HY. Anal Chem. 2012;84(8):3780–6. doi: 10.1021/ac3003616. [DOI] [PubMed] [Google Scholar]
- 30.a) Song JW, Cavnar SP, Walker AC, Luker KE, Gupta M, Tung YC, Luker GD, Takayama S. PLoS One. 2009;4(6):e5756. doi: 10.1371/journal.pone.0005756. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zheng X, Cheung LS, Schroeder JA, Jiang L, Zohar Y. Lab Chip. 2011;11(20):3431–9. doi: 10.1039/c1lc20455f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.a) Gossett DR, Weaver WM, Mach AJ, Hur SC, Tse HT, Lee W, Amini H, Di Carlo D. Anal Bioanal Chem. 2010;397(8):3249–67. doi: 10.1007/s00216-010-3721-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kersaudy-Kerhoas M, Sollier E. Lab Chip. 2013;13(17):3323–46. doi: 10.1039/c3lc50432h. [DOI] [PubMed] [Google Scholar]
- 32.Wilding P, Pfahler J, Bau HH, Zemel JN, Kricka LJ. Clin Chem. 1994;40(1):43–7. [PubMed] [Google Scholar]
- 33.Lee D, Sukumar P, Mahyuddin A, Choolani M, Xu G. J Chromatogr A. 2010;1217(11):1862–6. doi: 10.1016/j.chroma.2010.01.065. [DOI] [PubMed] [Google Scholar]
- 34.Sethu P, Sin A, Toner M. Lab Chip. 2006;6(1):83–9. doi: 10.1039/b512049g. [DOI] [PubMed] [Google Scholar]
- 35.Mohamed H, McCurdy LD, Szarowski DH, Duva S, Turner JN, Caggana M. IEEE Trans NanoBiosci. 2004;3(4):251–6. doi: 10.1109/tnb.2004.837903. [DOI] [PubMed] [Google Scholar]
- 36.Preira P, Grandne V, Forel JM, Gabriele S, Camara M, Theodoly O. Lab Chip. 2013;13(1):161–70. doi: 10.1039/c2lc40847c. [DOI] [PubMed] [Google Scholar]
- 37.Tan SJ, Yobas L, Lee GY, Ong CN, Lim CT. Biomed Microdevices. 2009;11(4):883–92. doi: 10.1007/s10544-009-9305-9. [DOI] [PubMed] [Google Scholar]
- 38.McFaul SM, Lin BK, Ma H. Lab Chip. 2012;12(13):2369–76. doi: 10.1039/c2lc21045b. [DOI] [PubMed] [Google Scholar]
- 39.a) Huang NT, Chen W, Oh BR, Cornell TT, Shanley TP, Fu J, Kurabayashi K. Lab Chip. 2012;12(20):4093–101. doi: 10.1039/c2lc40619e. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Chen W, Huang NT, Li X, Yu ZTF, Kurabayashi K, Fu J. Frontiers in Tumor Immunity. Cancer Immunotherapy & Immuno-monitoring: Mechanism, Treatment, Diagnosis, and Emerging Tools. 2003 In press. [Google Scholar]
- 40.a) Xu T, Lu B, Tai YC, Goldkorn A. Cancer Res. 2010;70(16):6420–6. doi: 10.1158/0008-5472.CAN-10-0686. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lin HK, Zheng S, Williams AJ, Balic M, Groshen S, Scher HI, Fleisher M, Stadler W, Datar RH, Tai YC, Cote RJ. Clin Cancer Res. 2010;16(20):5011–8. doi: 10.1158/1078-0432.CCR-10-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hosokawa M, Asami M, Nakamura S, Yoshino T, Tsujimura N, Takahashi M, Nakasono S, Tanaka T, Matsunaga T. Biotechnol Bioeng. 2012;109(8):2017–24. doi: 10.1002/bit.24471. [DOI] [PubMed] [Google Scholar]
- 42.a) Songjaroen T, Dungchai W, Chailapakul O, Henry CS, Laiwattanapaisal W. Lab Chip. 2012;12(18):3392–8. doi: 10.1039/c2lc21299d. [DOI] [PubMed] [Google Scholar]; b) Vella SJ, Beattie P, Cademartiri R, Laromaine A, Martinez AW, Phillips ST, Mirica KA, Whitesides GM. Anal Chem. 2012;84(6):2883–91. doi: 10.1021/ac203434x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Didar TF, Li K, Tabrizian M, Veres T. Lab Chip. 2013;13(13):2615–22. doi: 10.1039/c3lc50181g. [DOI] [PubMed] [Google Scholar]
- 44.Kim MS, Sim TS, Kim YJ, Kim SS, Jeong H, Park JM, Moon HS, Kim SI, Gurel O, Lee SS, Lee JG, Park JC. Lab Chip. 2012;12(16):2874–80. doi: 10.1039/c2lc40065k. [DOI] [PubMed] [Google Scholar]
- 45.Lin MX, Hyun KA, Moon HS, Sim TS, Lee JG, Park JC, Lee SS, Jung HI. Biosensors Bioelectron. 2013;40(1):63–7. doi: 10.1016/j.bios.2012.06.016. [DOI] [PubMed] [Google Scholar]
- 46.Di Carlo D. Lab Chip. 2009;9(21):3038–46. doi: 10.1039/b912547g. [DOI] [PubMed] [Google Scholar]
- 47.Di Carlo D, Irimia D, Tompkins RG, Toner M. Proc Natl Acad Sci U S A. 2007;104(48):18892–7. doi: 10.1073/pnas.0704958104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bhagat AA, Hou HW, Li LD, Lim CT, Han J. Lab Chip. 2011;11(11):1870–8. doi: 10.1039/c0lc00633e. [DOI] [PubMed] [Google Scholar]
- 49.Hou HW, Warkiani ME, Khoo BL, Li ZR, Soo RA, Tan DS, Lim WT, Han J, Bhagat AA, Lim CT. Sci Rep. 2013;3:1259. doi: 10.1038/srep01259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu L, Guan G, Hou HW, Bhagat AA, Han J. Anal Chem. 2012;84(21):9324–31. doi: 10.1021/ac302085y. [DOI] [PubMed] [Google Scholar]
- 51.Parichehreh V, Medepallai K, Babbarwal K, Sethu P. Lab Chip. 2013;13(5):892–900. doi: 10.1039/c2lc40663b. [DOI] [PubMed] [Google Scholar]
- 52.Tanaka T, Ishikawa T, Numayama-Tsuruta K, Imai Y, Ueno H, Matsuki N, Yamaguchi T. Lab Chip. 2012;12(21):4336–43. doi: 10.1039/c2lc40354d. [DOI] [PubMed] [Google Scholar]
- 53.a) Balbino TA, Aoki NT, Gasperini AAM, Oliveira CLP, Azzoni AR, Cavalcanti LP, de la Torre LG. Chem Eng J. 2013;226:423–433. [Google Scholar]; b) Hou S, Wang S, Yu ZTF, Zhu NQM, Liu K, Sun J, Lin WY, Shen CKF, Fang X, Tseng HR. Angew Chem Int Ed Engl. 2008;47(6):1072–1075. doi: 10.1002/anie.200704264. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Takayama S, McDonald JC, Ostuni E, Liang MN, Kenis PJA, Ismagilov RF, Whitesides GM. Proc Natl Acad Sci U S A. 1999;96(10):5545–5548. doi: 10.1073/pnas.96.10.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Torisawa YS, Mosadegh B, Luker GD, Morell M, O’Shea KS, Takayama S. Integr Biol. 2009;1(11–12):649–654. doi: 10.1039/b915965g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.a) Sethu P, Moldawer LL, Mindrinos MN, Scumpia PO, Tannahill CL, Wilhelmy J, Efron PA, Brownstein BH, Tompkins RG, Toner M. Anal Chem. 2006;78(15):5453–61. doi: 10.1021/ac060140c. [DOI] [PubMed] [Google Scholar]; b) Sethu P, Anahtar M, Moldawer LL, Tompkins RG, Toner M. Anal Chem. 2004;76(21):6247–53. doi: 10.1021/ac049429p. [DOI] [PubMed] [Google Scholar]
- 55.a) Choi S, Ku T, Song S, Choi C, Park JK. Lab Chip. 2011;11(3):413–8. doi: 10.1039/c0lc00148a. [DOI] [PubMed] [Google Scholar]; b) Choi S, Song S, Choi C, Park JK. Lab Chip. 2007;7(11):1532–8. doi: 10.1039/b705203k. [DOI] [PubMed] [Google Scholar]; c) Choi S, Song S, Choi C, Park JK. Small. 2008;4(5):634–41. doi: 10.1002/smll.200700308. [DOI] [PubMed] [Google Scholar]
- 56.Bernate JA, Liu C, Lagae L, Konstantopoulos K, Drazer G. Lab Chip. 2013;13(6):1086–92. doi: 10.1039/c2lc40927e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davis JA, Inglis DW, Morton KJ, Lawrence DA, Huang LR, Chou SY, Sturm JC, Austin RH. Proc Natl Acad Sci U S A. 2006;103(40):14779–84. doi: 10.1073/pnas.0605967103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beech JP, Holm SH, Adolfsson K, Tegenfeldt JO. Lab Chip. 2012;12(6):1048–51. doi: 10.1039/c2lc21083e. [DOI] [PubMed] [Google Scholar]
- 59.Huang R, Barber TA, Schmidt MA, Tompkins RG, Toner M, Bianchi DW, Kapur R, Flejter WL. Prenat Diagn. 2008;28(10):892–9. doi: 10.1002/pd.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holm SH, Beech JP, Barrett MP, Tegenfeldt JO. Lab Chip. 2011;11(7):1326–32. doi: 10.1039/c0lc00560f. [DOI] [PubMed] [Google Scholar]
- 61.Loutherback K, D’Silva J, Liu L, Wu A, Austin RH, Sturm JC. AIP Adv. 2012;2(4):42107. doi: 10.1063/1.4758131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou R, Chang HC. J Colloid Interface Sci. 2005;287(2):647–56. doi: 10.1016/j.jcis.2005.02.023. [DOI] [PubMed] [Google Scholar]
- 63.Browne AW, Ramasamy L, Cripe TP, Ahn CH. Lab Chip. 2011;11(14):2440–6. doi: 10.1039/c1lc20144a. [DOI] [PubMed] [Google Scholar]
- 64.Faivre M, Abkarian M, Bickraj K, Stone HA. Biorheology. 2006;43(2):147–59. [PubMed] [Google Scholar]
- 65.Shevkoplyas SS, Yoshida T, Munn LL, Bitensky MW. Anal Chem. 2005;77(3):933–7. doi: 10.1021/ac049037i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang S, Undar A, Zahn JD. Lab Chip. 2006;6(7):871–80. doi: 10.1039/b516401j. [DOI] [PubMed] [Google Scholar]
- 67.a) Kretzmer G, Schugerl K. Appl Microbiol Biotechnol. 1991;34(5):613–6. doi: 10.1007/BF00167909. [DOI] [PubMed] [Google Scholar]; b) Li YS, Haga JH, Chien S. J Biomech. 2005;38(10):1949–71. doi: 10.1016/j.jbiomech.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 68.Kotz KT, Xiao W, Miller-Graziano C, Qian WJ, Russom A, Warner EA, Moldawer LL, De A, Bankey PE, Petritis BO, Camp DG, 2nd, Rosenbach AE, Goverman J, Fagan SP, Brownstein BH, Irimia D, Xu W, Wilhelmy J, Mindrinos MN, Smith RD, Davis RW, Tompkins RG, Toner M. Nat Med. 2010;16(9):1042–7. doi: 10.1038/nm.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450(7173):1235–9. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, Floyd FP, Gilman AJ, Lord JB, Winokur D, Springer S, Irimia D, Nagrath S, Sequist LV, Lee RJ, Isselbacher KJ, Maheswaran S, Haber DA, Toner M. Proc Natl Acad Sci U S A. 2010;107(43):18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang S, Liu K, Liu J, Yu ZT, Xu X, Zhao L, Lee T, Lee EK, Reiss J, Lee YK, Chung LW, Huang J, Rettig M, Seligson D, Duraiswamy KN, Shen CK, Tseng HR. Angew Chem Int Ed Engl. 2011;50(13):3084–8. doi: 10.1002/anie.201005853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hughes AD, Mattison J, Powderly JD, Greene BT, King MR. J Vis Exp. 2012;(64):e4248. doi: 10.3791/4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sheng W, Chen T, Kamath R, Xiong X, Tan W, Fan ZH. Anal Chem. 2012;84(9):4199–206. doi: 10.1021/ac3005633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Choi S, Karp JM, Karnik R. Lab Chip. 2012;12(8):1427–30. doi: 10.1039/c2lc21225k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Launiere C, Gaskill M, Czaplewski G, Myung JH, Hong S, Eddington DT. Anal Chem. 2012;84(9):4022–8. doi: 10.1021/ac2033408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li P, Gao Y, Pappas D. Anal Chem. 2012;84(19):8140–8. doi: 10.1021/ac302002a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.a) Johnson JL, Jones MB, Ryan SO, Cobb BA. Trends Immunol. 2013;34(6):290–8. doi: 10.1016/j.it.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Varki A, Esko JD, Colley KJ. 2009 [Google Scholar]
- 78.a) Laderach DJ, Gentilini L, Jaworski FM, Compagno D. Prostate Cancer. 2013;2013:519436. doi: 10.1155/2013/519436. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Padler-Karavani V. Cancer Lett. 2013 doi: 10.1016/j.canlet.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 79.a) Simone G, Malara N, Trunzo V, Perozziello G, Neuzil P, Francardi M, Roveda L, Renne M, Prati U, Mollace V, Manz A, Di Fabrizio E. Small. 2013;9(12):2152–2161. doi: 10.1002/smll.201202867. [DOI] [PubMed] [Google Scholar]; b) Simone G, Neuzil P, Perozziello G, Francardi M, Malara N, Di Fabrizio E, Manz A. Lab Chip. 2012;12(8):1500–1507. doi: 10.1039/c2lc21217j. [DOI] [PubMed] [Google Scholar]
- 80.Zhu H, Stybayeva G, Macal M, Ramanculov E, George MD, Dandekar S, Revzin A. Lab Chip. 2008;8(12):2197–205. doi: 10.1039/b810244a. [DOI] [PubMed] [Google Scholar]
- 81.Gurkan UA, Anand T, Tas H, Elkan D, Akay A, Keles HO, Demirci U. Lab Chip. 2011;11(23):3979–89. doi: 10.1039/c1lc20487d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hou S, Zhao L, Shen Q, Yu J, Ng C, Kong X, Wu D, Song M, Shi X, Xu X, OuYang WH, He R, Zhao XZ, Lee T, Brunicardi FC, Garcia MA, Ribas A, Lo RS, Tseng HR. Angew Chem Int Ed Engl. 2013;52(12):3379–83. doi: 10.1002/anie.201208452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ariyasu S, Hanaya K, Watanabe E, Suzuki T, Horie K, Hayase M, Abe R, Aoki S. Langmuir. 2012;28(36):13118–26. doi: 10.1021/la302393p. [DOI] [PubMed] [Google Scholar]
- 84.a) Kang JH, Krause S, Tobin H, Mammoto A, Kanapathipillai M, Ingber DE. Lab Chip. 2012;12(12):2175–81. doi: 10.1039/c2lc40072c. [DOI] [PubMed] [Google Scholar]; b) Huang YY, Hoshino K, Chen P, Wu CH, Lane N, Huebschman M, Liu H, Sokolov K, Uhr JW, Frenkel EP, Zhang JX. Biomed Microdevices. 2013;15(4):673–81. doi: 10.1007/s10544-012-9718-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Casavant BP, Strotman LN, Tokar JJ, Thiede SM, Traynor AM, Ferguson JS, Lang JM, Beebe DJ. Lab Chip. 2013 doi: 10.1039/c3lc50912e. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Casavant BP, Guckenberger DJ, Berry SM, Tokar JT, Lang JM, Beebe DJ. Lab Chip. 2013;13(3):391–6. doi: 10.1039/c2lc41136a. [DOI] [PubMed] [Google Scholar]
- 85.Issadore D, Shao H, Chung J, Newton A, Pittet M, Weissleder R, Lee H. Lab Chip. 2011;11(1):147–51. doi: 10.1039/c0lc00149j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Saliba AE, Saias L, Psychari E, Minc N, Simon D, Bidard FC, Mathiot C, Pierga JY, Fraisier V, Salamero J, Saada V, Farace F, Vielh P, Malaquin L, Viovy JL. Proc Natl Acad Sci U S A. 2010;107(33):14524–9. doi: 10.1073/pnas.1001515107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Han KH, Frazier AB. IEE Proc Nanobiotechnol. 2006;153(4):67–73. doi: 10.1049/ip-nbt:20050019. [DOI] [PubMed] [Google Scholar]
- 88.Nam J, Huang H, Lim H, Lim C, Shin S. Anal Chem. 2013;85(15):7316–23. doi: 10.1021/ac4012057. [DOI] [PubMed] [Google Scholar]
- 89.Arifin DR, Yeo LY, Friend JR. Biomicrofluidics. 2007;1(1):14103. doi: 10.1063/1.2409629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kang YJ, Li DQ, Kalams SA, Eid JE. Biomed Microdevices. 2008;10(2):243–249. doi: 10.1007/s10544-007-9130-y. [DOI] [PubMed] [Google Scholar]
- 91.Shah GJ, Veale JL, Korin Y, Reed EF, Gritsch HA, Kim CJ. Biomicrofluidics. 2010;4(4) doi: 10.1063/1.3509457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.a) Gao J, Yin XF, Fang ZL. Lab Chip. 2004;4(1):47–52. doi: 10.1039/b310552k. [DOI] [PubMed] [Google Scholar]; b) Xuan X, Li D. Electrophoresis. 2005;26(18):3552–60. doi: 10.1002/elps.200500298. [DOI] [PubMed] [Google Scholar]
- 93.Wang MW. Electrophoresis. 2012;33(5):780–7. doi: 10.1002/elps.201100354. [DOI] [PubMed] [Google Scholar]
- 94.Jubery TZ, Dutta P. Electrophoresis. 2013;34(5):643–50. doi: 10.1002/elps.201200560. [DOI] [PubMed] [Google Scholar]
- 95.Gao J, Riahi R, Sin ML, Zhang S, Wong PK. Analyst. 2012;137(22):5215–21. doi: 10.1039/c2an35707k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vahey MD, Voldman J. Anal Chem. 2009;81(7):2446–2455. doi: 10.1021/ac8019575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Augustsson P, Magnusson C, Nordin M, Lilja H, Laurell T. Anal Chem. 2012;84(18):7954–62. doi: 10.1021/ac301723s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dykes J, Lenshof A, Astrand-Grundstrom IB, Laurell T, Scheding S. PLoS One. 2011;6(8):e23074. doi: 10.1371/journal.pone.0023074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lenshof A, Ahmad-Tajudin A, Jaras K, Sward-Nilsson AM, Aberg L, Marko-Varga G, Malm J, Lilja H, Laurell T. Anal Chem. 2009;81(15):6030–7. doi: 10.1021/ac9013572. [DOI] [PubMed] [Google Scholar]
- 100.Nordin M, Laurell T. Lab Chip. 2012;12(22):4610–6. doi: 10.1039/c2lc40629b. [DOI] [PubMed] [Google Scholar]
- 101.Burguillos MA, Magnusson C, Nordin M, Lenshof A, Augustsson P, Hansson MJ, Elmer E, Lilja H, Brundin P, Laurell T, Deierborg T. PLoS One. 2013;8(5):e64233. doi: 10.1371/journal.pone.0064233. [DOI] [PMC free article] [PubMed] [Google Scholar]