Abstract
The transcription factor signal transducer and activator of transcription 5 (STAT5) is constitutively activated in a wide range of leukemias and lymphomas, and drives the expression of genes necessary for proliferation, survival, and self-renewal. Thus, targeting STAT5 is an appealing therapeutic strategy for hematological malignancies. Given the importance of bromodomain-containing proteins in transcriptional regulation, we considered the hypothesis that a pharmacological bromodomain inhibitor could inhibit STAT5-dependent gene expression. We found that the small molecule bromodomain and extra-terminal (BET) bromodomain inhibitor JQ1 decreases STAT5-dependent (but not STAT3-dependent) transcription of both heterologous reporter genes and endogenous STAT5 target genes. JQ1 reduces STAT5 function in leukemia and lymphoma cells with constitutive STAT5 activation, or inducibly activated by cytokine stimulation. Among the BET bromodomain sub-family of proteins, it appears that BRD2 is the critical mediator for STAT5 activity. In experimental models of acute T cell lymphoblastic leukemias, where activated STAT5 contributes to leukemia cell survival, Brd2 knock-down or JQ1 treatment shows strong synergy with tyrosine kinase inhibitors in inducing leukemia cells apoptosis. By contrast, mononuclear cells isolated form umbilical cord blood, which is enriched in normal hematopoietic precursor cells, were unaffected by these combinations. These findings indicate a unique functional association between BRD2 and STAT5, and suggest that combinations of JQ1 and tyrosine kinase inhibitors may be an important rational strategy for treating leukemias and lymphomas driven by constitutive STAT5 activation.
Keywords: Targeted therapy, transcription factors, leukemias, gene expression, kinase inhibitors
Introduction
The transcription factor STAT5 is an essential mediator of the pathogenesis of many forms of leukemia and lymphoma. Under physiological conditions, STATs are activated rapidly and transiently. However, STAT5 is constitutively active in many forms of hematologic malignancies, including chronic myelogenous leukemia (CML), acute myelogenous leukemia (AML), acute lymphocytic leukemia (ALL) (1, 2), and Hodgkin lymphoma (HL) (3). STAT5 is an essential mediator in neoplasms induced by mutated tyrosine kinases (TKs) such as BCR-ABL and Jak2V617F (4) (5), and can be activated through the autocrine or paracrine secretion of cytokines that signal through Jak kinases.
Treatment of leukemias driven by constitutively activated oncogenic kinases has been greatly improved by the development of tyrosine kinases inhibitors (TKIs), especially in CML. However, patients with CML who initially respond well to TKIs may acquire resistance with progression of their disease. Indeed, increased activation of STAT5 has been associated with leukemia progression and TKI resistance (6). Moreover, despite the identification of various kinases as therapeutic targets in leukemia, TKIs used as a single agent have had only limited success beyond CML. For example, even in ALL containing the BCR-ABL translocation, TKIs have had only modest benefit. There are likely multiple mechanisms for the limited benefits of TKIs in ALL (7–9). For example, murine leukemia models suggest that activated tyrosine kinases may be important in initial lymphoid transformation, but dispensable once leukemia is established (10).
Since STAT5 is at a convergence point of a variety of kinases, and is one of the direct mediators of the abnormal gene expression underlying leukemic pathogenesis, STAT5 may be a critical therapeutic target. Reflecting this, CML cells with mutant BCR-ABL conferring resistance to TKIs show equal sensitivity to STAT5 inhibitors as cells with unmutated BCR-ABL (11). Also, dual inhibition of kinases and STAT5 leads to more efficient reduction of leukemia cell viability (11, 12). All of these data support the concept that STAT5 is a promising therapeutic target in hematological malignancy.
Although meaningful progress has been made in developing STAT inhibitors, transcription factors remain challenging drug targets. An alternate approach to inhibit the transcriptional function of STAT5 is to identify key transcriptional co-factors whose inhibition will decrease STAT5-dependent gene transcription. One such group of transcriptional regulators is the BET (bromodomains and extra-terminal domain) family of bromodomain-containing proteins, which includes BRD2, BRD3, BRD4 and BRDT. Members of this family bind specifically to poly-acetylated histone tails within active euchromatin, and recruit protein complexes that mediate chromatin remodeling and transcriptional elongation. Nuclear BET-protein interactome studies have indicated that BET proteins are integral components of a large number of nuclear protein complexes (13, 14). Therefore, we utilized the recently synthesized bromodomain inhibitor JQ1 to probe the role of BET family members in the STAT5-driven pathogenesis of hematologic cancers (15).
Materials and Methods
Cells
The T lymphocytic leukemia cell lines ALL-SIL, DU528, DND41 and TALL-1 were obtained from Thomas Look (Dana Farber Cancer Institute, Boston, MA), and cultured in RPMI media supplemented with 10% (for DND41) or 20% FBS (for ALL-SIL, DU528 and TALL-1). K562 and HEL cells (obtained from Daniel G. Tenen, Beth Israel-Deaconess Medical Center, Boston, MA) were cultured in RPMI media supplemented with 10% FBS. The malignant hematopoietic cell lines used, with their clinical and molecular characteristics, are summarized in Supplemental table 1. Cells were authenticated by short tandem repeat (STR) loci and the sex identity locus profiling using PowerPlex 16HS PCR Amplification Kit (Promega Corporation) at Genetica (April 2013). STAT3 and STAT5-dependent luciferase activities were measured in STAT-luc/U3A and NCAM2-luc/T47D cells, respectively (16). Luciferase activity was quantified using Bright-Glo Luciferase assay system (Promega, Madison, WI).
Bone marrow mononuclear cells from patients with untreated T cell acute lymphocytic leukemia were obtained through a Dana-Farber Cancer Institute Institutional Review Board-approved protocol for which patients gave written informed consent in accordance with the Declaration of Helsinki. Similarly, human umbilical cord blood was obtained through an Institutional Review Board-approved protocol and mononuclear cells was isolated by Ficoll-Paque density gradient centrifugation. The mononuclear cells layer was collected and cultured in RPMI 1640 medium supplemented with 10% FBS, L-glutamine, sodium pyruvate, essential amino acids and vitamins, beta-mercaptoethanol, penicillin, and streptomycin.
Compounds
JQ1 and iBET were synthesized as described (15, 17). Imatinib and nilotinib were from Novartis Pharma (Basel, Switzerland). Jak Inhibitor 1 was obtained from EMD Millipore (Billerica, MA). All drugs were dissolved in dimethyl sulfoxide (DMSO) and were diluted to a final concentration of 0.1% DMSO in all experiments.
Immunoblots and chromatin immunoprecipitation (ChIP)
Immunoblots were performed using the following antibodies: Phospho-specific STAT5 (9351) from Cell Signaling; BCLx (sc-7195), CIS (sc-15344), PIM1 (sc-28777), Myc (sc-764), total STAT5 (sc-835) and HSP90 (sc-13119) from Santa Cruz Biotechnology (Santa Cruz, CA); Brd2 (A302-583A) from Bethyl Antibody Laboratories, Inc. (Montgomery, TX). Band intensity was quantitated using Image J software (NIH).
ChIP was performed as described (18). Briefly, leukemia cells were formaldehyde fixed and sonicated, and lysates were immunoprecipitated with anti-STAT5 or anti-polymerase II (sc-9001, Santa Cruz Biotechnology) antibodies. Quantitative PCR was performed in triplicate on ChIP product or input using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) and region-specific primers (Supplemental table 2). ChIP values were normalized to input and expressed as mean fold change relative to a control DNA binding site known not to bind STAT5.
Quantitation of viable cell number
Viable cells were measured by adenosine triphosphate (ATP)–dependent bioluminescence using the CellTiter-Glo assay (Promega, Madison, WI). The combinatorial index to measure the effects of drug combinations on the number of viable cells was calculated using CalcuSyn software (Conservion, Ferguson, MO).
Quantitative RT-PCR (qRT-PCR)
RNA was harvested using an RNeasy Mini Kit from QIAGEN. cDNA was generated using the TaqMan Reverse Transcription kit (Applied Biosystems), and quantitative reverse transcription polymerase chain reaction (RT-PCR) was performed using primers as indicated (Supplemental table 2). Data are expressed as mean fold change ± standard deviation of three replicates.
Apoptosis assays
Cells were stained with annexin-V–fluorescein isothiocyanate and propidium iodide using the annexin-V–FLUOS Staining Kit (Roche Applied Science, Penzberg, Germany). Samples were analyzed on the BD FACSCanto II Flow Cytometer (BD Biosciences, San Jose, CA) using negative and single-color controls to adjust compensation.
Luciferase assays
Cells were electroporated with plasmids expressing firefly luciferase under the control of STAT5-responsive regulatory regions from the NCAM2 gene (NCAM-luc) or region B of the BCL6 gene (B-luc) (19, 20). Cells were also co-transfected with a plasmid expressing renilla luciferase under a constitutive promoter for normalization. Five hours after electroporation drugs were added, and dual luciferase assay was performed 24 hours after electroporation (Promega, Madison, WI).
In vitro kinase assay
Kinase inhibitory activity of JQ1 was analyzed using the SelectScreen Kinase Profiling service (Invitrogen).
Expression plasmids and shRNA
A constitutively activated mutant form of STAT5a, STAT5a1*6 (caSTAT5), was expressed in the green fluorescent protein (GFP)-expressing retroviral vector pMSCV-IRES-eGFP (pMIG). shRNA constructs targeting BET bromodomain proteins were obtained from the Broad Institute (Cambridge, MA), with the targeting sequences indicated (Supplemental table 3).
Drug-induced enrichment of leukemia cells
Leukemia cell lines were infected with the GFP-expressing retroviral vector pMIG or pMIG-caSTAT5. 24 hours after infection, imatinib, Jak inhibitor 1 or JQ1 were added, and 72 hours after drug treatment, cells were analyzed via flow cytometry to determine the percentage of GFP-positive cells.
Results
Bromodomain inhibitors block STAT5-dependent gene expression
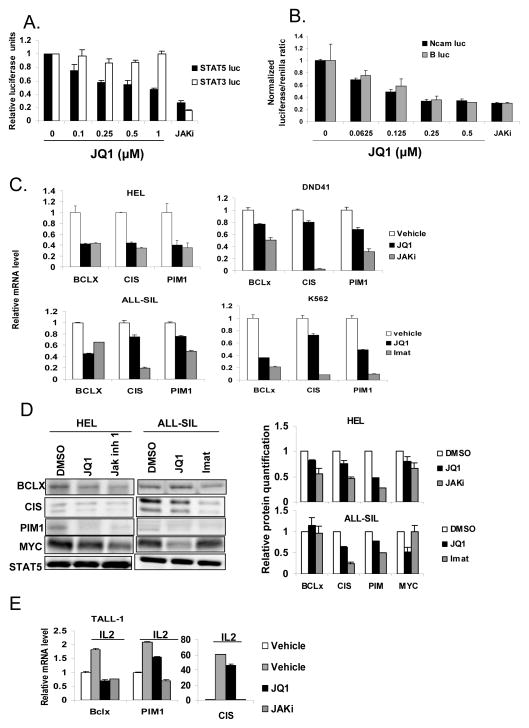
To determine whether BET bromodomain-containing proteins are critical to STAT5 transcriptional function, we first examined the effect of the bromodomain inhibitor JQ1 on reporter cell systems in which luciferase is regulated by individual transcription factors. JQ1 caused a dose-dependent inhibition of STAT5-dependent luciferase expression (Figure 1A). By contrast, JQ1 had no effect on transcription mediated by the highly related transcription factor STAT3. Interestingly, at maximally effective doses, JQ1 led to approximately a 60% decrease in reporter gene expression, suggesting that BET bromodomain proteins are not essential for basal STAT5 transcriptional activity, but may be required for maximal activity. By contrast, Jak inhibitor 1, which completely abolishes the activating STAT tyrosine phosphorylation, decreased STAT5-dependent transcription in this assay by approximately 80%.
Figure 1. JQ1 decreases STAT5 activity.
(A) Reporter cell lines were treated with the indicated doses of JQ1 or Jak inhibitor 1 (JAKi; 1 μM) for 1 hour, after which IL-6 or prolactin was added to activate STAT3 or STAT5, respectively. Six hours later, luciferase activity was quantitated by luminometry and normalized to cell viability. (B) HEL erythroleukemia cells were transfected with one of two luciferase constructs regulated by a STAT5-dependent promoter (NCAM-luc or B-luc). Cells were then treated with JQ1 at the indicated concentration, and normalized luciferase activity was determined. Jak inhibitor 1 (JAKi; 1 μM) was used as a positive control. (C) Expression of STAT5 target genes BCL-x, CIS, and PIM1 was measured by quantitative RT-PCR (normalized to 18S RNA) and presented relative to vehicle treatment control, in the indicated leukemia cell lines. Cells were treated for 6 hours with vehicle control, 0.5 μM JQ1 or 1 μM TKI (Jak inhibitor 1 for HEL and DND41, and imatinib for ALL-SIL and K562). (D) HEL erythroleukemia cells and ALL-SIL T acute lymphoblastic leukemia cells were treated with vehicle (DMSO), JQ1 (0.25 μM) or the indicated TKIs (Jak inhibitor 1 (0.5 μM) or Imatinib (0.0625 μM)) for 30 hours, after which cells were harvested and immunoblots were performed to the indicated proteins. Total STAT5 level was used as a loading control. Densitometric quantification of two separate experiments is shown on the right. (E) TALL-1 cells were pre-treated with vehicle, JQ1 (1 μM) or Jak inhibitor 1 (1 μM) for 1 hour, after which IL-2 (50 units/ml) was added to stimulate STAT5 activation. RNA was analyzed 90 minutes after IL-2 stimulation.
We next determined the effect of JQ1 in leukemia cell lines in which STAT5 is constitutively activated through a variety of mechanisms. We used two reporter constructs in which luciferase is regulated by distinct STAT5-dependent promoter sequences derived from the NCAM2 gene (NCAM-luc) or the BCL6 gene (B-luc). JQ1 treatment led to a dose-dependent reduction of STAT5-dependent luciferase activity mediated by both of these promoters in multiple lymphoid and myeloid leukemia cell types (Figure 1B and Supplemental Figure S1).
Constitutively activated STAT5 drives cancer pathogenesis by increasing expression of genes regulating cell cycle progression and promoting survival. Thus, we determined the effect of JQ1 on the expression levels of well-characterized endogenous STAT5 targets genes (Supplemental Figure S2), including BCL-x (21, 22), CIS (20), and PIM1 (23). JQ1 inhibited the expression of STAT5 target genes in leukemia cell lines with constitutively activated STAT5 driven by Jak2 (HEL and DND41) or Abl (ALL-SIL and K562) (Figure 1C). Protein expression of STAT5 target genes was also reduced by JQ1, as was the previously described target of JQ1, Myc (15) (Figure 1D). As these endogenous genes may also be regulated by other transcription factors, the response to JQ1 (and kinase inhibitors) was, as expected, more variable than that seen with the reporter systems. However, these results also suggest that JQ1 does not cause non-specific inhibition of transcription. Since autocrine or paracrine production of cytokines is an important mechanism of STAT5 activation, we next evaluated systems in which STAT5 phosphorylation is cytokine induced. JQ1 inhibited IL-2 induced STAT5 target gene expression in T lymphocytic leukemia cells (Figure 1E). Taken together, these data demonstrate that JQ1 inhibits STAT5-dependent transcriptional function, and this inhibition is independent of the mechanism driving STAT5 activation.
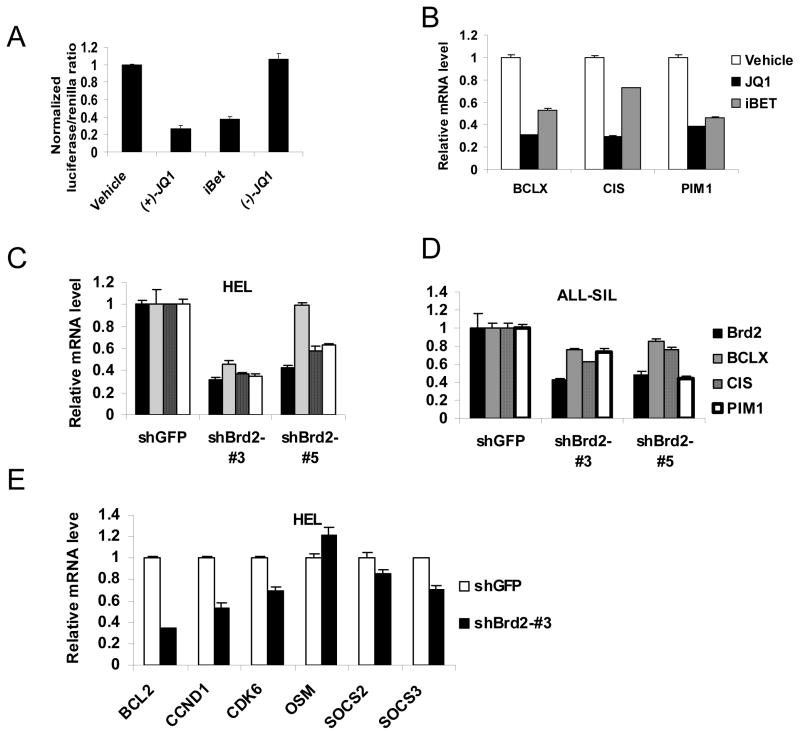
To further evaluate whether bromodomain inhibition blocks STAT5 transcriptional function, we tested whether a second BET bromodomain inhibitor I-BET, which is structurally distinct from JQ1, also inhibits STAT5 transcriptional activity. We also evaluated an inactive (−)-JQ1 enantiomer, which is structurally incapable of inhibiting BET bromodomains (15). We found that I-BET was as effective as JQ1 in inhibiting STAT5-dependent transcription using luciferase reporter cells (Figure 2A). As expected, the (−)-JQ1 enantiomer had no activity in this assay (Figure 2A). Furthermore, both JQ1 and I-BET reduced expression of endogenous STAT5 target genes in ALL cells (Figure 2B). These results indicate that structurally unrelated bromodomain inhibitors can inhibit STAT5 transcriptional function.
Figure 2. Inhibition of Brd2 reduces STAT5 transcriptional function.
(A) ALL-SIL cells were transfected with a STAT5-luciferase construct (NCAM-luc) and treated with vehicle, JQ1 (1 μM), iBET (1 μM) or the enantiomer (−)-JQ1 (1 μM), after which STAT5-dependent gene expression was determined by dual luciferase assay. (B) DU528 cells were treated with vehicle, JQ1 (1 μM) or iBET (1 μM) for 6 hours, after which expression of the indicated STAT5 target genes was determined by quantitative RT-PCR. HEL cells (C, E) or ALL-SIL cells (D) were treated with the indicated shRNA constructs targeting BRD2, after which expression of the indicated genes was determined by quantitative RT-PCR.
JQ1 inhibits STAT5 function by blocking BRD2
We next focused on determining which BET bromodomain proteins are necessary for STAT5 transcriptional function. In particular, we examined BRD2, BRD3 and BRD4, as BRDT is only expressed in testis and ovary. To do this, we used lentiviral vector-mediated shRNAs to knock-down each individual BET protein in leukemia cells, and determined the effect on expression of STAT5 target genes. The efficacy and specificity of shRNAs targeting BRD2, BRD3 and BRD4 was validated by RT-PCR analysis (Supplemental figure S3). Despite 80% knock-down of BRD3 or BRD4 by shRNAs, no reproducible decrease was seen in the expression of the STAT5 target genes Bcl-x, CIS and PIM1 (Supplemental figure S4). By contrast, knock-down of BRD2 led to a prominent reduction in expression of STAT5 target genes in multiple leukemia cell lines (Figure 2C, D, E). These experiments confirm a specific association between depletion of BRD2 and reduction of STAT5 target gene expression in all cell types examined, further supporting the hypothesis that reduction of STAT5 transcriptional function by JQ1 is mediated through inhibition of BRD2.
Since BRD2 is associated with transcription factor-dependent gene expression in the setting of hyperacetylated chromatin (24), we considered whether BRD2 regulates transcription of Bcl-x, CIS, and PIM1 independent of STAT5. To examine this hypothesis, we knocked down BRD2 in the T lymphoblastic leukemia cell lines TALL-1, which lacks constitutive STAT5 activation. Knock-down of BRD2 did not decrease expression of BCL-x, CIS, or PIM1 in these cell lines, in contrast to what was seen in cells having constitutively activated STAT5 (Supplemental figure S5). These results provide further evidence that down-regulation of BCL-x, CIS, and PIM1 expression after BRD2 depletion is due to inhibition of STAT5 transcriptional function.
BRD2 inhibition by JQ1 can inhibit STAT5 function without affecting STAT5 phosphorylation
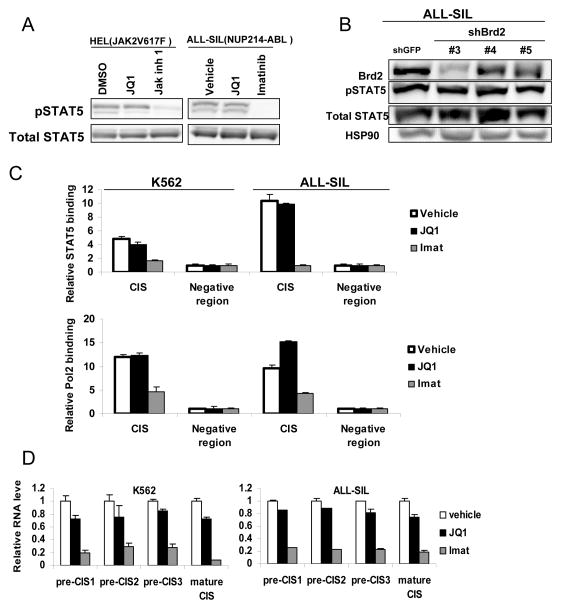
Although JQ1 has been found to be able to decrease STAT5 phosphorylation in a subset of acute B lymphocytic leukemia cells through down-regulation of the IL-7 receptor (IL7R) (25), we found JQ1 can inhibit STAT5 transcription in a wide variety of cell types in which IL7R is not involved in STAT5 activation, including breast cancer cells (Figure 1A). Therefore, we considered alternative hypotheses for how BRD2 inhibition by JQ1 reduces STAT5 function. First, we evaluated the effect of this drug on STAT5 protein expression and its activating tyrosine phosphorylation in systems not dependent on the IL7R. In both HEL myeloid leukemia cells (in which STAT5 is activated by the constitutively activated kinase JAK2V617F) and ALL-SIL T-ALL cells (in which STAT5 is activated by the fusion tyrosine kinase NUP214-ABL), JQ1 had no effect on STAT5 protein levels or tyrosine phosphorylation (Figure 3A). By contrast, the TKIs Jak inhibitor 1 and imatinib completely abrogated STAT5 phosphorylation, respectively. Similarly, knocking-down BRD2 in ALL-SIL with multiple distinct constructs did not influence STAT5 expression or phosphorylation (Figure 3B).
Figure 3. Brd2 inhibition by JQ1 or shRNA knock-down does not affect STAT5 phosphorylation or DNA binding.
(A) HEL erythroleukemia cells and ALL-SIL T acute lymphoblastic leukemia cells were treated with vehicle (DMSO), JQ1 (0.5 μM) or the indicated TKIs (1 μM) for 6 hours, after which cells were harvested and immunoblots were performed to STAT5 and phosphorylated STAT5. (B) ALL-SIL cells were infected with one of three shRNA constructs targeting BRD2 or GFP (as a control). Three days after puromycin selection, cells were harvested and immunoblots were performed with the indicated antibodies. (C) Cells were treated with vehicle, JQ1 (0.5 μM) or imatinib (1 μM) for 6 hours, after which cells were harvested and ChIP was performed. STAT5 and RNA polymerase II binding were analyzed relative to a known STAT5-negative binding region. (D) Comparison of CIS transcripts close to the transcriptional start site (designated as pre-CIS) and the mature form of CIS RNA level by quantitative RT-PCR (normalized to 18S RNA)
We next considered the possibility that JQ1 is inhibiting the interaction of STAT5 with its cognate genomic regulatory sequences. To examine this, we performed chromatin immunoprecipitation (ChIP) to evaluate if JQ1 inhibits STAT5 binding to the well-characterized regulatory sites for CIS. Despite reduction of CIS expression after 6 hours of treatment with JQ1, JQ1 did not significantly reduce STAT5 binding to its genomic binding site (Figure 3C). By contrast, the TKI imatinib, which inhibited STAT5 phosphorylation, significantly reduced DNA binding by STAT5. Given that STAT5 could still bind to the CIS promoter in the presence of JQ1, we evaluated whether the binding of RNA polymerase II was affected by this treatment. Kinase inhibitors, which blocked STAT5 phosphorylation and DNA binding, inhibited the recruitment of RNA polymerase II to the CIS promoter (Figure 3C). By contrast, JQ1 had little effect on RNA polymerase recruitment to this site. Thus, we investigated whether JQ1 reduces transcriptional initiation or elongation by measuring the abundance of sequences close to the transcriptional start site and comparing them with the mature form of CIS RNA. We found equivalent decreases in sequences located close to the transcriptional start sites as with mature RNA transcripts of CIS (Figure 3D), suggesting that whereas transcriptional initiation was inhibited, elongation appeared to be unaffected. Taken together, these results show that JQ1 can decrease STAT5 transcriptional function without affecting STAT5 phosphorylation, DNA binding, or recruitment of RNA polymerase II, and support the notion that JQ1 likely reduces STAT5 transcriptional initiation by targeting essential co-factors for STAT5, likely BRD2.
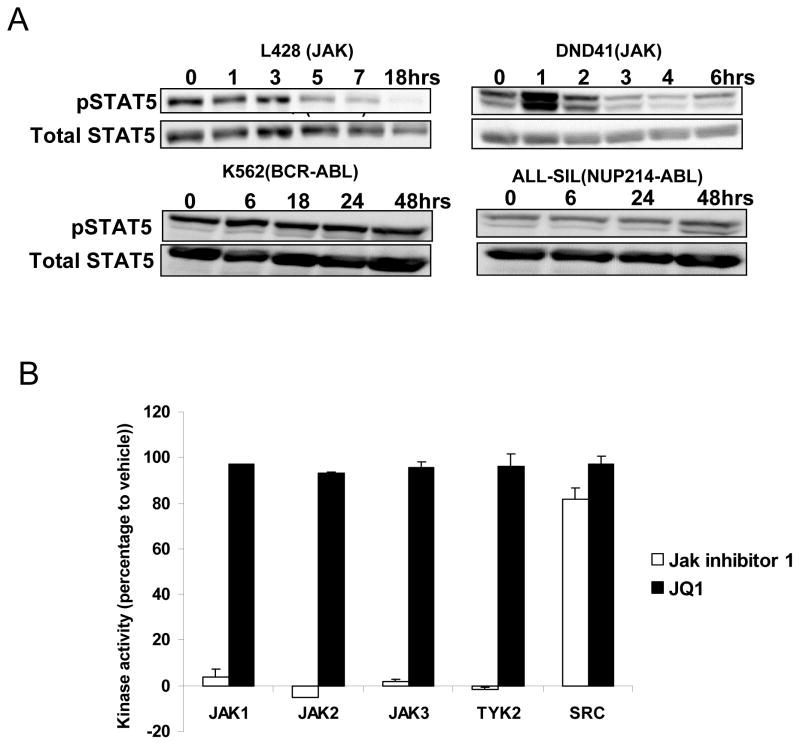
Although JQ1 had no effect on the phosphorylation of STAT5 in most cell lines examined, we did observe that JQ1 decreased STAT5 phosphorylation in lymphoid malignancies such as Hodgkin lymphoma and acute T lymphoblastic leukemia, in which STAT5 activation is mediated by signaling through Jak kinases (Figure 4A). This is consistent with what has been observed in a subset of B-ALL in which STAT5 activation is Jak-dependent (25). To exclude the possibility that JQ1 was acting directly as a kinase inhibitor in this setting, we tested it for activity against a range of kinases using in vitro kinase assays. At a concentration of 500 nM, JQ1 had no effect on the kinase activity of JAK1, JAK2, JAK3, or TYK2, or the unrelated tyrosine kinase SRC (Figure 4B).
Figure 4. JQ1 is not a direct JAK kinase inhibitor.
(A) The indicated cell line was treated with JQ1 (0.5 μM) and harvested at the indicated time points, after which immunoblots were performed for phosphorylated and total STAT5. (B) The effect of JQ1 (0.5 μM) on the activity of selected tyrosine kinases was analyzed using SelectScreen Kinase Profiling (Invitrogen). Jak inhibitor 1 (0.5 μM) was used as positive control for inhibition of Jak family kinases.
BRD2 inhibition strongly synergizes with TKIs to decrease leukemia cell survival
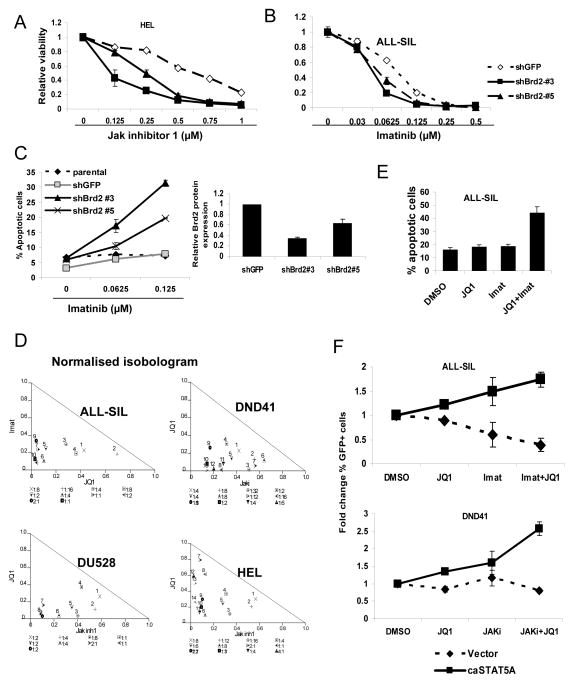
Leukemogenesis driven by a variety of oncogenic tyrosine kinases is dependent on STAT5 activation. Since BRD2 is essential for maximal STAT5 function, we next considered whether inhibition of BRD2 has an effect on leukemia cell survival. Reduction of BRD2 by itself does not significantly decrease the viability of leukemia cells (data not shown). However, when HEL cells, which are driven by a mutated form of the kinase Jak2, are co-treated with BRD2 knockdown and Jak inhibitor 1, there is a dramatic cooperative effect, with a decrease in the IC50 concentration of 50 to 75% (Figure 5A). Similarly, reduction of BRD2 in ALL-SIL cells, which express NUP-214-ABL, reduces the IC50 of the kinase inhibitor imatinib by 50% (Figure 5B). Consistent with this finding, induction of apoptosis of leukemia cells by kinase inhibitors is enhanced by knockdown of BRD2 proportional to the decrease in BRD2 expression (Figure 5C). These results suggest that dual inhibition of STAT5 function by blocking phosphorylation and inhibiting an important transcriptional co-regulator leads to maximal inhibition of STAT5 function and reduction of leukemia cell survival.
Figure 5. Inhibition of Brd2 increases sensitivity of leukemia cells to TKIs.
(A) HEL and (B) ALL-SIL cells were infected with lentiviral vectors containing distinct shRNAs targeting BRD2. After 4 days of selection in puromycin, cells were treated with TKIs as indicated. Cell viability was measured 72 hours after drug treatment. (C) Parental ALL-SIL cells or cells with Brd2 knock-down were treated with imatinib for 48 hours, after which apoptosis was quantitated by annexin V and PI staining followed by flow cytometry. (D) The indicated leukemia cell lines were treated for 72 hours with JQ1 or the indicated TKI, alone or in combination. Isobologram analysis was performed based on change in viable cell number after treatment. Data points below the line indicate synergy between the two agents. (E) ALL-SIL cells were treated with JQ1 (0.5 μM), imatinib (0.125 μM), or both for 48 hours, after which apoptosis was quantified by annexin V staining and flow cytometry. (F) ALL-SIL or DND41 T lymphocytic leukemia cells were infected with a vector expressing either GFP alone or GFP and constitutively activated STAT5a (caSTAT5a). Cells were then treated with JQ1 (0.25 μM), Jak inhibitor 1 (1 μM), or imatinib (0.125 μM), or the indicated combinations for 72 hours, and the fraction of GFP positive cells was quantified by flow cytometry and normalized to the vehicle (DMSO) control.
Having shown that a reduction of BRD2 decreases STAT5 transcriptional function in several leukemia cell types and that there is a BRD2-dependent mechanism of STAT5 inhibition by JQ1, we next explored the therapeutic implications of JQ1 in leukemia, particularly in aggressive T-ALL in which STAT5 is driven by constitutively activated tyrosine kinases. We found JQ1 used as a single agent only modestly decreased the viability of T-ALL cell lines with constitutive STAT5 activation, and required relatively high concentrations (Supplemental Figure S6A). This low activity occurred despite a prominent decrease in Myc expression (Supplemental Figure S6B), as has been reported for JQ1. This suggested that reduction in Myc expression alone is not sufficient for the therapeutic effect of JQ1 in T-ALL cells. By contrast, we found strong consistent synergy between JQ1 and TKIs, including imatinib and Jak inhibitor 1, in reducing leukemia cell viability across a broad range of doses in multiple cell lines (Figure 5D). Consistent with this finding, combination treatment of JQ1 and TKIs led to a synergistic induction of apoptosis (Figure 5E).
To determine whether STAT5 was the key target in leukemia cells treated with TKIs and JQ1, we examined the effect of a constitutively activated form of STAT5, STAT5a1*6. ALL-SIL or DND41 cells were transfected with a construct expressing GFP alone or with STAT5a1*6. Cells were then treated with vehicle, JQ1 and/or a TKI, and the relative proportion of GFP expressing cells was determined by flow cytometry. In the presence of constitutively activated STAT5, treatment with TKIs (imatinib for ALL-SIL or Jak inhibitor 1 for DND41) or JQ1 alone led to a slight enrichment of GFP positive cells. However, treatment with TKIs and JQ1 in combination led to a significant increase in the proportion of GFP+ cells (Figure 5F), suggesting that constitutively activated STAT5 rescues these leukemia cells from cell death induced by TKIs and JQ1. These results indicate that the effects of JQ1 and TKIs are, at least partially, mediated through STAT5 inhibition, supporting the importance of this transcription factor as a therapeutic target.
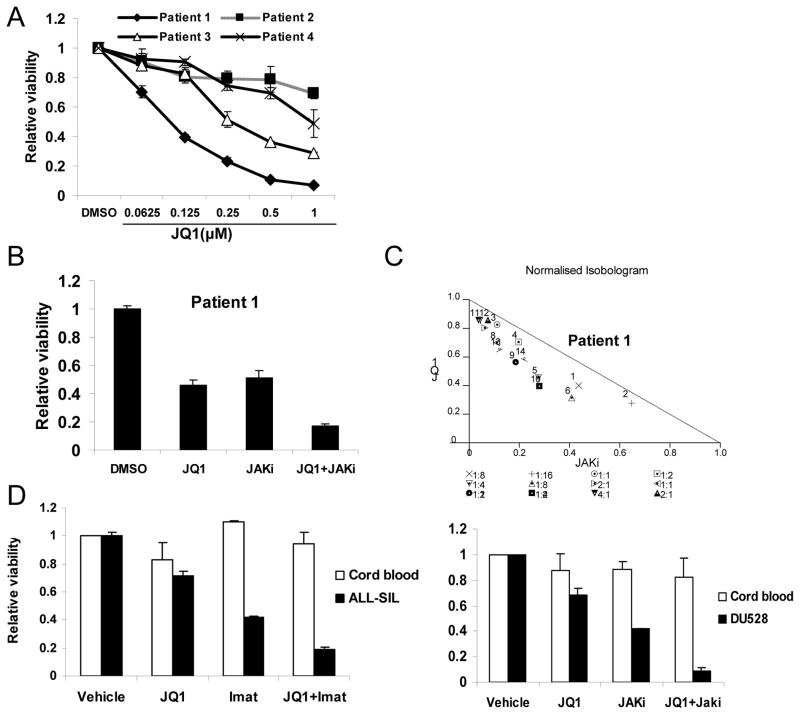
Finally, we wished to determine whether primary T-cell acute lymphoblastic leukemia (T-ALL) cells were also sensitive to JQ1 and TKIs. We obtained bone marrow mononuclear cells from four patients with untreated T-ALL in which greater than 90% of the cells were lymphoblasts. As the number of cells available was insufficient to assess STAT5 phosphorylation, we used sensitivity to kinase inhibitors as a surrogate marker for STAT5 activation. Of the four, none was sensitive to imatinib and one (Patient 1) was sensitive to JAK inhibition (data not shown). This is consistent with reports that the NUP214–ABL1 translocation is a rare event in T-ALL (26). We found that these primary T-ALL cells showed variable sensitivity to JQ1 treatment, though again cells from Patient 1 showed the greatest sensitivity to JQ1 (Figure 6A). Consistent with our observations in T-ALL cell lines, we observed synergistic reduction of viability of leukemia cells from patient 1 when JQ1 and JAK inhibitor I were combined (Figure 6B and C).
Figure 6. The combination of JQ1 and TKIs shows toxicity to primary T-ALL cells but not normal hematopoietic cells.
(A) Bone marrow mononuclear cells harvested from four untreated T-ALL patients were treated with JQ1 at the indicated concentrations for 72 hours, at which point viable cell number was determined by ATP-dependent bioluminescence. (B) Bone marrow mononuclear cells harvested from patient 1 with TALL patients were treated with JQ1 (0.125μM), JAK inhibitor 1 (1 μM), or the combination for 72 hours, after which viable cell number was quantitated by ATP-dependent bioluminescence. (C) Isobologram analysis was performed based on loss of cell viability for 14 different combinations of JQ1 and JAKi. Data points below the line indicate synergistic effects. (D) Mononuclear cells from umbilical cord blood (n=4), or ALL-SIL or DU528 T lymphocytic leukemia cells were treated for 72 hours with JQ1 (0.25 μM) or TKIs (imatinib 0.125 μM or JAKi 0.5 μM), alone or in combination, and viable cell number was quantitated by ATP-dependent bioluminescence.
To exclude the possibility that the combination of JQ1 with a kinase inhibitor is non-specifically cytotoxic to immature hematopoietic cells, we obtained mononuclear cells from umbilical cord blood, which is highly enriched in progenitor cells. In contrast to what is seen in leukemia cells, JQ1 and TKIs, either alone or in combination, had minimal effects on the viability of normal hematopoietic cells (Figure 6D and Supplemental figure S7). Together, these results suggest that the use of JQ1 combined with TKIs in ALL patients would not be associated with toxicity to normal hematopoietic cells.
Discussion
Constitutive activation of transcription factors of the STAT family can directly lead to cancer pathogenesis (27). In hematological malignancies in particular, inappropriate activation of STAT5 is a common event that leads to increased expression of genes regulating cell cycle progression and survival (1) (12, 28). In addition, constitutive STAT5 activation has been implicated in leukemia stem cell self-renewal (29–31). Since STAT5 is downstream of many kinases that are activated aberrantly, targeting STAT5 is an attractive approach for the therapy of leukemias. However, STAT5, like other transcription factors, is structurally more difficult to inhibit directly by small molecules. Since STATs recruit co-factors to activate transcription, an alternate approach is to target the interaction with these proteins. In particular, we considered the possibility that BET bromodomain proteins were key co-factors for STAT5 in leukemias. In fact, we found that the BET bromodomain inhibitor JQ1 inhibits STAT5-dependent expression of reporter genes as well as endogenous target genes. RNA interference-based experiments subsequently demonstrated that among the three BET bromodomain proteins expressed in these hematological malignancies and targeted by JQ1, only BRD2 is necessary for STAT5 transcriptional function.
In addition to STAT5, STAT3 is constitutively activated in a wide range of hematological and non-hematological cancers. However, although JQ1 significantly reduces the transcriptional function of STAT5, it had essentially no effects on STAT3-dependent gene expression. This lack of inhibition of STAT3 function is consistent with recently published data in which global transcriptional profiling and unbiased gene set enrichment analysis (GSEA) showed no inhibition of a STAT3 target gene signature by JQ1 in multiple myeloma (32). Given the structural similarity between STAT5 and STAT3, further genomic and structural studies are necessary to elucidate the mechanism of this selectivity.
Although JQ1 can inhibit STAT5 phosphorylation in a subset of B-ALL through downregulation of the IL7R, we found JQ1 treatment or BRD2 knock-down reduces STAT5 function through phosphorylation-independent mechanisms. JQ1 decreases STAT5 transcriptional function in a wide variety of cell types, regardless of the kinase driving STAT5 activation. Furthermore, chromatin immunoprecipitation (ChIP) analysis showed that JQ1 did not significantly reduce STAT5 binding to its genomic binding site. These results support the notion that JQ1 decreases STAT5 function through targeting a transcriptional co-activator rather than STAT5 itself. Consistent with this hypothesis, knocking-down BRD2 (but not BRD3 or BRD4) reduces STAT5 target gene expression without affecting STAT5 phosphorylation.
As a member of the BET family, BRD2 functions as a nuclear transcriptional regulator. BRD2 can mediate recruitment of E2 promoter binding factors (E2Fs) and histone acetyltransferase to regulate gene expression (24, 33). BRD2 is also associated preferentially in vivo with hyperacetylated chromatin along the entire length of transcribed genes (24). To eliminate the possibility that Brd2 may regulate STAT5 target genes independent of STAT5, we also knocked down Brd2 in leukemia cells that lacked STAT5 activation. We found that Brd2 inhibition did not reduce expression of these genes, indicating that they only require BRD2 in the context of STAT5-dependent expression. BRD2 is known to participate in multi-protein transcriptional complexes (34). Our data provide evidence that BRD2 likely participates in the STAT5 transcriptional complex, and acts as a critical co-activator for STAT5 function. The recruitment of STAT5 to its genomic binding sites is not dependent on BRD2, but rather maximal transcriptional initiation of these target genes requires BRD2. Genome-wide approaches will be useful to dissect the functional association between BRD2 and STAT5 in the transcriptional regulation of target genes.
Several lines of evidence suggest an important role for BRD2 in the pathogenesis of lymphocytic malignancies. Transgenic mice with constitutive expression of BRD2 in the lymphoid compartment develop B-cell lymphoma and leukemia (35). In addition, high levels of BRD2 phosphorylation have been observed in human lymphocytic leukemic cells compared to normal lymphocytes (36). Interestingly, both a wild-type BRD2 transgene and a pseudo-kinase-null point mutant lead to lymphomagenesis, indicating that it is the transcriptional regulation rather than cryptic kinase activity of BRD2 that is the basis for BRD2-driven neoplasia. Given our finding of the importance of BRD2 in STAT5 transcriptional function, we specifically explored the biological significance of BRD2 inhibition in lymphocytic leukemias with constitutively activated STAT5. We found that reduction of BRD2 by RNA interference has only modest effects on T lymphoblastic leukemia cell viability. This may be due to the fact that BRD2 is only necessary for maximal STAT5 transcriptional activity, and its inhibition does not completely abolish STAT5 function. Nonetheless, this reduction in STAT5 function is sufficient to render leukemia cells more sensitive to TKI-induced apoptosis. These results are also consistent with synergy seen between TKIs that inhibit STAT5 phosphorylation and drugs such as pimozide that target STAT5 independently (11, 37).
To explore the therapeutic implication of targeting STAT5 by dual BET bromodomain and tyrosine kinase inhibition, we focused on ALL, a clinically aggressive disease. Around 10% of ALL is characterized by tyrosine kinases activated through point mutation, or oncogenic fusions that drive STAT5 activation, including Bcr-Abl in B-ALL and NUP-214-ABL in T-ALL (26). However, unlike CML, targeting these tyrosine kinases in ALL confers very limited benefit, likely due to the co-activation of other pathways. In acute myeloid leukemia (AML), JQ1 can decrease Myc expression by blocking BRD4, leading to a significant reduction in cell viability (38). By contrast, we found that JQ1 used as a single agent only modestly reduces the viability of T-ALL cells with STAT5 activation, despite a reduction of MYC expression. This suggests that inhibition of Myc is not sufficient for a therapeutic response in this disease. However, there is strong synergy in inducing apoptosis in T-ALL cells when JQ1 is combined with TKIs. This synergistic effect is, at least partially, mediated through STAT5 inhibition, as over-expression of a constitutively activated STAT5 can rescue cell death induced by the combination of JQ1 and TKIs. These findings also reaffirm the important role of STAT5 activation in the pathogenesis of T-ALL.
In conclusion, we have found that STAT5 is another crucial target of JQ1, apart from the well known target Myc. Inhibition of STAT5 function by JQ1 was mediated by blockage of Brd2 and BRD2 may act as an important co-factor in STAT5 transcriptional function. Pharmacological or genetic inhibition of Brd2 decreases STAT5-transcriptional function and in combination with TKIs, significantly reduces leukemic cell survival. These findings suggest rational therapeutic strategies that may be particularly effective in aggressive forms of ALL where oncogenic kinases drive STAT5 activation and promote leukemia cell survival.
Supplementary Material
Acknowledgments
Financial support: This work was supported by NIH grant R01-CA160979, the Lymphoma Research Foundation, Gabrielle’s Angel Foundation, the Brent Leahey Fund, the Kittredge Foundation, and the Joan Harris Cancer Foundation (to D.A. Frank), and the Leukemia & Lymphoma Society and the American Society of Hematology (to J.E. Bradner).
Footnotes
Conflict of Interest: Derivatives of JQ1 have been licensed to Tensha Therapeutics by the Dana-Farber Cancer Institute, which consequently holds equity in Tensha (JQ and JEB).
References
- 1.Lin TS, Mahajan S, Frank DA. STAT signaling in the pathogenesis and treatment of leukemias. Oncogene. 2000;19:2496–504. doi: 10.1038/sj.onc.1203486. [DOI] [PubMed] [Google Scholar]
- 2.Sternberg DW, Gilliland DG. The role of signal transducer and activator of transcription factors in leukemogenesis. Journal of Clinical Oncology. 2004;22:361–71. doi: 10.1200/JCO.2004.10.124. [DOI] [PubMed] [Google Scholar]
- 3.Scheeren FA, Diehl SA, Smit LA, Beaumont T, Naspetti M, Bende RJ, et al. IL-21 is expressed in Hodgkin lymphoma and activates STAT5: evidence that activated STAT5 is required for Hodgkin lymphomagenesis. Blood. 2008;111:4706–15. doi: 10.1182/blood-2007-08-105643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoelbl A, Kovacic B, Kerenyi MA, Simma O, Warsch W, Cui Y, et al. Clarifying the role of Stat5 in lymphoid development and Abelson-induced transformation. Blood. 2006;107:4898–906. doi: 10.1182/blood-2005-09-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walz C, Ahmed W, Lazarides K, Betancur M, Patel N, Hennighausen L, et al. Essential role for Stat5a/b in myeloproliferative neoplasms induced by BCR-ABL1 and Jak2V617F in mice. Blood. 2012;119:3550–60. doi: 10.1182/blood-2011-12-397554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warsch W, Kollmann K, Eckelhart E, Fajmann S, Cerny-Reiterer S, Holbl A, et al. High STAT5 levels mediate imatinib resistance and indicate disease progression in chronic myeloid leukemia. Blood. 2011;117:3409–20. doi: 10.1182/blood-2009-10-248211. [DOI] [PubMed] [Google Scholar]
- 7.Duy C, Hurtz C, Shojaee S, Cerchietti L, Geng H, Swaminathan S, et al. BCL6 enables Ph+ acute lymphoblastic leukaemia cells to survive BCR-ABL1 kinase inhibition. Nature. 2011;473:384–8. doi: 10.1038/nature09883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soverini S, Gnani A, Colarossi S, Castagnetti F, Abruzzese E, Paolini S, et al. Philadelphia-positive patients who already harbor imatinib-resistant Bcr-Abl kinase domain mutations have a higher likelihood of developing additional mutations associated with resistance to second- or third-line tyrosine kinase inhibitors. Blood. 2009;114:2168–71. doi: 10.1182/blood-2009-01-197186. [DOI] [PubMed] [Google Scholar]
- 9.Iacobucci I, Lonetti A, Messa F, Cilloni D, Arruga F, Ottaviani E, et al. Expression of spliced oncogenic Ikaros isoforms in Philadelphia-positive acute lymphoblastic leukemia patients treated with tyrosine kinase inhibitors: implications for a new mechanism of resistance. Blood. 2008;112:3847–55. doi: 10.1182/blood-2007-09-112631. [DOI] [PubMed] [Google Scholar]
- 10.Hantschel O, Warsch W, Eckelhart E, Kaupe I, Grebien F, Wagner KU, et al. BCR-ABL uncouples canonical JAK2-STAT5 signaling in chronic myeloid leukemia. Nature Chemical Biology. 2012;8:285–93. doi: 10.1038/nchembio.775. [DOI] [PubMed] [Google Scholar]
- 11.Nelson EA, Walker SR, Weisberg E, Bar-Natan M, Barrett R, Gashin LB, et al. The STAT5 inhibitor pimozide decreases survival of chronic myelogenous leukemia cells resistant to kinase inhibitors. Blood. 2011;117:3421–9. doi: 10.1182/blood-2009-11-255232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bar-Natan M, Nelson EA, Walker SR, Kuang Y, Distel RJ, Frank DA. Dual inhibition of Jak2 and STAT5 enhances killing of myeloproliferative neoplasia cells. Leukemia. 2012;26:1407–10. doi: 10.1038/leu.2011.338. [DOI] [PubMed] [Google Scholar]
- 13.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–33. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dawson MA, Kouzarides T, Huntly BJ. Targeting epigenetic readers in cancer. New England Journal of Medicine. 2012;367:647–57. doi: 10.1056/NEJMra1112635. [DOI] [PubMed] [Google Scholar]
- 15.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–73. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson EA, Walker SR, Kepich A, Gashin LB, Hideshima T, Ikeda H, et al. Nifuroxazide inhibits survival of multiple myeloma cells by directly inhibiting STAT3. Blood. 2008;112:5095–102. doi: 10.1182/blood-2007-12-129718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–23. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelson EA, Walker SR, Alvarez JV, Frank DA. Isolation of unique STAT5 targets by chromatin immunoprecipitation-based gene identification. J Biol Chem. 2004;279:54724–30. doi: 10.1074/jbc.M408464200. [DOI] [PubMed] [Google Scholar]
- 19.Nelson EA, Walker SR, Li W, Liu XS, Frank DA. Identification of human STAT5-dependent gene regulatory elements based on interspecies homology. J Biol Chem. 2006;281:26216–24. doi: 10.1074/jbc.M605001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker SR, Nelson EA, Frank DA. STAT5 represses BCL6 expression by binding to a regulatory region frequently mutated in lymphomas. Oncogene. 2007;26:224–33. doi: 10.1038/sj.onc.1209775. [DOI] [PubMed] [Google Scholar]
- 21.Dumon S, Santos SC, Debierre-Grockiego F, Gouilleux-Gruart V, Cocault L, Boucheron C, et al. IL-3 dependent regulation of Bcl-xL gene expression by STAT5 in a bone marrow derived cell line. Oncogene. 1999;18:4191–9. doi: 10.1038/sj.onc.1202796. [DOI] [PubMed] [Google Scholar]
- 22.Gesbert F, Griffin JD. Bcr/Abl activates transcription of the Bcl-X gene through STAT5. Blood. 2000;96:2269–76. [PubMed] [Google Scholar]
- 23.Kim KT, Baird K, Ahn JY, Meltzer P, Lilly M, Levis M, et al. Pim-1 is up-regulated by constitutively activated FLT3 and plays a role in FLT3-mediated cell survival. Blood. 2005;105:1759–67. doi: 10.1182/blood-2004-05-2006. [DOI] [PubMed] [Google Scholar]
- 24.LeRoy G, Rickards B, Flint SJ. The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Mol Cell. 2008;30:51–60. doi: 10.1016/j.molcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ott CJ, Kopp N, Bird L, Paranal RM, Qi J, Bowman T, et al. BET bromodomain inhibition targets both c-MYC and IL7R in high-risk acute lymphoblastic leukemia. Blood. 2012;120:2843–52. doi: 10.1182/blood-2012-02-413021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quintas-Cardama A, Tong W, Manshouri T, Vega F, Lennon PA, Cools J, et al. Activity of tyrosine kinase inhibitors against human NUP214-ABL1-positive T cell malignancies. Leukemia. 2008;22:1117–24. doi: 10.1038/leu.2008.80. [DOI] [PubMed] [Google Scholar]
- 27.Frank DA. STAT signaling in cancer: Insights into pathogenesis and treatment strategies. Cancer Treat Res. 2003;115:267–91. doi: 10.1007/0-306-48158-8_11. [DOI] [PubMed] [Google Scholar]
- 28.Benekli M, Baumann H, Wetzler M. Targeting signal transducer and activator of transcription signaling pathway in leukemias. J Clin Oncol. 2009;27:4422–32. doi: 10.1200/JCO.2008.21.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heuser M, Sly LM, Argiropoulos B, Kuchenbauer F, Lai C, Weng A, et al. Modeling the functional heterogeneity of leukemia stem cells: role of STAT5 in leukemia stem cell self-renewal. Blood. 2009;114:3983–93. doi: 10.1182/blood-2009-06-227603. [DOI] [PubMed] [Google Scholar]
- 30.Yoshimoto G, Miyamoto T, Jabbarzadeh-Tabrizi S, Iino T, Rocnik JL, Kikushige Y, et al. FLT3-ITD up-regulates MCL-1 to promote survival of stem cells in acute myeloid leukemia via FLT3-ITD-specific STAT5 activation. Blood. 2009;114:5034–43. doi: 10.1182/blood-2008-12-196055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fatrai S, Wierenga AT, Daenen SM, Vellenga E, Schuringa JJ. Identification of HIF2alpha as an important STAT5 target gene in human hematopoietic stem cells. Blood. 2011;117:3320–30. doi: 10.1182/blood-2010-08-303669. [DOI] [PubMed] [Google Scholar]
- 32.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Denis GV, Vaziri C, Guo N, Faller DV. RING3 kinase transactivates promoters of cell cycle regulatory genes through E2F. Cell Growth & Differentiation. 2000;11:417–24. [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang YW, Veschambre P, Erdjument-Bromage H, Tempst P, Conaway JW, Conaway RC, et al. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:8538–43. doi: 10.1073/pnas.95.15.8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greenwald RJ, Tumang JR, Sinha A, Currier N, Cardiff RD, Rothstein TL, et al. E mu-BRD2 transgenic mice develop B-cell lymphoma and leukemia. Blood. 2004;103:1475–84. doi: 10.1182/blood-2003-06-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Denis GV, Green MR. A novel, mitogen-activated nuclear kinase is related to a Drosophila developmental regulator. Genes & Development. 1996;10:261–71. doi: 10.1101/gad.10.3.261. [DOI] [PubMed] [Google Scholar]
- 37.Nelson EA, Walker SR, Xiang M, Weisberg E, Bar-Natan M, Barrett R, et al. The STAT5 Inhibitor Pimozide Displays Efficacy in Models of Acute Myelogenous Leukemia Driven by FLT3 Mutations. Genes & Cancer. 2012;3:503–11. doi: 10.1177/1947601912466555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–8. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.