Abstract
Triple-negative breast cancer (TNBC) is a subclass of breast cancers (i.e. estrogen receptor negative, progesterone receptor negative, and HER2 negative) that have poor prognosis and very few identified molecular targets. Strikingly, a high percentage of TNBC’s overexpress the epidermal growth factor receptor (EGFR), yet EGFR inhibition has yielded little clinical benefit. Over the last decade, advances in EGFR biology have established that EGFR functions in two distinct signaling pathways: 1) classical membrane-bound signaling, and 2) nuclear signaling. Previous studies have demonstrated that nuclear EGFR (nEGFR) can enhance resistance to anti-EGFR therapies and is correlated with poor overall survival in breast cancer. Based on these findings we hypothesized that nEGFR may promote intrinsic resistance to cetuximab in TNBC. To examine this question, a battery of TNBC cell lines and human tumors were screened and found to express nEGFR. Knockdown of EGFR expression demonstrated that TNBC cell lines retained dependency on EGFR for proliferation, yet all cell lines were resistant to cetuximab. Further, Src Family Kinases (SFKs) influenced nEGFR translocation in TNBC cell lines and in vivo tumor models, where inhibition of SFK activity led to potent reductions in nEGFR expression. Inhibition of nEGFR translocation led to a subsequent accumulation of EGFR on the plasma membrane, which greatly enhanced sensitivity of TNBC cells to cetuximab. Collectively, these data suggest that targeting both the nEGFR signaling pathway, through the inhibition of its nuclear transport, and the classical EGFR signaling pathway with cetuximab may be a viable approach for the treatment of TNBC patients.
Keywords: triple-negative breast cancer, nuclear EGFR, cetuximab, resistance
INTRODUCTION
Approximately 15–20% of all breast cancers lack expression of the estrogen receptor, progesterone receptor, and epidermal growth factor receptor 2 (HER2), and are thus considered to be triple-negative breast cancers (TNBCs) (1, 2). While a high percentage of TNBC patients initially respond to conventional chemotherapy, they tend to have a higher rate of relapse and worse prognosis as compared to other breast cancer subtypes (1, 2). In efforts to identify new molecular targets in TNBC, various groups have performed gene expression profiling studies and identified that the epidermal growth factor receptor (EGFR) is commonly overexpressed (3–6). While inhibition of EGFR activity has yielded modest clinical success in TNBC, substantial gains in clinical response rates have not been achieved (7, 8). Thus, improving the efficacy of anti-EGFR therapy in TNBC is imperative.
Classically, EGFR functions as a plasma membrane-bound receptor tyrosine kinase (RTK) that initiates growth and survival signals (9). However, studies over the last 15 years have identified that EGFR can be localized and function from intracellular organelles, one of which includes the nucleus (10, 11). Within the nucleus, EGFR can function as a co-transcription factor to regulate genes involved in tumor progression (10, 11), in addition to functioning as a nuclear kinase to enhance DNA replication and repair, (12–14). These nuclear functions have been linked to three parameters of tumor biology: 1) inverse correlation with overall survival in numerous cancers (15–20), 2) resistance to therapeutic agents including radiation (12, 21–24), chemotherapy (12, 13, 24), and anti-EGFR therapies gefitinib (25) and cetuximab (26), and 3) enhanced tumor growth (27, 28). These findings suggest that tumors rely on two distinct compartments of EGFR signaling to sustain their oncogenic phenotype: 1) classical membrane-bound EGFR signaling, and 2) nEGFR signaling.
Previous work from our laboratory has identified that non-small cell lung cancer (NSCLC) cells that have acquired resistance to cetuximab express increased nEGFR and Src Family Kinase (SFK) activity (26, 29). SFK inhibition blocked nEGFR translocation in cetuximab resistant cells, and led to an increase in plasma membrane EGFR expression and enhanced sensitivity to cetuximab (26, 30). Further, the SFK dependent phosphorylation site on EGFR, tyrosine 1101 (Y1101), was identified to play a critical role in initiating EGFR’s nuclear transport (30). These studies suggest that nEGFR is a critical molecular determinant for cetuximab resistance and that SFK’s play an important role in regulating nEGFR translocation.
Based on these previous studies, we hypothesized that nEGFR may promote intrinsic resistance to cetuximab in TNBC. To examine this question, a battery of TNBC cell lines and human tumors were screened and found to express nEGFR. While TNBC cell lines were notably resistant to cetuximab therapy, all lines retained dependency on EGFR for proliferation. Further, SFKs influenced nEGFR transport in TNBC, where the overexpression of a negative regulator of Src decreased EGFR activity at tyrosine 1101 and inhibited nEGFR translocation. Interestingly, the creation of stable cell lines overexpressing each SFK demonstrated that all SFKs could promote nEGFR translocation. Treatment of TNBC cell lines and xenograft tumors with the anti-SFK therapeutic dasatinib inhibited nEGFR translocation, and enhanced surface level EGFR accumulation. Importantly, pre-treatment of TNBC cell lines with dasatinib greatly enhanced the sensitivity of cetuximab-resistant TNBC cell lines to cetuximab. Collectively, our data suggest that abrogating nEGFR translocation with SFK inhibitors may greatly enhance the efficacy of cetuximab in TNBC.
MATERIALS AND METHODS
Cell Lines
The human breast cancer cell lines SKBr3, BT474, BT549, MDAMB231 and MDAMB468, MCF-7, and the Chinese hamster ovary cell line CHOK1 were purchased from ATCC in November 2010 (Manassas, VA, USA). SUM149, SUM229, and SUM159 were purchased from Asterand in November 2010 (Detroit, MI, USA). All cell lines were authenticated by the indicated source and not by our laboratory. All cell lines were maintained in their respective media (Mediatech Inc., VA, USA) with 1% penicillin and streptomycin; SKBr3, BT549, and MDAMB231, Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum (FBS); BT474, RPMI 1640 with 10% FBS; SUM149, SUM229, and SUM159, F12K medium with 5% FBS, 1 μg/mL hydrocortisone and 5 μg/mL insulin; MDAMB468 and MCF-7, DMEM/F12K medium with 10% FBS; CHOK1 F12K medium with 10% FBS.
Antibodies, Compounds and TMAs
All antibodies were obtained from the following sources: EGFR (SC-03), pEGFR-1173 (SC-10168), HER2 (SC-284), SLAP (SC-1215), Histone H3 (SC-8654), HRP-conjugated goat-anti-rabbit IgG, goat-anti-mouse IgG, donkey-anti-goat IgG, EGFR blocking peptide (SC-03 P) purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA, USA). SFK (CS2123), pSFK-Y419 (CS2101), pEGFR-Y1045 (CS2237), pEGFR-Y1068 (CS3777), pHER2-Y1221/1222 (CS2243), c-Cbl (CS2747), GAPDH (CS2118), calnexin (CS2679), and anti-Flag (CS8146) purchased from Cell Signaling Technology (Beverly, MA, USA). pEGFR-Y1101 (ab76195) and EGFR (ab52894) purchased from Abcam (Cambridge, MA, USA). α-Tubulin purchased from Calbiochem (San Diego, CA, USA). Dasatinib (BMS-354825, Sprycel) was purchased from LC Laboratories (Wobum, MA, USA) and cetuximab (C225, Erbitux™) was purchased from University of Wisconsin Pharmacy. EGF was purchased from Millipore (Billerica, MA, USA). Two human TNBC TMAs (#695711112B and #69572306) were purchased from TriStar Technology Group (Rockville, MD, USA).
Cellular fractionation and immunoblotting analysis
Cellular fractionation and whole cell lysis were performed and quantitated as previously described (26, 31). ECL chemiluminescence detection system was used to visualize proteins. α-Tubulin, calnexin, and Histone H3 were used as loading and purity controls, respectively.
Immunoprecipitation
Cells were processed for IP as previously described (31). 250 μg of protein and 2 μg of SLAP primary antibody were used for IP.
Plasmids constructs, transfection, and siRNA technology
The following vectors were kindly supplied: pcDNA3.0-caSrc, -wtSRC and –EGFR wild type (WT) and –EGFRY1101F, Dr. J. Boerner (Wayne State University School of Medicine, Karmanos Cancer Institute, MI, USA); pcDNA3-SLAP, Dr. S. Roche (Centre de Recherche de Biochimie Macromoléculaire, Montpellier France); pTRE2pur-HA-Fyn, -Hck, and -Lck, Dr. P.S. Mischel (University of California, San Diego). WT human pDONR223-FGR (Plasmid 23877) pDONR223-Blk (Plasmid 23940) were purchased from Addgene (Cambridge, MA, USA). pQCXIP-YES, and –LYN as previously described (30). All SFK’s were sub-cloned into the PAC1/AGEI restriction sites of the pQCXIP expression vector (Clontech, Mountain View, CA, USA). Both transient and stable transfections were performed using Lipofectamine LTX and Opti-MEM I (Life Technology, Grand Island, NY). Stable transfection commenced 48 hr post transfection via addition of 500 ng/ml puromycin to the growth media. Single cell clones were chosen for expansion and validation for specific SFK expression.
For siRNAs, cells were transfected with 30 nM siEGFR (ON-TARGETplus, SMART pool #L-003114-00, Dharmacon, Lafayette, CO, USA) or siNon-targeting (NT) (ON-TARGETplus Non-targeting Pool, D-001810, Dharmacon) using Lipofectamine RNAiMAX (Life Technology) according to the manufacturer’s instructions. Vehicle (Veh) treated cells were treated with RNAiMAX only.
Cell Proliferation Assay
Crystal violet assay and Cell Counting Kit-8 (Dojindo Molecular Technologies, Rockville, MD, USA) were performed as previously described (26, 32). Cellular proliferation was measured 72–96 hr post siRNA and 96 hour post drug treatment.
Transmission Electron Microscopy
Cells were plated on glass cover slips at ~ 90% confluency. The pre-embedding labeling method was used for processing as previously described (33). Specifically, 0.8% Triton-X-100 was used for permeabilization and 7 ug/mL of EGFR primary antibody was used (SC-03, Santa Cruz). Cells were silver enhanced for 1.5 hr. Cells were sectioned onto copper grids at ~90 nm slices.
Immunofluorescence
Cells were processed for IF staining of EGFR as previously described (31). Primary antibody: EGFR-SC-03, 1:100. Secondary antibody (Life technologies): Alex Fluor 546 at 1:600 for 30 min-1 hr. All cells were mounted with ProLong® Gold Antifade Reagent with DAPI (Life Technologies). Confocal IF microscopy was performed using an A1 Nikon confocal microscope (600X). Z-slices were taken at 150 nm slices.
Nuance imaging Analysis
For image analysis, EGFR (ab52894, 1:50) and anti-E Cadherin antibody (NCH-38, Dako at 1:100 dilution) were used for IF staining. Images were acquired on the Nuance Multispectral Imaging System (Caliper Life Sciences, 200X). A spectral library comprised of the fluorescent spectrum of each fluorophore was constructed from vehicle treated cells stained with each fluorophore individually. Images were analyzed on the inForm Image Analysis Software (Caliper Life Sciences) as previously described (34) by pathologist D.Y. Relative expression of EGFR in each compartment was expressed as a ratio of proportion of counts in the high intensity bins (bins 6 to 10) divided by the proportion of counts in the low intensity bins (bins 1 to 5).
Immunohistochemistry
Cells were processed for IHC as previously described (32). EGFR antibody (SC-03) was used at a 1:100 dilution. The nEGFR staining pattern was scored by pathologist (D.Y) analysis at 5% increments by visual estimation at 20X magnification. Cases with at least one replicate core containing at least 5% of tumor cells demonstrating strong nuclear EGFR IHC protein expression were scored as nEGFR positive.
Flow Cytometry
Cells were processed as previously described (26). Cells were analyzed using a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA). Propidium iodide was added to each sample at a final concentration of 5mg/ml. Histogram analysis was performed using FlowJo software (Tree Star Inc. Ashland, OR, USA).
Mouse xenograft model and tumor collection
Athymic nude mice (4–6 week-old females) were obtained from Harlan Laboratories (Indianapolis, IN, USA). All animal procedures and maintenance were conducted in accordance with the institutional guidelines of the University of Wisconsin. 12 mice were injected in the dorsal flank with 2×106 MDAMB468 cells. Once tumors reached 100 mm3, mice were randomized into treatment groups: vehicle (sodium citrate monobasic buffer) or dasatinib (50 mg/kg/day). Mice were treated once daily for 4 days via oral gavage. Tumor volume measurements were evaluated by digital calipers and calculated by the formula (p)/6 x (large diameter) x (small diameter)2. Tumors were collected, processed, and stained as previously described (32, 35).
Statistical Analysis
Student t-tests were used to evaluate the significance in proliferation rate between vehicle and siEGFR or drug treated cells. Student t-tests were also used to evaluate significance in nEGFR expression levels by Nuance imaging analysis between vehicle and dasatinib treated cells. Differences were considered statistically significant if *P<0.05. Pearson’s correlation coefficient and Manders’ overlap coefficient for co-localization were calculated using Nikon NIS-Elements software. Significance of strong interaction is considered for values ≥0.5(36).
RESULTS
TNBC cell lines and human tumors express nuclear localized EGFR
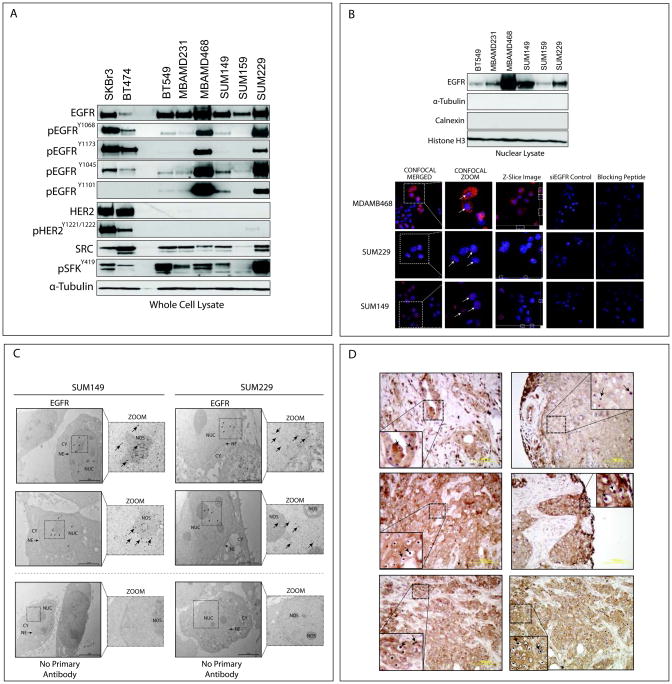
Six established TNBC cell lines were evaluated for EGFR expression (Figure 1A). All cell lines expressed total and activated forms of EGFR, in which the autophosphorylation status of EGFR at tyrosine 1068, 1173, and 1045 as well as the SFK specific phosphorylation site, tyrosine 1101, were evaluated. All TNBC cell lines expressed activated SFKs, as observed in previous studies (37, 38) (Figure 1A). Total and activated HER2 expression levels were low in all TNBC cell lines compared to HER2 positive cell lines SKBr3 and BT474.
Figure 1. TNBC cell lines and human tumors express nuclear localized EGFR.
(A) TNBC cells express total and activated forms of EGFR. Whole cell lysate was harvested from six TNBC cell lines and two HER2 positive cell lines. α-Tubulin was used as a loading control. (B) TNBC cells express nEGFR. Cell lines were harvested for nuclear proteins. α-Tubulin, calnexin, and Histone H3 were used as loading and purity controls, respectively. Confocal IF microscopy depicts nEGFR expression. Merged images were magnified to depict nEGFR (Confocal zoom, white arrows). A single Z-Slice image depicts overlap between blue and red signal (white dashed-line boxes). Magnification 600X. EGFR primary antibody specificity was validated with siEGFR and blocking peptides. (C) Immunogold labeling of nEGFR. TNBC cells were fixed and processed for transmission EM. CY, cytoplasm; NE, nuclear envelope; NUC, nucleus; NOS, nucleolus. Images were digitally zoomed to highlight gold particles in the nucleus (black arrows). (D) Human TNBC tumors express nEGFR. IHC staining for EGFR was performed on a total of 74 TNBC patient tumor sections. Representative cases demonstrating nEGFR expression are depicted (black arrows).
Since TNBC cell lines expressed EGFR, we hypothesized that some cell lines may also express nEGFR. Variant levels of nEGFR expression were observed in TNBC cell lines by nuclear fractionation analysis (Figure 1B). The harvested nuclear lysate was free from contaminating cytoplasmic and ER associated proteins, as indicated by lack of α-Tubulin and calnexin. The nuclear protein Histone H3 was used as a loading and nuclear protein purity control. In addition, confocal immunofluorescent (IF) microscopy indicated strong nEGFR immunofluorescent staining in MDAMB468, SUM229, and SUM149 cells (Figure 1B) by merging DAPI and Alexa Fluor 546 labeled EGFR (white arrows, magnified image). Statistical significance of co-localization was analyzed by Pearson’s and Manders’ correlation coefficients (significance of a strong interaction is ≥0.5) (36). For MDAMB468, SUM229, and SUM149 the Pearson’s coefficients were 0.52±0.04, 0.58±0.01, and 0.65±0.02, and the Manders’ overlap coefficients were 0.70±0.02, 0.78±0.03, and 0.84±0.01, (n=50 cells). While homogenous nEGFR staining was observed in SUM149 and SUM229 cells by IF, nEGFR staining in MDAMB468 cells was more heterogeneous. Knockdown of EGFR using siRNA or pre-incubation of primary antibody with blocking peptides led to dramatic decreases in EGFR signal. There was no signal detected from cells incubated with secondary antibody only (data not shown). We further validated nEGFR expression using transmission electron microscopy (EM) (Figure 1C). EGFR labeled with immunogold conjugated secondary antibodies indicated that EGFR was indeed localized in the nucleus, with localization in the nucleolus and around the nuclear envelope.
Given that nEGFR was expressed in established TNBC cell lines, we probed a human tissue microarray (TMA) containing 74 TNBC patient tumors for EGFR expression and localization. Pathologist analysis of tumors stained for EGFR via immunohistochemistry (IHC) indicated that 19% of the tumors expressed nEGFR (Figure 1D). Interestingly, nEGFR was highly localized to the nucleolus in greater than 5% of nEGFR positive tumors. Additionally, some tumor sections contained concentrated nEGFR, while other areas of the same tumor lacked nEGFR expression. There was no signal detected from cores stained with secondary antibody only (data not shown). Collectively, these data demonstrate that TNBC cell lines and human tumors express nEGFR.
TNBC cells are resistant to cetuximab therapy, but dependent on EGFR for proliferation
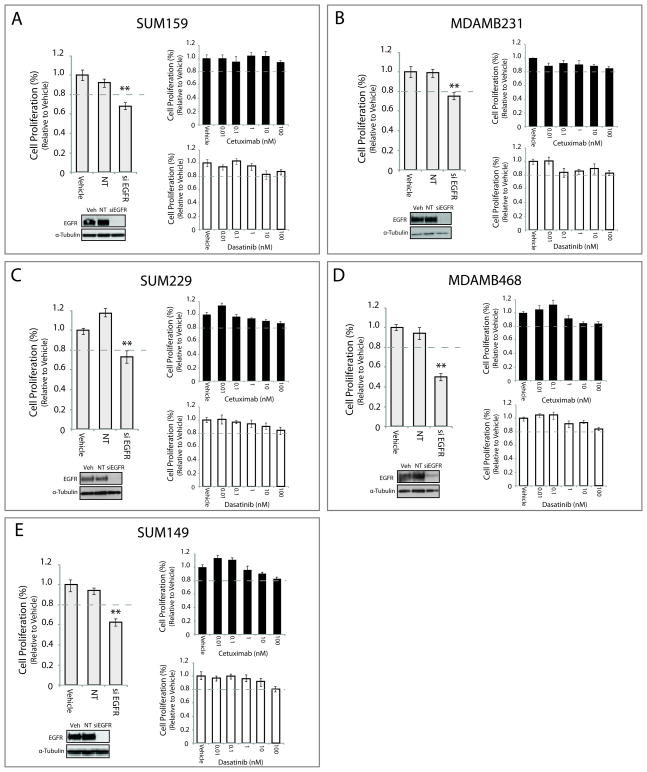
To determine the role of EGFR in TNBC proliferation, studies were performed to knock down EGFR expression in various TNBC cell lines using an EGFR directed siRNA pool. Loss of EGFR expression led to a 23–50% reduction in cell proliferation as compared to cells treated with vehicle or non-targeting (NT) siRNA (Figure 2). Each cell line challenged with increasing doses of cetuximab (from 0.01nM to 100nM) demonstrated only minor reductions in proliferation. The cell lines MDAMB231 (Figure 2B) and MDAMB468 (Figure 2D) demonstrated a 15% reduction in proliferation upon treatment with 100nM of cetuximab, whereas the SUM159 (Figure 2A), SUM229 (Figure 2C) and SUM149 (Figure 2E) were unaffected at this dose. Additionally, TNBC cell lines treated with increasing doses of dasatinib (0.01nM–100nM) were relatively resistant to growth inhibition. These results indicate that TNBC cell lines depend on EGFR for proliferation but are relatively resistant to cetuximab.
Figure 2. TNBC cell lines are dependent on EGFR for proliferation, but are intrinsically resistant to cetuximab and dasatinib.
Cell lines were incubated with siEGFR, non-targeting (NT) siRNA, or vehicle for 72–96 hr prior to performing proliferation assays (A–E). Cells were treated with cetuximab or dasatinib at indicated doses for the same time course. Proliferation is plotted as a percentage of growth relative to vehicle treated cells (n=3). Whole cell lysate was harvested from all cell lines at the same time point to confirm knockdown of EGFR. Data points are represented as mean±s.e.m. **p<0.01.
SFKs mediate the nuclear translocation of EGFR in TNBC
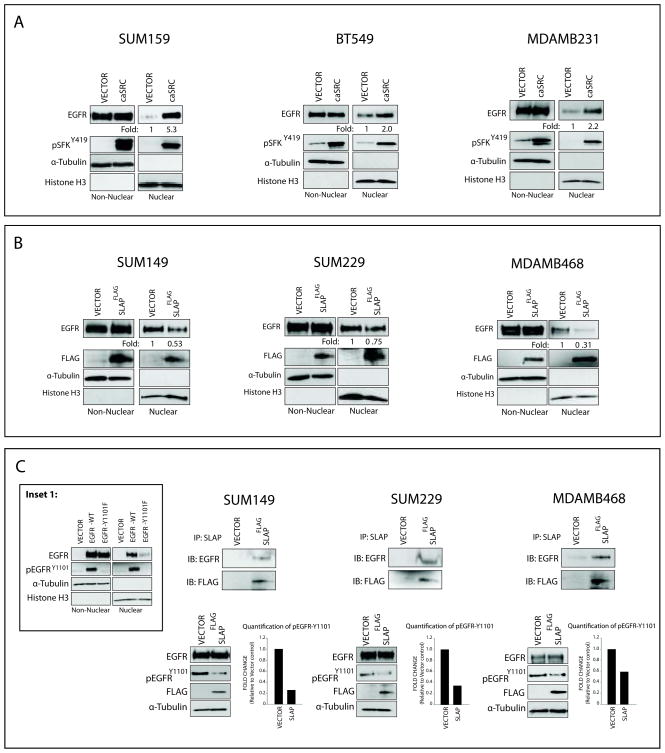
Previous studies from our laboratory indicate that SFKs influence nEGFR translocation in lung cancer (26, 30). To investigate if SFKs influence EGFR translocation from the plasma membrane to the nucleus in TNBC, a constitutively active Src (caSrc) was overexpressed in SUM159, BT549, and MDAMB231 cells. The overexpression of caSrc, indicated by enhanced pSFK-Y419, led to increases in nEGFR expression (Figure 3A). Next, a negative regulator of Src, Src-like adaptor protein (SLAP) (39), was overexpressed in SUM149, SUM229, and MDAMB468 cells. The overexpression of SLAP, indicated by the expression of the Flag tag, led to decreases in nEGFR levels (Figure 3B). These studies indicate that modulation of SFK activity can influence nEGFR expression in TNBC cell lines.
Figure 3. Src Family Kinases mediate nEGFR translocation in TNBC.
(A) Constitutively active Src (caSrc) enhances nEGFR translocation in TNBC cell lines. Cells were transfected with caSrc or an empty vector control for 48 hr prior to stimulation with EGF (5 nM, 45 min) to induce nEGFR translocation. Non-nuclear and nuclear proteins were harvested. nEGFR expression was quantitated using ImageJ software (B) A negative regulator of Src, src-like adaptor protein (SLAP), blocks nEGFR translocation in TNBC cell lines. Cells were transfected with SLAP-FLAG or an empty vector control for 48 hr prior to harvesting non-nuclear and nuclear proteins. nEGFR expression was analyzed. (C) SLAP can interact with EGFR and decrease EGFR activation at Tyrosine 1101. Cells were transfected with SLAP-FLAG or an empty vector control for 48 hr prior to harvesting whole cell lysate. 250 ug of cell lysate was immunoprecipitated with an anti-SLAP antibody. The same lysate was subjected to immunoblot analysis for activation of EGFR at Tyrosine 1101. pEGFR-Y1101 activity was quantitated using ImageJ software. Inset 1: EGFR mutated at Tyrosine 1101 is deficient in nuclear localization. Vector, EGFR-WT, and EGFR-Y1101F were transfected into CHOK1 cells for 48 hr prior to stimulation with EGF (5 nM, 45 min). Non-nuclear and nuclear proteins were harvested, and nEGFR expression was analyzed.
Previous studies elucidating the functions of SLAP have identified that SLAP functions as an antagonist for Src induced mitogenesis partly through the binding of Src substrates and effector molecules (39). Overexpression of SLAP resulted in the association with EGFR in three TNBC cell lines by co-immunoprecipitation (IP) analysis (Figure 3C). IP with an IgG control yielded no signal (data not shown). Since EGFR deficient in tyrosine 1101 (Y1101) phosphorylation is hindered in nuclear translocation (Figure 3C, Inset 1) (30), we probed for phosphorylated EGFR at Y1101 post SLAP transfection. Indeed, TNBC cell lines overexpressing SLAP had decreased phosphorylation of EGFR at Y1101 (Figure 3C), which correlated with decreased nEGFR (Figure 3B). These data demonstrate that SFK phosphorylation of EGFR at Y1101 can influence nEGFR translocation in TNBC.
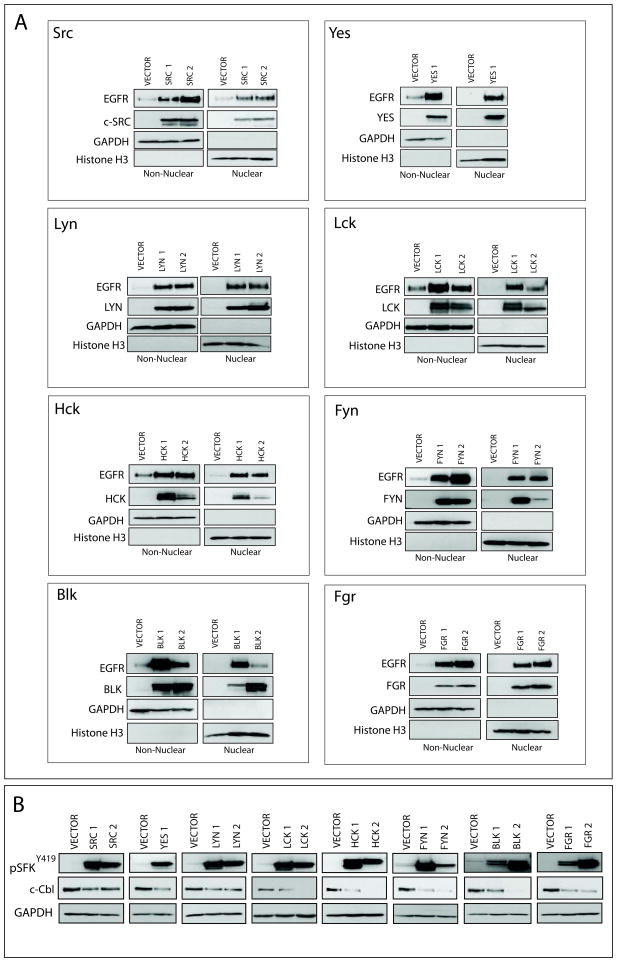
SFKs exhibit functional redundancy in their ability to influence nEGFR translocation
Previous reports suggest that the SFKs Yes and Lyn play a role in the nuclear translocation of EGFR (30). However, experiments in Figure 3 indicated that caSrc and SLAP could influence nEGFR translocation in TNBC cells suggesting that global increased activity of SFKs may influence nEGFR expression. To test this hypothesis, stable clones of individual SFKs (Src, Yes, Lyn, Lck, Hck, Fyn, Blk, and Fgr) were engineered in the breast cancer cell line MCF-7. One or two stable clones were chosen for each SFK for comparison to an empty vector stable cell line (Figure 4A). The overexpression of each SFK led to the enhanced expression and nuclear translocation of EGFR. All cell lines were stimulated with 5 nM EGF to promote the nuclear translocation of EGFR, however a basal level of nEGFR was detected in non-stimulated SFK stable cells (data not shown). Additionally, the stable overexpression of each SFK led to their increased activation, corresponding to a downregulation of the E3 ubiquitin ligase, c-Cbl (Figure 4B). This result may explain why an increase in total EGFR was observed in Figure 4A. Collectively, these data suggest that SFKs play functional redundant roles in promoting nEGFR translocation.
Figure 4. SFKs exhibit functional redundancy in their ability to influence nEGFR translocation.
(A) The stable overexpression of SFKs increase nEGFR expression. Eight different SFK’s were stably overexpressed in the breast cancer cell line MCF-7. SFK stable clones or an empty vector stable cell line were stimulated with EGF (5 nM, 45 min) to induce nEGFR translocation, prior to harvesting non-nuclear and nuclear proteins. GAPDH and Histone H3 were used as loading and purity controls, respectively. (B) The stable overexpression of SFKs downregulate c-Cbl. Whole cell lysate was harvested from SFK stable clones or an empty vector stable cell line. GAPDH was used as loading control.
Therapeutic inhibition of SFKs can block nEGFR translocation in vitro and in vivo TNBC tumor models
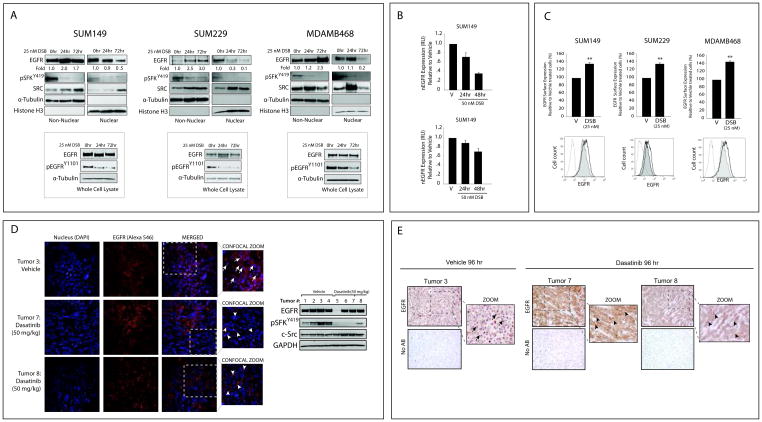
Since the modulation of SFK activity influenced nEGFR, the SFK inhibitor dasatinib was utilized to determine if it could abrogate EGFR translocation from the membrane to nucleus. Treatment of TNBC cells with dasatinib led to potent decreases in nEGFR levels (at 24 and 72 hours in SUM149 and SUM229, and at 72 hours in MDAMB468 cells) (Figure 5A). Analysis of whole cell lysate indicated that EGFR activity on Y1101 was inhibited by dasatinib at both time points. Additionally, dasatinib treatment led to subsequent increases in non-nEGFR levels (Figure 5A). Nuance imaging and Inform software was further used to analyze nEGFR levels post dasatinib treatment (Figure 5B). Cells were stained for EGFR, E-Cadherin and DAPI; E-Cadherin and DAPI were used to create a spectral library that segmented each cell into cytoplasm and nucleus as previously described (34). InForm software analysis of each cell line (n=2) demonstrated that dasatinib treated cells trended towards less nEGFR staining as compared to vehicle treated cells (p=0.08 at 48 hr).
Figure 5. Therapeutic inhibition of SFKs can block nEGFR translocation in TNBC cell lines and tumor models.
(A) Dasatinib can inhibit nEGFR translocation and enhance non-nuclear EGFR levels. Cells were treated with vehicle or dasatinib (25 nM) for 24 and 72 hr prior to harvesting whole cell, non-nuclear and nuclear proteins. (B) Dasatinib can block nEGFR translocation measured by Nuance imaging analysis. Cells were treated with vehicle or dasatinib (50 nM) for 24 and 48 hr prior to staining for EGFR, E-Cadherin, and DAPI. Nuclear EGFR fluorescence detected from dasatinib treated cells was normalized to nEGFR fluorescence detected from vehicle treated cells using InForm software (n=2). (C) Dasatinib can enhance plasma membrane-bound EGFR levels measured by flow cytometry. Cells were treated with dasatinib (25 nM) for 24 hr prior to EGFR surface level analysis. Surface level EGFR expression of dasatinib treated cells was normalized to vehicle treated cells (n=3). Shaded histogram= vehicle treated cells, Non-shaded histograms= dasatinib treated cells. IgG treated cells are used as a control (dotted line). (D, E) Dasatinib can block nEGFR translocation in MDAMB468 xenograft tumors. Mice with established MDAMB468 tumors were treated with 50 mg/kg of dasatinib or vehicle once a day for 4 days. Tumors were analyzed by confocal IF (D) and IHC (E) for EGFR expression. IF: merged images were magnified to depict nEGFR (arrows) and non-nEGFR (triangle). Magnification 600X for IF and 400X for IHC. Four tumors from vehicle (tumor # 1–4) or dasatinib treated mice (tumor # 5–8) were harvested for protein and analyzed for the indicated proteins. Data points are represented as mean±s.e.m. **p<0.01.
To further characterize the effect of dasatinib on non-nEGFR expression, surface level EGFR was analyzed by flow cytometry. TNBC cells treated with dasatinib for 24 hours contained 30–42% more plasma membrane-bound EGFR as compared to vehicle treated cells (Figure 5C). There was no additional increase in EGFR surface expression 72 hours post treatment (data not shown). Together, these data suggest that EGFR accumulates on the plasma membrane when nEGFR translocation is blocked by dasatinib.
To investigate if therapeutic inhibition of SFKs can abrogate nEGFR translocation in vivo, MDAMB468 cells were established as xenograft tumors in female athymic nude mice. Mice were randomized into two groups receiving 50 mg/kg of dasatinib or vehicle once daily for 4 days. Figure 5D represents confocal IF analyses of representative tumor sections harvested from either vehicle or dasatinib treated mice stained for EGFR. EGFR was highly nuclear localized in tumors from vehicle treated mice. However, tumors harvested from dasatinib treated mice harbored much less nEGFR, with a noticeable increase in surface EGFR expression. Immunoblot analysis of harvested tumors validated that dasatinib inhibited SFK activity; one dasatinib treated tumor (#5) contained less total EGFR expression. The inhibition of nEGFR translocation was also visualized by IHC staining of tumors harvested from dasatinib treated mice (Figure 5E). Interestingly, we found that dasatinib treatment of mice harboring colorectal tumors also contained less nEGFR expression within the tumor (Supplemental Figure 1), suggesting that SFKs may influence nEGFR translocation in different tumor types. Collectively, these data indicate that SFK inhibition prevents nEGFR translocation and enhances membrane accumulation of EGFR in vivo.
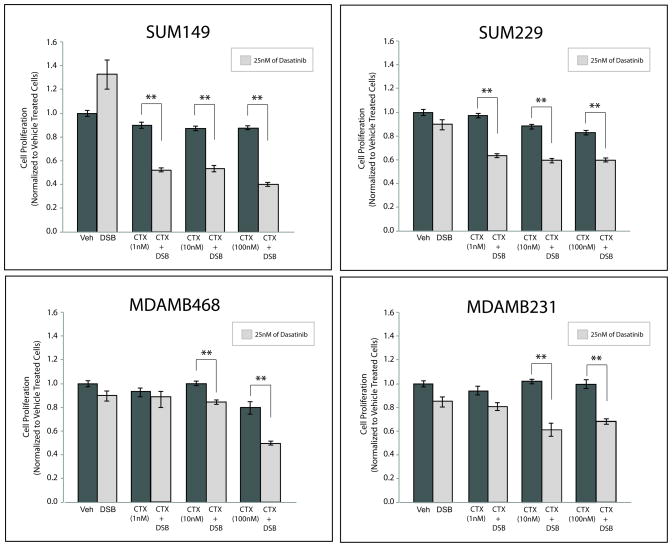
SFK inhibition can sensitize TNBC cells to cetuximab growth inhibition
Since SFK inhibition enhanced plasma membrane-bound EGFR expression, we hypothesized that TNBC cells may become more sensitive to cetuximab upon pre-treatment with dasatinib. To investigate this, we performed proliferation assays after pre-treating TNBC cells with dasatinib or vehicle for 24 hours, the time point at which an increase in surface level EGFR was detected, and subsequently treating cells with increasing doses of cetuximab for an additional 72 hours (Figure 6). All cell lines pre-treated with vehicle and subsequently treated with increasing doses of cetuximab demonstrated minor reductions in proliferation, consistent with data in Figure 2. Additionally, cells treated with 25 nM dasatinib monotherapy did not exhibit significant inhibition of proliferation. However, TNBC cell lines that received dasatinib for 24 hours prior to cetuximab treatment demonstrated significant reductions in proliferation over a wide range of cetuximab doses (1 nM to 100 nM). SUM149 and SUM229 cells demonstrated significant reductions in proliferation at low doses of cetuximab (1 nM), while MDAMB468 and MDAMB231 cells exhibited proliferation inhibition at higher doses of cetuximab (10 nM and 100 nM). Collectively, these data suggest that the blockade of nEGFR translocation via SFK inhibition can increase TNBC cell sensitivity to cetuximab.
Figure 6. Therapeutic inhibition of SFK activity can sensitize TNBC cells to cetuximab.
Cells were pretreated with vehicle or dasatinib (25 nM) for 24 hr prior to adding cetuximab to the growth medium at the indicated doses (1 nM, 10 nM, and 100 nM) for an additional 72 hr. Proliferation assays were performed and plotted as a percentage of growth relative to vehicle treated cells (n=3). Data points are represented as mean±s.e.m. **p<0.01.
DISCUSSION
TNBC is a subset of breast cancers that commonly overexpress the EGFR (3–6). Unfortunately, clinical trials targeting EGFR with cetuximab have yielded minimal benefit in TNBC (7, 8), even with the addition of platinum based chemotherapies (1, 5, 36). Thus, understanding why TNBCs are intrinsically resistant to cetuximab has become an important clinical question. Over the last decade, numerous studies have identified a role for nEGFR in resistance to anti-EGFR agents (25, 26). Previous studies from our laboratory demonstrated that NSCLC cell lines that had acquired resistance to cetuximab relied on nEGFR signaling to maintain their resistant phenotype (26). Based on these studies, we hypothesized that nEGFR may be a critical molecular determinant for cetuximab resistance in TNBC.
In the current study, nEGFR was detected in a panel of established TNBC cell lines and human tumors (Figure 1). In prior studies, 38% of a 130-breast cancer patient cohort (15) and 40% of a 113–breast cancer patient cohort (19) stained positive for nEGFR, which was further correlated with worse overall survival. The heterogeneity observed in nEGFR expression in the current study of TNBC tumors highlights the importance of simultaneously targeting both nEGFR and non-nEGFR cell populations. Another interesting observation lies in the localization of EGFR in the nucleolus, functions that have yet to be investigated and may be playing important roles in TNBC pathogenesis. Collectively, the preclinical data presented in the current study suggest that nEGFR may be indicative of cetuximab resistant tumors warranting further investigation for its role as a predictive marker for cetuximab response in TNBC.
Recent work from our laboratory has found that SFK dependent phosphorylation of EGFR on Y1101 is a necessary and early event for EGFR translocation from the plasma membrane to the nucleus (30). The current study aimed to identify if this mechanism of nuclear translocation was present in TNBC. We found that three TNBC cell lines (MDAMB468, SUM149 and SUM229) with the highest levels of phosphorylated Y1101 also expressed the highest levels of nEGFR (Figure 1A and 1B). Additionally, inhibition of SFK activity led to decreased phosphorylation of EGFR on Y1101 and reduced nEGFR levels (Figure 3C and 5A, 5B). Interestingly, Figure 5C indicates that surface level EGFR was enhanced within 24 hours of dasatinib treatment, even though a decrease in nEGFR expression was more prominent at later time points post treatment (Figure 5A and 5B); this suggests that the rate of nEGFR export, via its nuclear export sequence (28), varies between cell lines. Collectively, these data suggest that SFK phosphorylation of EGFR on Y1101 may be a critical step for EGFR nuclear translocation in TNBC.
SFKs consist of 11 intracellular tyrosine kinases that are differentially expressed in a variety of cancers (40). In the current study, eight individual SFKs were stably overexpressed, and found to function similarly in their ability to influence 1) the steady state expression of total EGFR, 2) nEGFR translocation, and 3) degradation of c-Cbl (Figure 4). These data suggest that SFKs exhibit functional redundancy in their ability to influence nEGFR translocation, and thus the use of broad-spectrum SFK inhibitors, such as dasatinib, may be highly beneficial in nEGFR positive cancers.
In the current study, SFK inhibition of nEGFR translocation led to an accumulation of plasma membrane-bound EGFR and sensitization to cetuximab therapy (Figure 5 and 6). Recent studies support our findings, where anti-tumorigenic effects of both cetuximab and dasatinib dual treatment with chemotherapy (41) and the use of non-competitive monoclonal antibodies degrading the EGFR (42) have been documented in TNBC. Additionally, a recent report demonstrated that targeting PCNA, a nEGFR substrate, could delay TNBC tumor growth (43). In the current study, sensitization to cetuximab was observed after pre-treatment of TNBC cells with dasatinib for 24 hours, the time point at which EGFR accumulation was detected on the plasma membrane due to the inhibition of nEGFR translocation. We speculate that the inhibition of nEGFR translocation drives TNBC cells to rely solely on classical membrane-bound EGFR signaling for sustained proliferation and survival signals; thus, TNBC cells become sensitized to cetuximab because cetuximab can abrogate classical EGFR signaling pathways. Previous studies in EGFR expressing NSCLC and HNSCC cell lines support this, where cell lines that lacked nEGFR expression were found to be more sensitive to cetuximab monotherapy (26, 30). Currently, the growth inhibitory effect of cetuximab and dasatinib therapy is being accessed in vivo TNBC models in our laboratory, a critical step for the movement of this proposed drug combination into clinical trials. Collectively, the data presented herein indicates that the dual targeting of both nEGFR and plasma membrane-bound EGFR is necessary for the complete inhibition of EGFR’s oncogenic functions, a therapeutic strategy that can be readily translated for the treatment of nEGFR expressing TNBC patients.
Supplementary Material
Acknowledgments
Financial Support: grant UL1TR000427 from the Clinical and Translational Science Award program, through the NIH National Center for Advancing Translational Sciences (D.L. Wheeler), grant RSG-10-193-01-TBG from the American Cancer Society (D.L. Wheeler), grant W81XWH-12-1-0467 from United States Army Medical Research and Materiel Command (D.L. Wheeler), Mary Kay Foundation grant MSN152261 (D.L. Wheeler) and NIH grant Q3 T32 GM08.1061-01A2 from Graduate Training in Cellular and Molecular Pathogenesis of Human Diseases (T.M. Brand).
We would like to thank Dr. Roche, Dr. Mischel, and Dr. Boerner for kindly sharing their expression vectors. We would like to acknowledge Lance Rodenkirch and the Keck Laboratory for Biological Imaging for their expertise in confocal microscopy, and Ben August and Amanda Thoma for their training and expertise in electron microscopy.
Footnotes
Conflicts of Interest: None
References
- 1.Schneider BP, Winer EP, Foulkes WD, Garber J, Perou CM, Richardson A, et al. Triple-negative breast cancer: risk factors to potential targets. Clin Cancer Res. 2008;14:8010–8. doi: 10.1158/1078-0432.CCR-08-1208. [DOI] [PubMed] [Google Scholar]
- 2.Stevens KN, Vachon CM, Couch FJ. Genetic susceptibility to triple-negative breast cancer. Cancer Res. 2013;73:2025–30. doi: 10.1158/0008-5472.CAN-12-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corkery B, Crown J, Clynes M, O’Donovan N. Epidermal growth factor receptor as a potential therapeutic target in triple-negative breast cancer. Ann Oncol. 2009;20:862–7. doi: 10.1093/annonc/mdn710. [DOI] [PubMed] [Google Scholar]
- 4.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–74. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 6.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–67. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masuda H, Zhang D, Bartholomeusz C, Doihara H, Hortobagyi GN, Ueno NT. Role of epidermal growth factor receptor in breast cancer. Breast Cancer Res Treat. 2012;136:331–45. doi: 10.1007/s10549-012-2289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gelmon K, Dent R, Mackey JR, Laing K, McLeod D, Verma S. Targeting triple-negative breast cancer: optimising therapeutic outcomes. Ann Oncol. 2012;23:2223–34. doi: 10.1093/annonc/mds067. [DOI] [PubMed] [Google Scholar]
- 9.Yarden Y, Pines G. The ERBB network: at last, cancer therapy meets systems biology. Nat Rev Cancer. 2012;12:553–63. doi: 10.1038/nrc3309. [DOI] [PubMed] [Google Scholar]
- 10.Brand TM, Iida M, Li C, Wheeler DL. The nuclear epidermal growth factor receptor signaling network and its role in cancer. Discov Med. 2011;12:419–32. [PMC free article] [PubMed] [Google Scholar]
- 11.Han W, Lo HW. Landscape of EGFR signaling network in human cancers: biology and therapeutic response in relation to receptor subcellular locations. Cancer Lett. 2012;318:124–34. doi: 10.1016/j.canlet.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dittmann K, Mayer C, Fehrenbacher B, Schaller M, Raju U, Milas L, et al. Radiation-induced epidermal growth factor receptor nuclear import is linked to activation of DNA-dependent protein kinase. J Biol Chem. 2005;280:31182–9. doi: 10.1074/jbc.M506591200. [DOI] [PubMed] [Google Scholar]
- 13.Hsu SC, Miller SA, Wang Y, Hung MC. Nuclear EGFR is required for cisplatin resistance and DNA repair. Am J Transl Res. 2009;1:249–58. [PMC free article] [PubMed] [Google Scholar]
- 14.Wang SC, Nakajima Y, Yu YL, Xia W, Chen CT, Yang CC, et al. Tyrosine phosphorylation controls PCNA function through protein stability. Nat Cell Biol. 2006;8:1359–68. doi: 10.1038/ncb1501. [DOI] [PubMed] [Google Scholar]
- 15.Lo HW, Xia W, Wei Y, Ali-Seyed M, Huang SF, Hung MC. Novel prognostic value of nuclear epidermal growth factor receptor in breast cancer. Cancer Res. 2005;65:338–48. [PubMed] [Google Scholar]
- 16.Psyrri A, Yu Z, Weinberger PM, Sasaki C, Haffty B, Camp R, et al. Quantitative determination of nuclear and cytoplasmic epidermal growth factor receptor expression in oropharyngeal squamous cell cancer by using automated quantitative analysis. Clin Cancer Res. 2005;11:5856–62. doi: 10.1158/1078-0432.CCR-05-0420. [DOI] [PubMed] [Google Scholar]
- 17.Li CF, Fang FM, Wang JM, Tzeng CC, Tai HC, Wei YC, et al. EGFR nuclear import in gallbladder carcinoma: nuclear phosphorylated EGFR upregulates iNOS expression and confers independent prognostic impact. Ann Surg Oncol. 2012;19:443–54. doi: 10.1245/s10434-011-1942-6. [DOI] [PubMed] [Google Scholar]
- 18.Xia WY, Wei YK, Du Y, Liu JS, Chang B, Yu YL, et al. Nuclear Expression of Epidermal Growth Factor Receptor is a Novel Prognostic Value in Patients With Ovarian Cancer. Mol Carcinogen. 2009;48:610–7. doi: 10.1002/mc.20504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hadzisejdic I, Mustac E, Jonjic N, Petkovic M, Grahovac B. Nuclear EGFR in ductal invasive breast cancer: correlation with cyclin-D1 and prognosis. Mod Pathol. 2010;23:392–403. doi: 10.1038/modpathol.2009.166. [DOI] [PubMed] [Google Scholar]
- 20.Traynor AM, Weigel TL, Oettel KR, Yang DT, Zhang C, Kim K, et al. Nuclear EGFR protein expression predicts poor survival in early stage non-small cell lung cancer. Lung Cancer. 2013;81:138–41. doi: 10.1016/j.lungcan.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dittmann K, Mayer C, Fehrenbacher B, Schaller M, Kehlbach R, Rodemann HP. Nuclear EGFR shuttling induced by ionizing radiation is regulated by phosphorylation at residue Thr654. FEBS Lett. 2010;584:3878–84. doi: 10.1016/j.febslet.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Dittmann K, Mayer C, Fehrenbacher B, Schaller M, Kehlbach R, Rodemann HP. Nuclear epidermal growth factor receptor modulates cellular radio-sensitivity by regulation of chromatin access. Radiother Oncol. 2011;99:317–22. doi: 10.1016/j.radonc.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 23.Dittmann K, Mayer C, Rodemann HP. Inhibition of radiation-induced EGFR nuclear import by C225 (Cetuximab) suppresses DNA-PK activity. Radiother Oncol. 2005;76:157–61. doi: 10.1016/j.radonc.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 24.Liccardi G, Hartley JA, Hochhauser D. EGFR nuclear translocation modulates DNA repair following cisplatin and ionizing radiation treatment. Cancer Res. 2011;71:1103–14. doi: 10.1158/0008-5472.CAN-10-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang WC, Chen YJ, Li LY, Wei YL, Hsu SC, Tsai SL, et al. Nuclear translocation of epidermal growth factor receptor by Akt-dependent phosphorylation enhances breast cancer-resistant protein expression in gefitinib-resistant cells. J Biol Chem. 2011;286:20558–68. doi: 10.1074/jbc.M111.240796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li C, Iida M, Dunn EF, Ghia AJ, Wheeler DL. Nuclear EGFR contributes to acquired resistance to cetuximab. Oncogene. 2009;28:3801–13. doi: 10.1038/onc.2009.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lo HW, Cao X, Zhu H, Ali-Osman F. Cyclooxygenase-2 is a novel transcriptional target of the nuclear EGFR-STAT3 and EGFRvIII-STAT3 signaling axes. Mol Cancer Res. 2010;8:232–45. doi: 10.1158/1541-7786.MCR-09-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gururaj AE, Gibson L, Panchabhai S, Bai M, Manyam G, Lu Y, et al. Access to the nucleus and functional association with c-Myc is required for the full oncogenic potential of DeltaEGFR/EGFRvIII. J Biol Chem. 2013;288:3428–38. doi: 10.1074/jbc.M112.399352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wheeler DL, Iida M, Kruser TJ, Nechrebecki MM, Dunn EF, Armstrong EA, et al. Epidermal growth factor receptor cooperates with Src family kinases in acquired resistance to cetuximab. Cancer Biol Ther. 2009;8:696–703. doi: 10.4161/cbt.8.8.7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iida M, Brand TM, Campbell DA, Li C, Wheeler DL. Yes and Lyn play a role in nuclear translocation of the epidermal growth factor receptor. Oncogene. 2012;32:759–67. doi: 10.1038/onc.2012.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brand TM, Iida M, Luthar N, Wleklinski MJ, Starr MM, Wheeler DL. Mapping C-terminal transactivation domains of the nuclear HER family receptor tyrosine kinase HER3. PLoS One. 2013;8:e71518. doi: 10.1371/journal.pone.0071518. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Li C, Brand TM, Iida M, Huang S, Armstrong EA, van der Kogel A, et al. Human epidermal growth factor receptor 3 (HER3) blockade with U3-1287/AMG888 enhances the efficacy of radiation therapy in lung and head and neck carcinoma. Discov Med. 2013;16:79–92. [PMC free article] [PubMed] [Google Scholar]
- 33.Yi H, Leunissen JLM, Shi GM, Gutekunst CA, Hersch SM. A novel procedure for pre-embedding double immunogold-silver labeling at the ultrastructural level. J Histochem Cytochem. 2001;49:279–83. doi: 10.1177/002215540104900301. [DOI] [PubMed] [Google Scholar]
- 34.Huang W, Hennrick K, Drew S. A colorful future of quantitative pathology: validation of Vectra technology using chromogenic multiplexed immunohistochemistry and prostate tissue microarrays. Hum Pathol. 2013;44:29–38. doi: 10.1016/j.humpath.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 35.Iida M, Brand TM, Starr MM, Li C, Huppert EJ, Luthar N, et al. Sym004, a Novel EGFR Antibody Mixture, Can Overcome Acquired Resistance to Cetuximab. Neoplasia. 2013;15:1196–206. doi: 10.1593/neo.131584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dunn KW, Kamocka MM, McDonald JH. A practical guide to evaluating colocalization in biological microscopy. American journal of physiology Cell physiology. 2011;300:C723–42. doi: 10.1152/ajpcell.00462.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tryfonopoulos D, Walsh S, Collins DM, Flanagan L, Quinn C, Corkery B, et al. Src: a potential target for the treatment of triple-negative breast cancer. Ann Oncol. 2011;22:2234–40. doi: 10.1093/annonc/mdq757. [DOI] [PubMed] [Google Scholar]
- 38.Elsberger B, Tan BA, Mitchell TJ, Brown SB, Mallon EA, Tovey SM, et al. Is expression or activation of Src kinase associated with cancer-specific survival in ER-, PR- and HER2-negative breast cancer patients? Am J Pathol. 2009;175:1389–97. doi: 10.2353/ajpath.2009.090273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manes G, Bello P, Roche S. Slap negatively regulates Src mitogenic function but does not revert Src-induced cell morphology changes. Mol Cell Biol. 2000;20:3396–406. doi: 10.1128/mcb.20.10.3396-3406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sen B, Johnson FM. Regulation of SRC family kinases in human cancers. Journal of signal transduction. 2011;2011:865819. doi: 10.1155/2011/865819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim EM, Mueller K, Gartner E, Boerner J. Dasatinib is synergistic with cetuximab and cisplatin in triple-negative breast cancer cells. The Journal of surgical research. 2013;185:231–9. doi: 10.1016/j.jss.2013.06.041. [DOI] [PubMed] [Google Scholar]
- 42.Ferraro DA, Gaborit N, Maron R, Cohen-Dvashi H, Porat Z, Pareja F, et al. Inhibition of triple-negative breast cancer models by combinations of antibodies to EGFR. Proc Natl Acad Sci U S A. 2013;110:1815–20. doi: 10.1073/pnas.1220763110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu YL, Chou RH, Liang JH, Chang WJ, Su KJ, Tseng YJ, et al. Targeting the EGFR/PCNA signaling suppresses tumor growth of triple-negative breast cancer cells with cell-penetrating PCNA peptides. PLoS One. 2013;8:e61362. doi: 10.1371/journal.pone.0061362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.