Abstract
The bacterial signaling molecule cyclic-di-GMP stimulates the synthesis of bacterial cellulose, frequently found in biofilms. Bacterial cellulose is synthesized and translocated across the inner membrane by a complex of the cellulose synthase BcsA and BcsB subunits. Here we present crystal structures of the cyclic-di-GMP-activated BcsA–B complex. The structures reveal that cyclic-di-GMP releases an auto-inhibited state of the enzyme by breaking a salt bridge which otherwise tethers a conserved gating loop that controls access to and substrate coordination at the active site. Disrupting the salt bridge by mutagenesis generates a constitutively active cellulose synthase. Additionally, the cyclic-di-GMP activated BcsA–B complex contains a nascent cellulose polymer whose terminal glucose unit rests at a novel location above BcsA’s active site where it is positioned for catalysis. Our mechanistic insights are the first examples of how cyclic-di-GMP allosterically modulates enzymatic functions.
Introduction
Biofilms are sessile multi-cellular bacterial communities that are encased in a 3-dimensional meshwork of biopolymers, such as polysaccharides, proteinaceous filaments and nucleic acids1–3. The biofilm matrix provides protection against mechanical stress4,5 and controls the diffusion of signaling molecules, nutrients and toxic compounds. In fact, biofilm communities exhibit increased tolerance towards conventional anti-microbial treatments and sterilization techniques and are responsible for many chronic infections associated with cystic fibrosis and endocarditis6,7 as well as nosocomial infections8. In many cases, biofilm formation occurs in response to an elevated cytosolic concentration of cyclic-di-GMP (c-di-GMP)9, a bacterial signaling molecule recognized by a wide range of effector proteins, including transcription factors, flagellar components, riboswitches and exopolysaccharide synthases1. Therefore, targeting c-di-GMP–binding effectors has emerged as an attractive new route for the development of urgently needed novel anti-microbial therapeutics.
C-di-GMP activates the synthesis of bacterial cellulose1,10, an extracellular polysaccharide often found in biofilms11. C-di-GMP monomers and dimers12,13 are both recognized by effector proteins via PilZ domains, first identified as regulatory components of cell motility14, which comprise an “RxxxR” motif in a flexible linker region followed by a β-sheet or β-barrel that contains a “DxSxxG” motif15. Both sequence motifs have been shown to interact with c-di-GMP in structures of isolated PilZ domains16,17. However, the mechanism by which c-di-GMP binding at PilZ domains modulates enzymatic functions is completely unknown to date.
Extracellular polysaccharides of the biofilm matrix, such as cellulose, alginate and poly-N-acetylglucosamine (PNAG), are likely synthesized and secreted by a conserved mechanism18–22. Bacterial cellulose synthase polymerizes glucose molecules via β-1,4 glycosidic linkages in a multi-step process which requires the presence of a divalent cation, mostly magnesium23. First, upon stimulation by c-di-GMP, the enzyme binds its substrate UDP-Glc (donor) at an intracellular glycosyltransferase (GT) domain. Second, the donor glucose is transferred to the 4′ hydroxyl group at the non-reducing end of the growing polysaccharide chain (acceptor), thereby extending the polymer and forming UDP as a second reaction product23,24. Third, following glycosyl transfer, the elongated polymer has to be translocated by one glucose unit into a transmembrane (TM) channel so that the newly added glucose unit occupies the acceptor site and UDP must be replaced with UDP-Glc for another round of catalysis.
The membrane-integrated bacterial cellulose synthase contains the inner membrane components BcsA and BcsB as well as the outer membrane protein BcsC25,26. BcsA, together with the periplasmic membrane-anchored BcsB subunit, forms a complex that is sufficient for cellulose synthesis and translocation23,27. BcsA is homologous to eukaryotic cellulose synthases28 and contains eight TM helices and a cytosolic GT domain between TM helices four and five27. The enzyme is a processive family-2 GT29 that elongates the non-reducing end of the growing polysaccharide chain. This reaction requires a general base, which is likely provided by the Asp residue of a “TED” motif found at the beginning of a short helix within the GT domain and in close proximity to the acceptor’s 4′ hydroxyl27. BcsA also forms a polysaccharide channel across the membrane, directly above the active site, thereby allowing the coupling of cellulose synthesis and translocation27,30.
Bacterial cellulose and alginate synthases are activated by c-di-GMP via PilZ domains15,31. BcsA forms a PilZ domain within its C-terminal intracellular extension, which consists of a six-stranded β-barrel and a preceding linker region15,27. The β-barrel rests against the intracellular GT domain and is connected to BcsA’s C-terminal TM helix (TM8) via a linker (TM8-β-barrel linker) harboring the “RxxxR” motif involved in c-di-GMP binding15.
The TM8-β-barrel linker also interacts with BcsA’s “gating loop”, which runs across the opening of the GT domain towards the cytosol, thereby blocking access to the catalytic pocket in the non-stimulated or “resting” state of the enzyme27. It was speculated that substrate binding to the active site requires the repositioning of the gating loop, perhaps induced by c-di-GMP27. This model is supported by biochemical studies indicating that increasing c-di-GMP concentrations do not alter KM, but instead increase the fraction of catalytically active enzymes23.
In order to unravel the mechanism by which c-di-GMP activates bacterial cellulose synthase, we determined c-di-GMP-bound structures of the Rhodobacter sphaeroides BcsA–B complex at intermediate states during cellulose synthesis and translocation. The c-di-GMP-bound structures reveal the architecture of the activated BcsA–B complex and provide unique insights into the mechanism of c-di-GMP signaling. These include the identification of a conserved regulatory salt bridge that auto-inhibits BcsA in the absence of c-di-GMP and the UDP-dependent repositioning of a gating loop to either open the catalytic pocket or to coordinate the nucleotide at the active site. Furthermore, the structures reveal the movement of a “finger helix” of BcsA, which interacts with the acceptor end of the translocating cellulose polymer, towards the TM channel entrance, correlating with the translocation of the cellulose polymer into the channel by one glucose unit. Thus, our data provide the first insights into the mechanism by which c-di-GMP modulates enzymatic functions and represent novel snapshots of cellulose synthesis and membrane translocation.
Results
Architecture of BcsA–B in complex with c-di-GMP
We purified Rhodobacter sphaeroides BcsA–B from E. coli, crystallized it in complex with c-di-GMP by the bicelle crystallization method,32,33 and solved the structure by molecular replacement at a resolution of 2.65 Å. Additionally, we obtained a c-di-GMP- and UDP-bound structure of BcsA–B by soaking crystals with UDP and refining at 3.2 Å resolution (Table 1). Both structures contain a translocating cellulose polymer 17 glucose units in length that co-purifies with the BcsA–B complex.
Table 1.
Crystallographic data collection and refinement statistics.
| UDP-free | UDP-bound | |
|---|---|---|
| Data collection | ||
| Space group | P212121 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 67.6, 214.7, 220.4 | 67.5, 216.8, 219.6 |
| α β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 34.75–2.65 (2.70–2.65) * | 49.62–3.20 (3.30–3.20) |
| Rpim | 0.056 (0.577) | 0.079 (0.499) |
| CC1/2^ | 0.995 (0.553) | 0.992 (0.566) |
| Mean I/σI | 7.8 (1.3) | 8.7 (2.1) |
| Completeness (%) | 92.3 (94.3) | 98.7 (99.5) |
| Redundancy | 4.5 (4.1) | 4.7 (4.7) |
| Refinement | ||
| Resolution (Å) | 20–2.65 | 49–3.2 |
| No. reflections | ||
| Total | 85,948 | 53,241 |
| Rfree | 4,307 | 2,714 |
| Rwork/Rfree | 19.9/23.0 | 20.6/23.8 |
| No. atoms | ||
| Protein | 10,673 | 10,709 |
| β-1,4 glucan | 187 | 187 |
| c-di-GMP | 92 | 92 |
| UDP | – | 25 |
| Mg2+ | 1 | 1 |
| Lipids | 89 | 58 |
| B-factors | ||
| Chain A | 74.5 | 74.6 |
| Chain B | 66.3 | 68.3 |
| Chain D | 77.2 | 91.3 |
| β-1,4 glucan | 78.1 | 81.2 |
| c-di-GMP-A | 59.2 | 61.5 |
| c-di-GMP-B | 64.1 | 58.8 |
| UDP | – | 80.8 |
| Lipids | 85.6 | 113.6 |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.003 | 0.002 |
| Bond angles (°) | 0.754 | 0.760 |
Values in parentheses refer to the highest-resolution shell.
Correlation between intensities from random half-data sets50.
Overall, the c-di-GMP-bound BcsA–B structure is consistent with the previously reported structure obtained from detergent-solubilized complexes (RMSD≈1Å for all atoms)27 (Fig. 1). Two register shifts by one residue were identified in regions that were poorly ordered in the previously reported structure of BcsA (residues 171–190) and BcsB (residues 268–280). The corrected register in BcsA positions Asp179 of the “DDG” motif in hydrogen bonding distance with the conserved Tyr216 and Asp180 in hydrogen bonding distance with the uracil moiety of UDP and Arg219 (Supplementary Fig. 1).
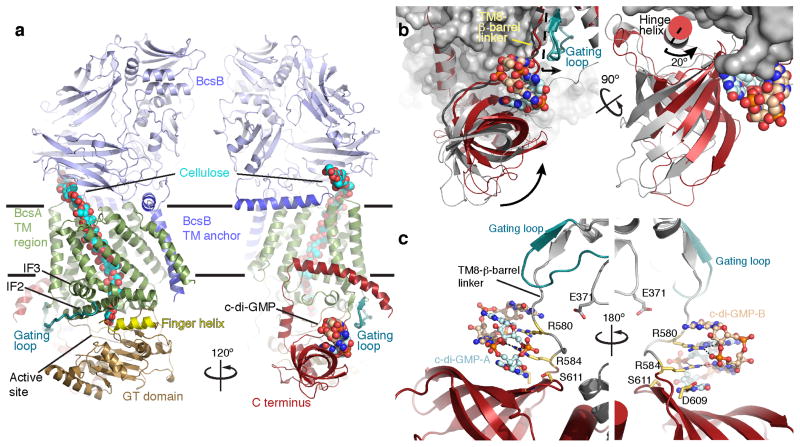
Figure 1.
Structure of the c-di-GMP-activated BcsA–B complex. (a) Cartoon representation of the BcsA–B structure in the presence of a c-di-GMP homo-dimer. BcsA is colored brown, green and red for its GT domain, TM region, and C terminus, respectively. The 6-stranded β-barrel within BcsA’s C terminus forms a c-di-GMP binding PilZ domain. BcsB is shown in blue. The c-di-GMP dimer and translocating cellulose polymer are shown in spheres. BcsA’s finger helix and gating loop are colored yellow and steel blue, respectively. IF: Amphipathic interface helices that surround the cytosolic entrance to BcsA’s TM channel. Horizontal bars indicate the putative membrane boundaries. (b) Comparison of BcsA’s PilZ positions in the presence and absence of c-di-GMP. BcsA is shown as a pale gray surface, and BcsA’s C terminus is shown as a red cartoon. The position of the β-barrel in the c-di-GMP-free state (pdb 4HG6) is shown as a gray cartoon and c-di-GMP is shown as spheres. (c) Interactions of the “RxxxR” and “DxSxxG” motifs with the c-di-GMP dimer. Residues of each motif are shown as yellow sticks and c-di-GMP is shown in sticks and spheres.
BcsA binds a c-di-GMP dimer on the β-barrel surface
BcsA’s C-terminal PilZ domain binds an intercalated c-di-GMP dimer12 (Fig. 1 and Supplementary Fig. 2). The guanine groups of the c-di-GMP dimer stack parallel to the β-barrel surface and perpendicular to the TM8-β-barrel linker. One c-di-GMP molecule (c-di-GMP-A) interacts with the “DxSxxG” motif on the β-barrel surface, while the second (c-di-GMP-B) is stabilized by π–π stacking interactions with c-di-GMP-A as well as by residues within the TM8-β-barrel linker (Fig. 1c).
All of the conserved PilZ domain residues mediate interactions with the c-di-GMP dimer (Supplementary Fig. 2b) as also observed with isolated PilZ domains16,17. Of note is the interaction of the “RxxxR” motif within the TM8-β-barrel linker with c-di-GMP (Fig. 1c). The N-terminal Arg of this motif (Arg580) runs co-planar to the second guanylate of c-di-GMP-A and forms hydrogen bonds via its guanidinium group with the guanine’s N7 and carbonyl oxygen. The C-terminal Arg (Arg584) of the “RxxxR” motif also interacts with c-di-GMP-A by stacking on top of the first and forming a salt bridge with the phosphate group of the second guanylate moiety, (Fig. 1c and Supplementary Fig. 2b). In the absence of c-di-GMP, Arg580 is rotated by almost 180° towards BcsA’s GT domain and forms a salt bridge with Glu37127. This interaction is broken upon c-di-GMP binding, leading to increased flexibility of BcsA’s gating loop as described below.
The non-conserved Arg579, directly preceding the “RxxxR” motif, runs co-planar to the guanine group of c-di-GMP-B and stacks on top of c-di-GMP-A (Supplementary Fig. 2b). A basic residue in this position is likely necessary to stabilize the interaction with a c-di-GMP dimer, as demonstrated by mutagenesis studies on isolated PilZ domains16,34.
Most structures of β-barrel-containing PilZ domains contain a short α-helix that follows the last strand of the β–barrel and lays flat across its opening16,17. In BcsA, this helix (termed hinge helix) is sandwiched at the interface between the β-barrel and the GT domain (Fig. 1b). When the β-barrel interacts with c-di-GMP, it rotates by approximately 20° around the hinge helix towards the GT domain (Fig. 1b). This rotation closes a groove between the β–barrel and the GT domain that accommodates a short stretch of BcsA’s non-conserved extreme C terminus in the c-di-GMP-free state27, leading to the disorder of BcsA’s C-terminal residues past Arg740.
Conformational changes of the gating loop
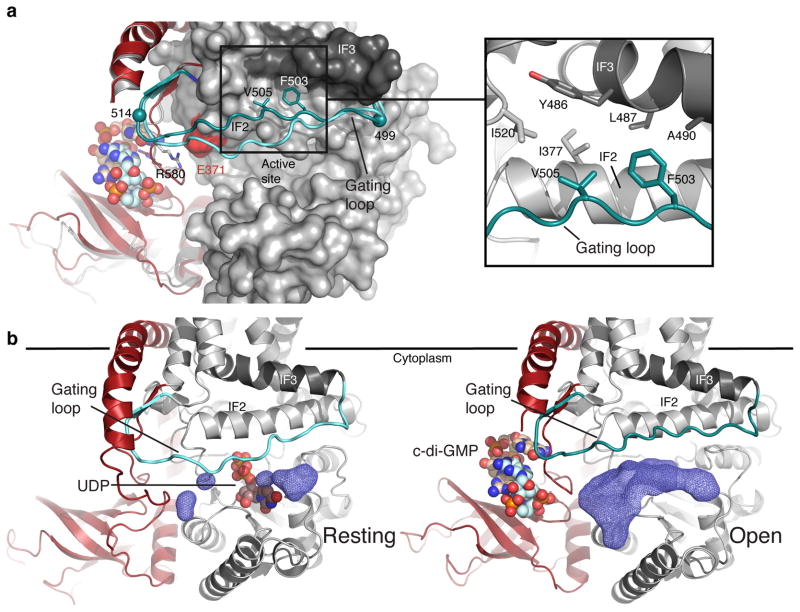
C-di-GMP-binding allows BcsA’s conserved gating loop (residues 499 to 517) to adopt a new conformation, away from the active site cleft and near the water-lipid interface, (Fig. 2a and Supplementary Fig. 3). In this “open” state, the gating loop is stabilized by hydrophobic interactions with BcsA’s amphipathic interface helices (IF), which run parallel to the plane of the membrane at the cytosolic water-lipid boundary (Fig. 1 and 2a) and form the entrance to BcsA’s TM channel27. Phe503 and Val505 of the gating loop’s “FxVTxK” motif (Supplementary Fig. 3a) pack into a conserved hydrophobic pocket formed by Ile377 from IF2, Tyr486, Leu487 and Ala490 of IF3 and Ile520 at the beginning of TM helix 7 (Fig. 2a).
Figure 2.
Conformational changes of BcsA’s gating loop. (a) Gating loop positions in the absence and the presence of c-di-GMP (shown in cyan and steel blue, respectively). Phe503 and Val505 of the “FxVTxK” motif are shown as sticks and the gating loop’s pivots, Arg499 and Glu514, are shown as spheres. The C terminus is colored as in Fig. 1. Inset: residues involved in stabilizing the gating loop in the “open” position are shown as sticks. (b) Accessible volume at the active site entrance (dark blue mesh) in the absence (left) and the presence (right) of c-di-GMP, calculated with a 3.5 Å probe sphere. UDP in the resting (pdb 4HG6) and c-di-GMP in the open BcsA–B structure are shown as spheres.
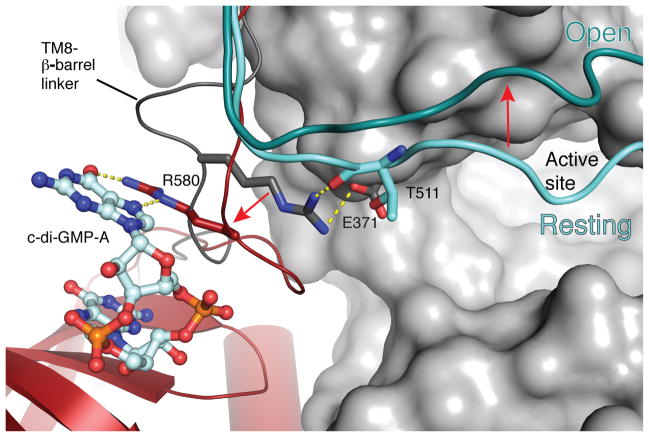
The transition of the gating loop from the previously observed resting to the open state is supported by c-di-GMP-induced conformational changes of the PilZ domain. In the absence of c-di-GMP, the gating loop rests in front of the GT domain entrance, thereby blocking the active site (Fig. 2 and Supplementary Fig. 3). This “resting” state is stabilized by Arg580 of the “RxxxR” motif, which contacts the backbone carbonyl of Thr511 near the C-terminal end of the gating loop (Fig. 3). Arg580 is positioned in close proximity to Thr511 because it also forms a salt bridge with Glu371 of the GT domain, right next to the gating loop’s Thr511 (Fig. 3). The gating loop further interacts with the PilZ domain via its C-terminal end, which forms a two-stranded β-sheet with the c-di-GMP-binding TM8-β-barrel linker (Fig. 1 and 2). Accordingly, in the presence of c-di-GMP, the TM8-β-barrel linker together with the interacting gating loop rotates by about 2 Å towards the water-lipid interface (Fig. 1b) and, importantly, Arg580 rotates by 180° away from the GT domain to coordinate c-di-GMP, thereby breaking its interaction with the gating loop and Glu371 (Fig. 3). This transition releases the gating loop, allowing it to pivot around Arg499 and Glu514 and to swing from its resting state towards the membrane interface. The movement of the gating loop, particularly of residues 504–510, creates a large window at the GT domain entrance approximately 22.5 by 12.5 Å wide, which is sufficient to allow UDP-Glc to enter and UDP to exit the active site (Fig. 2b).
Figure 3.
Stabilization of the gating loop by Arg580. A comparison of the Arg580 position in the absence and presence of c-di-GMP. Arg580 is shown as dark gray and red in the absence and presence of c-di-GMP, respectively. The gating loop is shown in cyan and steel blue representing the “resting” and “open” states, respectively. Glu371 is shown in sticks, and putative interactions are indicated. BcsA’s PilZ domain is colored red and the TM8-β-barrel linker in the resting state is shown as a dark gray cartoon.
In order to mimic a substrate-bound state of BcsA, we soaked crystals with UDP, a product and competitive inhibitor of BcsA23. In the presence of UDP the gating loop is found in another conformation, inserted deep into the substrate-binding pocket (Fig 4a and Supplementary Fig. 3c and d). The loop swings by approximately 15 Å towards the active site, thereby closing the large window formed in its open conformation (Fig. 4a and Supplementary Fig. 3e). In this inserted state, each residue of the loop’s “FxVTxK” motif is involved in coordinating UDP at the active site. Phe503 and Val505 rest on opposing sides above the uracil moiety while Thr506 and Lys508 coordinate its pyrophosphate (Fig. 5b). The pyrophosphate is further stabilized by Gln379 and Arg382 of the “QxxRW” motif, as well as a Mg2+ ion coordinated in turn by Asp246 and Asp248 of the “DxD” motif and His249. Thus, the insertion of the gating loop is likely important for positioning the donor for catalysis. Indeed, in the absence of c-di-GMP, its insertion into the active site is prevented due to steric clashes of its backbone with the Arg580-Glu371 salt bridge (Supplementary Fig. 4b), further ensuring a catalytically inactive state.
Figure 4.

Insertion of the gating loop into the catalytic pocket. (a) A comparison of the gating loop positions of c-di-GMP-bound BcsA–B in the absence and presence of UDP (shown in steel blue and green, respectively). The inserted gating loop is colored green, IF2 is shown as a gray cartoon helix, and UDP as well as the translocating cellulose polymer are shown as sticks. Trp383 of the “QxxRW” motif at the entrance to the TM channel is shown as gray sticks. (b) Coordination of UDP at the active site by the gating loop’s “FxVTxK” motif. The gating loop is colored green, representing the “inserted” state. UDP, the conserved residues of the gating loop as well as Gln379 and Arg382 of the “QxxRW” motif are shown as sticks. The terminal glucose of the cellulose polymer and the putative catalytic base (Asp343) are shown as cyan and gray sticks, respectively.
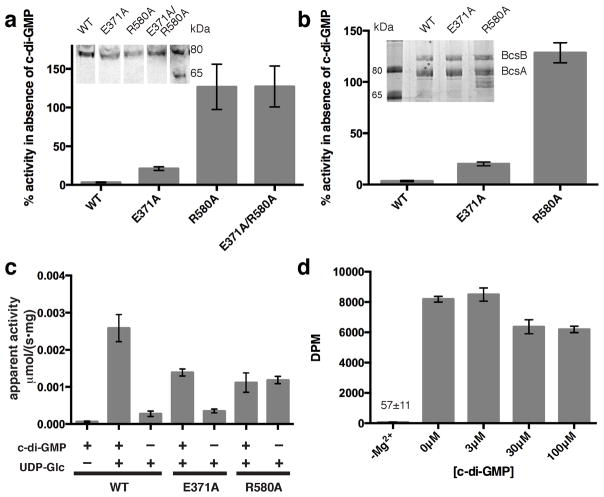
Figure 5.
Comparison of BcsA-catalyzed in vitro cellulose synthesis in the absence and the presence of c-di-GMP. (a and b) Inverted membrane vesicles and proteoliposomes (PL), respectively, containing BcsA–B with the indicated mutations in BcsA were used for cellulose synthesis assays. (WT: wild type). The activity in the absence of c-di-GMP is quantified relative to the activity in the presence of 30 μM c-di-GMP. Insets: Western analysis of IMVs against the C-terminal poly-histidine tag on BcsA (a) and Coomassie-stained SDS-PAGE of the purified BcsA–B complexes (b). (c) Catalytic rates of the indicated PL-reconstituted BcsA–B mutants in the presence and absence of 30 μM c-di-GMP as measured by quantifying the formation of UDP. (d) Activity of the PL-reconstituted BcsA R580A mutant at increasing c-di-GMP concentrations. No activity is observed in the presence of 30 μM c-di-GMP when magnesium is depleted with 25 mM EDTA (–Mg2+). DPM: Disintegrations per minute. (All data represent the means ± SD for 3 technical replicates).
C-di-GMP releases an auto-inhibited state of BcsA
Activation by c-di-GMP is a characteristic of prokaryotic cellulose synthases10,35. Arg580 within the TM8-β-barrel linker either interacts with c-di-GMP or, in the absence of the allosteric activator, is stabilized towards the GT domain by forming a salt bridge with Glu371, thereby tethering the gating loop in the resting position (Fig. 3). Although belonging to the evolutionarily conserved GT domain, Glu371 is only conserved among prokaryotic, c-di-GMP-responsive cellulose synthases, suggesting a regulatory function for the Arg580-Glu371 interaction (Supplementary Fig. 4a).
Indeed, disrupting this salt bridge by replacing Glu371 with Ala increases the enzyme’s catalytic activity in the absence of c-di-GMP approximately 6-fold compared to the wild type enzyme, (Fig. 5a and b). Under these conditions, Arg580 may still be able to interact with the gating loop’s backbone. However, replacing Arg580 with Ala, either in the wild type or E371A background, renders BcsA constitutively active as observed by quantifying the formation of each reaction product, cellulose (Fig. 5a and b and Supplementary Fig. 5a) or UDP (Fig. 5c). Importantly, the R580A mutant still binds c-di-GMP, although with slightly reduced affinity (Supplementary Fig. 5b); yet, even at a c-di-GMP concentration more than 50-fold above its dissociation constant, no further stimulation of cellulose biosynthesis is observed (Fig. 5d). These observations suggest that the Arg580-Glu371 salt bridge and the subsequent interaction of Arg580 with the gating loop are responsible for auto-inhibiting the synthase. This inhibition is then released when Arg580 rotates away from the GT domain to interact with c-di-GMP.
The TM channel entrance forms the acceptor-binding site
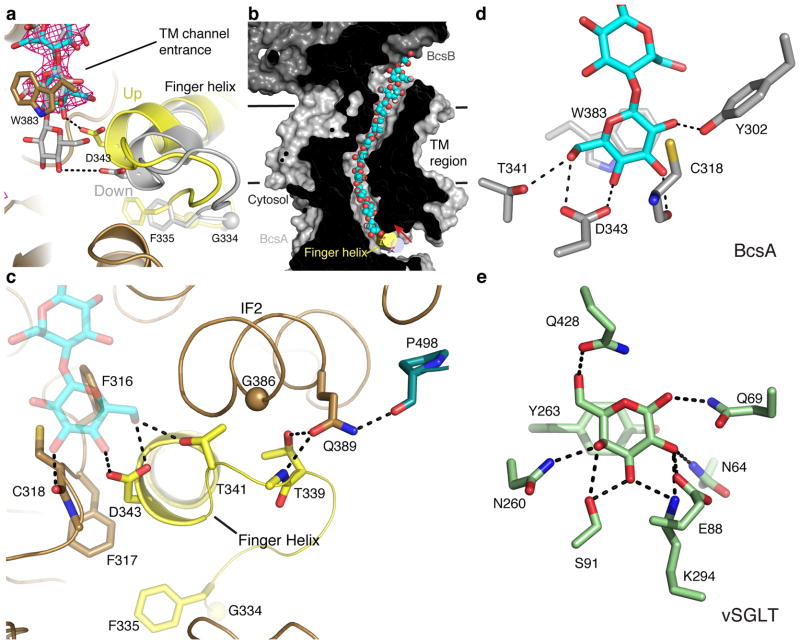
Following sugar transfer, processive GTs, including cellulose, chitin, alginate and hyaluronan synthases, must translocate the elongated polysaccharide, such that the newly formed product sits in a position where it can serve as the acceptor in a subsequent glycosyl transfer reaction. BcsA contacts the acceptor end of the translocating cellulose polymer via a “finger helix” that belongs to the conserved GT domain (Fig. 1). The finger helix contains the “TED” motif at its N terminus, of which Asp343 most likely forms the general base for catalysis27. In contrast to our previously reported structure in which the finger helix points away from the TM channel entrance (“down” state)27, this helix is bent towards the entrance to the channel in both of our new structures and the cellulose polymer is moved into the channel by one glucose unit (Fig. 6a). The finger helix bends near its last C-terminal helical turn and around the conserved His351 (Supplementary Fig. 6a), which is stabilized via side chain interactions with the conserved Ser357 and Tyr410, thereby forming a pivot (Supplementary Fig. 6b). Asp343 at the tip of the finger helix moves by approximately 5 Å towards the TM channel entrance, which is in agreement with the length of a glucopyranose unit (Fig. 6a and b).
Figure 6.
Movement of the finger helix, cellulose translocation, and the acceptor position. (a) Comparison of the positions of BcsA’s finger helix and translocating glucan in the resting (colored gray, pdb 4HG6) and c-di-GMP-bound states. An unbiased SigmaA-weighted mFo–DFc difference electron density of the translocating cellulose polymer in the UDP-free, c-di-GMP-bound state is contoured at 3.5σ and shown as a magenta mesh. The positions of the glucan as observed in the resting state and the c-di-GMP-bound structure are shown as gray and cyan sticks, respectively. BcsA’s finger helix and the preceding small loop are colored yellow. (b) Cut-away view of BcsA’s TM channel with the position of the finger helix in the c-di-GMP-bound and resting states shown as yellow and gray solid cylinders, respectively. The translocating glucan is shown as cyan and red spheres and Asp343 is shown as sticks. Membrane boundaries are indicated by horizontal black lines. (c) Conserved residues involved in stabilizing the finger helix in the “up” position are shown as sticks. (d) Residues coordinating the polymer’s terminal glucose are shown as sticks. (e) Stabilization of a single galactose molecule by the sodium-dependent sugar transporter vSGLT. Residues coordinating galactose are shown as sticks (pdb 3DH4).
A network of conserved hydrophilic and hydrophobic interactions stabilizes the “up” position of the finger helix near the TM channel entrance, including residues from the gating loop, IF2 and the TM channel. Phe316 and Phe317 of the “FFCGS” motif (Supplementary Fig. 7) at the TM channel entrance straddle the helix and additional van-der-Waals interactions are mediated via Gly386 and Met390 of IF2 and Tyr410 within the N-terminal amphipathic section of TM helix 5 (Fig. 6c and Supplementary Fig. 6b). In addition, Thr339, preceding the “TED” motif of the finger helix, hydrogen bonds with the conserved Gln389 of IF2, which in turn interacts with Pro498 at the N-terminal end of the gating loop (Fig. 6c).
The transition of the finger helix towards the channel entrance is supported by a small peripheral loop (residues 333 to 338) that precedes the finger helix. The loop carries a conserved Gly residue (Gly334) at its midpoint, which is followed by a bulky hydrophobic residue, mostly Phe or Ile (Fig. 6a and c and Supplementary Fig. 6a). In the c-di-GMP-bound state, the loop moves towards the GT domain and Phe335 plugs into a conserved hydrophobic pocket beneath the active site, where it is surrounded by the side chains of Met230, Leu234, Val244, Phe317, Leu324 and Leu329 (Fig. 6c and Supplementary Fig. 6c).
The position of the translocating glucan’s terminal glucose unit in the c-di-GMP-bound BcsA–B complex suggests that the acceptor coordination site is located just inside the entrance to the TM channel (Fig. 6a and d). One face of the acceptors glucopyranose ring forms CH-π stacking interactions36 with Trp383 of the “QxxRW” motif, which is characteristic of processive GTs, while its opposite side contacts the carbonyl oxygen of Cys318 of the “FFCGS” motif (Fig. 6c and Supplementary Fig. 7). The N-terminal part of the finger helix contacts the acceptor via the “TED” motif, of which Thr341 and Asp343 form hydrogen bonds with the acceptor’s 2′ or 6′ (depending on its orientation) and 4′ hydroxyl groups (Fig. 6c and d). In particular, the side chain carboxylate of Asp343 is within 2.6 Å of the acceptor’s 4′ hydroxyl group, consistent with its putative role as the catalytic base during glycosyl transfer27. The acceptor further interacts with Tyr302, also located at the entrance to the TM channel (Fig. 6d). Accordingly, this implies that cellulose synthase forms the acceptor coordination site with the invariant “QxxRW” and “FFCGS” motifs at the entrance to the TM channel (Supplementary Fig. 7). The stabilization of the terminal glucose unit at this position resembles the coordination of a single galactose molecule in the sodium-coupled sugar transporter vSGLT37, (Fig. 6e), suggesting that BcsA’s acceptor binding site might also suffice to coordinate a single glucose molecule to initiate cellulose synthesis.
Discussion
Comparing the structures of the c-di-GMP-activated and resting states of the BcsA–B complex, at intermediate states during cellulose translocation provides unique insights into the mechanism of cellulose biosynthesis. In the absence of c-di-GMP, BcsA is catalytically inactive and its gating loop blocks the entrance to the active site23,27. Allosteric activation by c-di-GMP displaces the gating loop from the active site, thereby forming a large opening towards the substrate-binding pocket, wide enough for substrate diffusion. However, opening and closing the active site is unlikely to be the only function of BcsA’s gating loop. When UDP binds to the active site, the gating loop inserts deeply into the catalytic pocket and coordinates the nucleotide via conserved residues. Most likely, this also reflects how BcsA interacts with its substrate UDP-Glc, positioning it for catalysis, excluding water from the active site and perhaps also stabilizing the UDP leaving group during glycosyl transfer. A similar mechanism of substrate-dependent loop insertion and de-insertion has been described for non-processive galactosyltransferases38,39. The functional importance of the gating loop is further underlined by its sequence homology with the location of the isoxaben resistance mutation in Arabidopsis thaliana cellulose synthase 3 (Supplementary Fig. 3a). Here, Thr942 of the “FxVTxK” motif is mutated to Ile, thereby allowing growth in the presence of the herbicide isoxaben40. However, because pro- and eukaryotic cellulose synthases differ in their predicted TM topologies28, further experimental analyses are required to confirm a similar eukaryotic gating loop function.
UDP, the second reaction product of many GTs24,41, competitively inhibits BcsA, which has also been observed for hyaluronan synthases23,42. BcsA binds UDP and UDP-Glc with similar affinities23, however, the large excess of UDP-Glc over UDP under physiological conditions would favor substrate binding upon gating loop opening43. Presumably during or after UDP-Glc binding, the gating loop inserts into the active site to initiate catalysis. Following glycosyl transfer and with the newly extended glucan at the active site, the gating loop may retract from the GT domain, thereby allowing UDP to UDP-Glc exchange. Because the gating loop undergoes its full range of motion in the presence of c-di-GMP, it is likely that the allosteric activator remains bound during catalysis. In vivo, c-di-GMP-stimulated cellulose biosynthesis may terminate upon depletion of the activator, whose cytosolic concentration is in turn controlled by the synergy of diguanylate cyclases and diesterases1.
The BcsA–B complex contains a translocating cellulose polymer that spans the distance from the GT domain to the periplasmic BcsA–B interface. In the c-di-GMP activated structure, the polymer’s acceptor terminus rests at the entrance to the TM channel, one glucose unit further into the pore compared to its position in the absence of c-di-GMP27. Thus, while our previously reported structure likely represents a state post glycosyl transfer but prior to translocation, the c-di-GMP-activated BcsA–B structure is consistent with a state after polymer translocation. Cellulose translocation may be accomplished by BcsA’s finger helix, which hydrogen bonds with the acceptor glucose and pivots towards the TM channel entrance in the c-di-GMP-activated complex. In this position, Asp343 of the finger helix is at an ideal distance to facilitate catalysis. Perhaps the finger helix returns to the “down” position after glycosyl transfer to interact with the new polymer terminus. A similar mechanism involving a flexible loop or helical domain has been postulated for the processive translocation of unfolded polypeptide chains44,45.
C-di-GMP stimulates the biosynthesis of several extracellular polysaccharides important for biofilm formation, including alginate and PNAG46–48. While the mechanism for activating PNAG biosynthesis most likely differs from BcsA48, alginate and cellulose synthases share a strikingly similar organization49. Alginate is a major component of Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients18,49. In contrast to BcsA–B, the alginate synthase’s c-di-GMP-binding PilZ domain is located at the intracellular N terminus of Alg44, the non-catalytic subunit that resembles BcsB and likely interacts with the catalytic Alg8 subunit. Thus, c-di-GMP could exert control by a similar mechanism in alginate synthase as revealed for bacterial cellulose synthase.
Our analyses provide the first insights into how enzymatic functions can be modulated by c-di-GMP. A detailed mechanistic characterization of this bacterial signaling system is required for the development of novel anti-microbial therapeutics.
Online Methods
Protein purification
BcsA–B was purified as previously described 27 with the exception that gel filtration was carried out in 20 mM Tris pH 7.5, 100 mM NaCl, 5 mM MgCl2, 5 mM cellobiose, 10% glycerol, 5 mM N,N-Dimethyl-N-dodecylamine N-oxide (LDAO), and 0.3 mM LysoFosCholine Ether 12 (LFCE12), (GF buffer). Peak fractions containing BcsA and BcsB were collected and concentrated to ~10 mg/ml and spun at 180,000 g for 15 min at 4°C. Bicelles were prepared by mixing 250 μl water with 100 mg of 1,2-dimyristoyl-sn-glycero-3-phosphocholine:1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine:3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate, (DMPC:POPE:CHAPS) at a molar ratio of ~2.34:0.05:1. The concentrated protein was mixed with the bicelles at a 4:1 (v:v) ratio and allowed to equilibrate on ice for at least 1 hour. 2 mM UDP and 1 mM c-di-GMP were added before incubating on ice overnight.
Crystallization
Crystals belonging to space group P212121 were grown by sitting-drop vapor diffusion at 30°C in 1.65–1.9 M sodium acetate and 100 mM sodium citrate, pH 3–3.5, (final pH 5.6) at a 1:1 (v:v) ratio of protein/bicelle and well solution. Crystals appeared within 3 days and reached their final size within 14 days. For the UDP-free structure, cryo-protection and dilution of UDP were achieved by gradual addition of a solution containing 20 mM sodium citrate pH 3, 100 mM NaCl, 5 mM MgCl2, 1.95 M sodium acetate, 20% glycerol, and 20 % bicelles to the drop. For the UDP-bound structure, cryo-protection and soaking with UDP were achieved by gradual addition of a solution containing 20 mM sodium citrate pH 3.5, 50 mM MgCl2, 1.95 M sodium acetate, 20% glycerol, 12% bicelles, and 10 mM UDP to the drop. Crystals were flash-cooled in liquid nitrogen for data collection.
Data Collection and Processing
Diffraction data were collected at 100K at a wavelength of 1.0Å. Data in the absence of UDP were collected at the Advanced Photon Source SER-CAT beamline 22-ID and data in the presence of UDP were collected at GM/CA-CAT beamline 23-ID. The data were integrated using Mosflm51 and scaled in Aimless as part of the CCP4 program suite52.
Structure Determination
Initial phases for the UDP-free structure were determined by molecular replacement (MR) in Phaser53 using ligand-free pdb 4HG6 with BcsA residues 499–512 (gating loop) and 574–758 (C terminus) truncated as a search model. MR-phases for the UDP-bound structure were determined in MOLREP54 using the UDP-free structure without the gating loop as search model.
The models were refined by rigid body and restrained refinement in Refmac552 as well as simulated annealing in Phenix55. Phases were improved using density modification in Parrot56,57, and model building was performed in Coot58. Iterative rounds of model building, refinement, and density modification resulted in a map of sufficient quality to place the missing domains and ligands. In order to minimize model bias, simulated annealing composite omit maps, prime and switch maps, and kicked maps55,59 were calculated and evaluated throughout the model building process. Additionally, TLS parameters determined from the TLSMD server60 were utilized in later rounds of refinement. The model contains residues 13–740 of BcsA and residue 54–720 of BcsB. Residue 532–543 of BcsB are disordered as previously observed27 and were omitted from the model. A 10 residue long unidentified peptide likely belonging to either the extended N terminus of BcsB or the C terminus of BcsA is sandwiched by BcsB’s flavodoxin-like domain 2 and carbohydrate-binding domain 227 between crystallographic symmetry mates. This peptide was modeled as a poly-alanine with chain identifier “D”. The UDP-free model contains 5 partially ordered lipids, one modeled as 1,2-diacyl-sn-glycero-3-phosphoethanolamine and 4 modeled as 1,2-diacyl-sn-glycero-3-phosphocholine. Two of these lipids were also observed in the UDP-bound structure.
The UDP-free BcsA–B structure was refined to an R/Rfree of 19.9/23.0. 95.8% of residues lie in the favored regions of the Ramachandran plot with no outliers. The UDP-bound complex structure was refine to an R/Rfree of 20.6/23.8. 95.1% of residues lie in the favored region on a Ramachandran plot with 0.7% outliers. Figures were prepared in Pymol61 and the solvent accessible surface analysis was performed in HOLLOW62. Crystallographic software support is provided by SBGrid63.
Inverted Membrane Vesicle Preparation
Inverted membrane vesicles (IMVs) containing wild type BcsB and the indicated BcsA mutants were prepared as previously described30. Control IMVs were prepared from E. coli transformed with an empty pETDuet vector. In brief, the constructs were expressed as described27 and the cells were resuspended in RB buffer containing 20 mM sodium phosphate pH 7.3, 100 mM NaCl, and 10% glycerol using 20 ml RB per cell pellet from a 1 L culture. The cells were lysed in a microfluidizer and spun at 12,000 g for 20 min to clear the cell debris. The supernatant was applied on the surface of a 1.8 M sucrose cushion and spun at 150,000 g for 2 hr at 4°C. IMVs were harvested, diluted 3-fold in RB, and spun at 150,000 g overnight. The pellet from a 3 L culture was resuspended in 1 ml RB, homogenized in a dounce and stored in aliquots at −80°C.
Proteoliposome Preparation
Purified BcsA–B complex containing the indicated mutations were reconstituted into proteoliposomes (PLs) as previously described23. Briefly, BcsA–B was purified as described above with the exception that 1 mM LysoFosCholine Ether 14 (LFCE14) was used instead of LDAO. The protein was concentrated to 5 μM, incubated with 4 mg/ml E. coli total lipid extract (diluted from a 20 mg/ml stock solution in 40 mM LDAO) and allowed to equilibrate on ice for at least 20 min. Bio-Beads (BioRad) were added, and the solution was rotated until it became turbid, indicating the formation of PLs. The samples were then aliquoted, snap-frozen in liquid nitrogen, and stored at −80°C. The final protein concentration for all mutants was determined by UV absorbance and SDS-PAGE followed by Coomassie staining.
In vitro Cellulose Synthesis Assay
IMVs or PLs were added to a solution containing 20 mM sodium phosphate, 100 mM NaCl, 20 mM MgCl2, 5 mM UDP-glucose, 12.5 μCi/ml UDP-[3H]-glucose as well as 30μM c-di-GMP unless indicated otherwise. The reaction was incubated at 37°C for 45 min with shaking at 350 rpm. 2% SDS was added to terminate the reaction and dissolve the vesicles. The mixture was then spun at 21,000 g for at least 20 min to pellet the insoluble cellulose. The supernatant was carefully removed, and the pellet was resuspended in 50 mM Tris pH 7.5 and 100 mM NaCl and spotted on Whatman 3 mm grid paper. The product was purified in 60% ethanol by descending paper chromatography, with the insoluble cellulose remaining at the origin, and quantified by scintillation counting30. All measurements were performed at least in triplicate and error bars represent standard deviations.
Western Analysis
10 μL IMVs were analyzed by SDS-PAGE and transferred to a nitrocellulose membrane using a BioRad Mini-Transfer Cell according to the manufacturer’s specifications. The nitrocellulose membrane was blocked in 5% milk/TBS-Tween solution for 30 min and incubated overnight with a mouse anti-penta-His (Qiagen) antibody. The membranes were washed three times in 5% milk/TBS-Tween before incubating with an IRDye800-conjugated anti-mouse secondary antibody (Rockland) for 45 min at RT. After washing, the membranes were scanned on an Odyssey Infrared Imager (Licor).
Enzyme-coupled activity assay
Enzyme-coupled kinetic assays were carried out as previously described23 with the exception that the protein was reconstituted into PLs instead of nanodiscs, the experiments were performed in 150 μl reaction volume in 96-well flat bottom Microplates (Greiner), and 3 mM UDP-Glc was used.
Isothermal Titration Calorimetry
The protein was purified in 1 mM LFCE14 as described above. Measurements were carried out at 25° C in a MicroCal iTC200 system (GE Healthcare) with 250μl of BcsA–B in the cell at 9.3 μM for WT and 11 μM for the BcsA–Arg580Ala complex and 400 μM c-di-GMP in the syringe. An initial 0.5 μl injection was followed by 39 1 μl injections spaced 180s apart with stirring at 700 rpm. The data were fit using Origin 7.0 as provided by the manufacturer.
Supplementary Material
Acknowledgments
We are grateful to O. Pornillos, D. Cosgrove, J. Casanova, and T. Rapoport for critical comments on the manuscript. X-ray diffraction data were collected at GM/CA- and Southeast Regional-Collaborative Access Team beamlines at the Advanced Photon Source (APS), Argonne National Laboratory. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. DE-AC02-06CH11357 and W-31-109-Eng-38. GM/CA at APS has been funded in whole or in part with funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Sciences (Y1-GM-1104). J.L.W.M. is supported by a National Science Foundation Graduate Research Fellowship, Grant No. DGE-1315231. The research was supported by NIH grant R01GM101001 (J.Z.).
Footnotes
Accession codes. Atomic coordinates and structure factors for the UDP-free and UDP-bound complexes have been deposited at the Protein Data Bank under accession number 4P02 and 4P00, respectively.
Author contributions
J.Z. designed the experiments. J.L.W.M. crystallized the BcsA–B complex and solved the structures and performed biochemical experiments. J.L.W.M., J.T.M. and J.Z. analyzed the data and wrote the manuscript.
References
- 1.Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the First 25 Years of a Universal Bacterial Second Messenger. Microbiol Mol Biol Rev. 2013;77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gloag ES, Turnbull L, Huang A, Vallotton P, Wang H, Nolan LM, Mililli L, Hunt C, Lu J, Osvath SR, Monahan LG, Cavaliere R, Charles IG, Wand MP, Gee ML, Prabhakar R, Whitchurch CB. Self-organization of bacterial biofilms is facilitated by extracellular DNA. Proc Natl Acad Sci U S A. 2013;110:11541–11546. doi: 10.1073/pnas.1218898110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCrate OA, Zhou X, Reichhardt C, Cegelski L. Sum of the parts: composition and architecture of the bacterial extracellular matrix. J Mol Biol. 2013;425:4286–4294. doi: 10.1016/j.jmb.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilking JN, Zaburdaev V, De Volder M, Losick R, Brenner MP, Weitz DA. Liquid transport facilitated by channels in Bacillus subtilis biofilms. Proc Natl Acad Sci U S A. 2013;110:848–852. doi: 10.1073/pnas.1216376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358:135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 6.Romling U, Balsalobre C. Biofilm infections, their resilience to therapy and innovative treatment strategies. J Intern Med. 2012;272:541–561. doi: 10.1111/joim.12004. [DOI] [PubMed] [Google Scholar]
- 7.Pritt B, O’Brien L, Winn W. Mucoid Pseudomonas in cystic fibrosis. Am J Clin Pathol. 2007;128:32–34. doi: 10.1309/KJRPC7DD5TR9NTDM. [DOI] [PubMed] [Google Scholar]
- 8.Gomes F, Teixeira P, Oliveira R. Mini-review: Staphylococcus epidermidis as the most frequent cause of nosocomial infections: old and new fighting strategies. Biofouling. 2014;30:131–141. doi: 10.1080/08927014.2013.848858. [DOI] [PubMed] [Google Scholar]
- 9.Cotter PA, Stibitz S. c-di-GMP-mediated regulation of virulence and biofilm formation. Curr Op Microbiol. 2007;10:17–23. doi: 10.1016/j.mib.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 10.Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, Benziman M. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 11.Zogaj X, Bokranz W, Nimtz M, Römling U. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect Immun. 2003;71:4151–4158. doi: 10.1128/IAI.71.7.4151-4158.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z, Kim S, Gaffney BL, Jones RA. Polymorphism of the signaling molecule c-di-GMP. J Am Chem Soc. 2006;128:7015–7024. doi: 10.1021/ja0613714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gentner M, Allan MG, Zaehringer F, Schirmer T, Grzesiek S. Oligomer formation of the bacterial second messenger c-di-GMP: reaction rates and equilibrium constants indicate a monomeric state at physiological concentrations. J Am Chem Soc. 2012;134:1019–1029. doi: 10.1021/ja207742q. [DOI] [PubMed] [Google Scholar]
- 14.Christen M, Christen B, Allan MG, Folcher M, Jeno P, Grzesiek S, Jenal U. DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc Natl Acad Sci U S A. 2007;104:4112–4117. doi: 10.1073/pnas.0607738104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amikam D, Galperin MY. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics. 2006;22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 16.Ko J, Ryu KS, Kim H, Shin JS, Lee JO, Cheong C, Choi BS. Structure of PP4397 reveals the molecular basis for different c-di-GMP binding modes by Pilz domain proteins. J Mol Biol. 2010;398:97–110. doi: 10.1016/j.jmb.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, Forouhar F, Neely H, Seetharaman J, Camilli A, Hunt JF. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 2007;26:5153–5166. doi: 10.1038/sj.emboj.7601918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hay ID, Ur Rehman Z, Moradali MF, Wang Y, Rehm BH. Microbial alginate production, modification and its applications. Microb Biotechnol. 2013;6:637–650. doi: 10.1111/1751-7915.12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hay ID, Wang Y, Moradali MF, Rehman ZU, Rehm BH. Genetics and regulation of bacterial alginate production. Environ Microbiol. 2014 doi: 10.1111/1462-2920.12389. [DOI] [PubMed] [Google Scholar]
- 20.Whitney JC, Howell PL. Synthase-dependent exopolysaccharide secretion in Gram-negative bacteria. Trends Microbiol. 2013;21:63–72. doi: 10.1016/j.tim.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Preston JFr, Romeo T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol. 2004;186:2724–2734. doi: 10.1128/JB.186.9.2724-2734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merzendorfer H. Insect chitin synthases: a review. J Comp Physiol B, Biochem Syst Environ Physiol. 2006;176:1–15. doi: 10.1007/s00360-005-0005-3. [DOI] [PubMed] [Google Scholar]
- 23.Omadjela O, Narahari A, Strumillo J, Melida H, Mazur O, Bulone V, Zimmer J. BcsA and BcsB form the catalytically active core of bacterial cellulose synthase sufficient for in vitro cellulose synthesis. Proc Natl Acad Sci U S A. 2013;110:17856–17861. doi: 10.1073/pnas.1314063110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown C, Leijon F, Bulone V. Radiometric and spectrophotometric in vitro assays of glycosyltransferases involved in plant cell wall carbohydrate biosynthesis. Nat Protoc. 2012;7:1634–1650. doi: 10.1038/nprot.2012.089. [DOI] [PubMed] [Google Scholar]
- 25.Mayer R, Ross P, Weinhouse H, Amikam D, Volman G, Ohana P, Calhoon RD, Wong HC, Emerick AW, Benziman M. Polypeptide composition of bacterial cyclic diguanylic acid-dependent cellulose synthase and the occurrence of immunologically crossreacting proteins in higher plants. Proc Natl Acad Sci USA. 1991;88:5472–5476. doi: 10.1073/pnas.88.12.5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saxena IM, Kudlicka K, Okuda K, Brown RM. Characterization of genes in the cellulose-synthesizing operon (acs operon) of Acetobacter xylinum: implications for cellulose crystallization. J Bacteriol. 1994;176:5735–5752. doi: 10.1128/jb.176.18.5735-5752.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morgan JL, Strumillo J, Zimmer J. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature. 2013;493:181–186. doi: 10.1038/nature11744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slabaugh E, Davis JK, Haigler CH, Yingling YG, Zimmer J. Cellulose synthases: new insights from crystallography and modeling. Trends Plant Sci. 2013 doi: 10.1016/j.tplants.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 29.Cantarel BL, Coutinho PM, Rancurel C, Bernard T. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–8. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hubbard C, McNamara JT, Azumaya C, Patel MS, Zimmer J. The hyaluronan synthase catalyzes the synthesis and membrane translocation of hyaluronan. J Mol Biol. 2012;418:21–31. doi: 10.1016/j.jmb.2012.01.053. [DOI] [PubMed] [Google Scholar]
- 31.Merighi M, Lee VT, Hyodo M, Hayakawa Y, Lory S. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol Microbiol. 2007;65:876–895. doi: 10.1111/j.1365-2958.2007.05817.x. [DOI] [PubMed] [Google Scholar]
- 32.Faham S, Bowie JU. Bicelle crystallization: a new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure. J Mol Biol. 2002;316:1–6. doi: 10.1006/jmbi.2001.5295. [DOI] [PubMed] [Google Scholar]
- 33.Faham S, Boulting GL, Massey EA, Yohannan S, Yang D, Bowie JU. Crystallization of bacteriorhodopsin from bicelle formulations at room temperature. Protein Science. 2005;14:836–840. doi: 10.1110/ps.041167605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujiwara T, Komoda K, Sakurai N, Tajima K, Tanaka I, Yao M. The c-di-GMP recognition mechanism of the PilZ domain of bacterial cellulose synthase subunit A. Biochem Biophys Res Commun. 2013;431:802–807. doi: 10.1016/j.bbrc.2012.12.103. [DOI] [PubMed] [Google Scholar]
- 35.Aloni Y, Delmer DP, Benziman M. Achievement of high rates of in vitro synthesis of 1,4-beta-D-glucan: activation by cooperative interaction of the Acetobacter xylinum enzyme system with GTP, polyethylene glycol, and a protein factor. Proc Natl Acad Sci USA. 1982;79:6448–6452. doi: 10.1073/pnas.79.21.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumari M, Sunoj RB, Balaji PV. Exploration of CH···π mediated stacking interactions in saccharide: aromatic residue complexes through conformational sampling. Carbohydr Res. 2012;361:133–140. doi: 10.1016/j.carres.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 37.Faham S, Watanabe A, Besserer GM, Cascio D, Specht A, Hirayama BA, Wright EM, Abramson J. The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport. Science. 2008;321:810–814. doi: 10.1126/science.1160406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qasba PK, Ramakrishnan B, Boeggeman E. Structure and Function of β-1,4-Galactosyltransferase. Curr Drug Targets. 2008;4:292–309. doi: 10.2174/138945008783954943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramakrishnan B, Ramasamy V, Qasba PK. Structural snapshots of beta-1,4-galactosyltransferase-I along the kinetic pathway. J Mol Biol. 2006;357:1619–1633. doi: 10.1016/j.jmb.2006.01.088. [DOI] [PubMed] [Google Scholar]
- 40.Scheible WR, Eshed R, Richmond T, Delmer D, Somerville C. Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis Ixr1 mutants. Proc Natl Acad Sci U S A. 2001;98:10079–10084. doi: 10.1073/pnas.191361598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 42.Tlapak-Simmons VL, Baron CA, Weigel PH. Characterization of the purified hyaluronan synthase from Streptococcus equisimilis. Biochemistry. 2004;43:9234–9242. doi: 10.1021/bi049468v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buckstein MH, He J, Rubin H. Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J Bacteriol. 2008;190:718–726. doi: 10.1128/JB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Erlandson KJ, Miller SB, Nam Y, Osborne AR, Zimmer J, Rapoport TA. A role for the two-helix finger of the SecA ATPase in protein translocation. Nature. 2008;455:984–987. doi: 10.1038/nature07439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin A, Baker TA, Sauer RT. Pore loops of the AAA+ ClpX machine grip substrates to drive translocation and unfolding. Nature Struct Mol Biol. 2008;15:1147–1151. doi: 10.1038/nsmb.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rehm BHA. Alginate Production: Precursor Biosynthesis, Polymerization and Secretion. Microbiology Monographs. 2009;13:55–71. [Google Scholar]
- 47.Itoh Y, Rice JD, Goller C, Pannuri A, Taylor J, Meisner J, Beveridge TJ, Preston JFr, Romeo T. Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-beta-1,6-N-acetyl-D-glucosamine. J Bacteriol. 2008;190:3670–3680. doi: 10.1128/JB.01920-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steiner S, Lori C, Boehm A, Jenal U. Allosteric activation of exopolysaccharide synthesis through cyclic di-GMP-stimulated protein-protein interaction. EMBO J. 2013;32:354–368. doi: 10.1038/emboj.2012.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keiski CL, Harwich M, Jain S, Neculai AM, Yip P, Robinson H, Whitney JC, Riley L, Burrows LL, Ohman DE, Howell PL. AlgK is a TPR-containing protein and the periplasmic component of a novel exopolysaccharide secretin. Structure. 2010;18:265–273. doi: 10.1016/j.str.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leslie AG. The integration of macromolecular diffraction data. Acta Crystallogr D Biol Crystallogr. 2006;62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 52.CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 53.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 55.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cowtan K. Recent developments in classical density modification. Acta Crystallogr D Biol Crystallogr. 2010;66:470–478. doi: 10.1107/S090744490903947X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cowtan KD, Zhang KY. Density modification for macromolecular phase improvement. Prog Biophys Mol Biol. 1999;72:245–270. doi: 10.1016/s0079-6107(99)00008-5. [DOI] [PubMed] [Google Scholar]
- 58.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 59.Terwilliger TC. Using prime-and-switch phasing to reduce model bias in molecular replacement. Acta Crystallogr D Biol Crystallogr. 2004;60:2144–2149. doi: 10.1107/S0907444904019535. [DOI] [PubMed] [Google Scholar]
- 60.Painter J. TLSMD web server for the generation of multi-group TLS models. J Appl Cryst. 2006;39:109–111. [Google Scholar]
- 61.PyMol. The PYMOL Molecular Graphics System. DeLano Scientific. 2000 [Google Scholar]
- 62.Ho BK, Gruswitz F. HOLLOW: generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct Biol. 2008;8:49. doi: 10.1186/1472-6807-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morin A, Eisenbraun B, Key J, Sanschagrin PC, Timony MA, Ottaviano M, Sliz P. Collaboration gets the most out of software. Elife. 2013;2:e01456. doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.