Abstract
Puromycin is an aminonucleoside antibiotic with structural similarity to aminoacyl tRNA. This structure allows the drug to bind the ribosomal A-site and incorporate into nascent polypeptides causing chain termination, ribosomal subunit dissociation, and widespread translational arrest at high concentrations. In contrast, at sufficiently low concentrations, puromycin incorporates primarily at the C-terminus of proteins. While a number of techniques utilize puromycin incorporation as a tool for probing translational activity in vivo, these methods cannot be applied in yeasts that are insensitive to puromycin. Here, we describe a mutant strain of the yeast Saccharomyces cerevisiae that is sensitive to puromycin and characterize the cellular response to the drug. Puromycin inhibits the growth of yeast cells mutant for erg6Δ, pdr1Δ, and pdr3Δ (EPP) on both solid and liquid media. Puromycin also induces the aggregation of the cytoplasmic processing body component Edc3 in the mutant strain. We establish that puromycin is rapidly incorporated into yeast proteins and test the effects of puromycin on translation in vivo. This work establishes the EPP strain as a valuable tool for implementing puromycin-based assays in yeast, which will enable new avenues of inquiry into protein production and maturation.
Keywords: puromycin, translation, p-bodies, ERG6, PDR1, PDR3
Introduction
Recent work in a variety of organisms has underscored the importance of post-transcriptional regulation in many biological processes [McCarthy, 1998]; [Kim & Kyung Lee, 2012]; [Anderson, 2010]; [Freeman & Espinosa, 2013]; [Nousch & Eckmann, 2013]. The ability to identify and characterize the subset of transcripts actively translated at a given time is central to understanding post-transcriptional regulation. Techniques such as ribosome footprinting [Ingolia et al., 2009], polyribosome profiling [Pospisek & Valasek, 2013], and ribosome affinity purification [Halbeisen et al., 2009] have been used to differentiate translating from non-translating RNAs and gain unprecedented insight into translational control. While robust and powerful, a common limitation to these approaches is their focus on ribosome-associated mRNA without direct measurement of the protein products of translation. In contrast, approaches that label translational products directly by feeding cells isotopically labeled or chemically derivatized amino acids [Ong et al., 2002]; [Hinz et al., 2012] require special media conditions, expensive or radioactive amino acids, and labeling times that preclude their application to the study of global translational dynamics. For example, while posttranscriptional events such as p-body induction or tRNA sequestration to the nucleus can occur within minutes [Brengues et al., 2005]; [Whitney et al., 2007], intrinsic metabolic labeling of proteins in vivo (e.g. stable isotopic labeling by amino acids in cell culture, SILAC) can take several hours to be detected following a pulse [Schwanhausser et al., 2009], presumably due to the time necessary for the isotopic amino acid to be taken up by the cell and incorporated into aminoacylated tRNA pools.
The aminonucleoside puromycin has been used as a tool for studying translation in vitro for many years [Hansen et al., 1994]. Puromycin has characteristics of both amino acids and nucleic acids and is structurally similar to aminoacylated tRNA. This structure allows puromycin to bind the A-site of actively translating ribosomes [Schmeing et al., 2002], where it becomes a substrate for ribosomal peptidyl transferase activity and is incorporated into nascent polypeptide chains. Puromycin incorporation is a chain terminating event, leading to ribosomal subunit dissociation and, at high levels, general translational arrest [Eulalio et al., 2007]. Global translational arrest is the mechanism by which puromycin inhibits cell growth, however at sublethal concentrations the drug is preferentially incorporated at the C-terminus of nascent proteins [Miyamoto-Sato et al., 2000]. This property makes puromycin useful for the study of translation and protein dynamics, both in vitro and in vivo. Recently several techniques have exploited this property of puromycin including mRNA display [Tabuchi, 2003]; [Roberts & Szostak, 1997], the generation of fluorescently labeled proteins for protein interaction arrays [Starck et al., 2004], proteomic measurements of translation state by purification and mass spectrometric identification of ex vivo puromycinylated proteins (PUNCH-P) [Aviner et al., 2013], and the global detection of translation in vivo through the visualization of puromycin labeled proteins by immunocytochemistry (SUnSET) [Schmidt et al., 2009]. In a recent paper, Liu et al. demonstrated the ability to both visualize protein translation in vivo and enrich puromycin-incorporated proteins using an alkyne analog of puromycin and subsequent copper-catalyzed azide-alkyne cycloaddition (CuAAC) “click” chemistry [Liu et al., 2012]. Puromycin-based assays take advantage of the fact that the drug is non-radioactive, relatively inexpensive, and rapidly incorporated. Protein labeling with puromycin can also be accomplished without the use of amino acid auxotrophies or limiting media conditions, permitting a wider range of experimental designs. However, as yeasts and Gram negative bacteria exhibit little or no uptake of the drug and are generally insensitive to its effects on growth [Schindler & Davies, 1975]; [Melcher, 1971]; [Halegoua et al., 1976], these recent technologies are inaccessible to researchers studying these organisms.
Here we report a strain of Saccharomyces cerevisiae that is sensitive to puromycin at the level of growth on both liquid and solid media. We also characterize a common cellular response to puromycin treatment, the induction of cytoplasmic processing bodies (p-bodies). Finally, we demonstrate the utility of puromycin as a probe for translation by showing that yeast proteins rapidly incorporate puromycin in vivo.
Materials and methods
Yeast strains and media
The genotypes of the S. cerevisiae strains used in this study are listed Table 1. The EPP strain was not amenable to transformation using standard protocols many of the strains were constructed by crossing the EPP mutant strain (14339, gift of Dr. Julian Simon) to a strain containing the construct of interest, e.g. EDC3-GFP::HIS3MX or the galactose inducible reporter proteins of various lengths. Strains containing pairwise combinations of the EPP mutations (YAD267, YAD269 and YAD271) were generated by crossing an EPP parental strain (14339) with the Edc3-GFP strain from the GFP collection (YAD50) [Huh et al., 2003]. YAD270 and YAD273 were also generated by this cross. To enable the re-use of the NAT selectable drug marker for tagging of the galactose-driven 3x HA tagged reporter constructs, YAD270 was crossed to the KAN-marked pdr1 deletion strain (YAD336) to yield YAD337 and YAD338. Strains containing the three galactose driven 3xHA reporter genes (pGal1:3xHA-ADE17::NatNT2, pGal1:3xHA-HSP104::NatNT2, pGal1:3xHA-NEW1::NatNT2) were constructed in two steps. First, the reporter genes were constructed by PCR as described [Longtine et al., 1998] and transformed into BY4741 to generate (YAD445, YAD446 a YAD447). Next, to introduce those reporter constructs into the EPP background, these strains were crossed to YAD338 (yielding YAD517, YAD518 and YAD519).
Table 1.
S. cerevisiae strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | [Brachmann, et al., 1998] |
| 14339 | MATα met15Δ0 leu2Δ0 ura3Δ0 erg6Δ::LEU2 pdr1Δ::natMX pdr3Δ::hphMX | [Dunstan, et al., 2002] |
| YAD50 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 EDC3-GFP(S65T)::HIS3MX6 | [Huh, et al., 2003] |
| YAD337 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 erg6Δ::LEU2 pdr3Δ::hphMX pdr1Δ::KanMX4 | This study |
| YAD336 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pdr1Δ::KanMX4 | [Giaever, et al., 2002] |
| YAD521 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pdr3Δ::KanMX4 | [Giaever, et al., 2002] |
| YAD522 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 erg6Δ::KanMX4 | [Giaever, et al., 2002] |
| YAD267 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 erg6Δ::Leu pdr1Δ::natMX EDC3-GFP(S65T)::HIS3MX6 | This study |
| YAD269 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 erg6Δ::LEU2 pdr3Δ::Hyg EDC3-GFP(S65T)::HIS3MX6 | This study |
| YAD271 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pdr1Δ::Nat pdr3Δ::hphMX EDC3-GFP(S65T)::HIS3MX6 | This study |
| YAD273 | MATα met15Δ0 ura3Δ0 erg6Δ::Leu pdr3Δ::hphMX pdr1Δ::Nat EDC3-GFP(S65T)::HIS3MX6 | This study |
| YAD338 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 erg6Δ::LEU2 pdr3Δ::hphMX pdr1Δ::KanMX4 | This study |
| YAD445 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pGal1:3xHA-ADE17::NatNT2 | This study |
| YAD446 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pGal1:3xHA-HSP104::NatNT2 | This study |
| YAD447 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pGal1:3xHA-NEW1::NatNT2 | This study |
| YAD517 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 erg6Δ::LEU2 pdr3Δ::hphMX pdr1Δ::KanMX4 pGal1:3xHA-ADE17::NatNT2 | This study |
| YAD518 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 erg6Δ::LEU2 pdr3Δ::hphMX pdr1Δ::KanMX4 pGal1:3xHA-HSP104::NatNT2 | This study |
| YAD519 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 erg6Δ::LEU2 pdr3Δ::hphMX pdr1Δ::KanMX4 pGal1:3xHA-NEW1::NatNT2 | This study |
Unless noted, standard media and methods were used for growth and genetic manipulation of yeast [Rose MD, 1990]. Puromycin (FW 544.43, A.G. Scientific, Inc., Product Number: P-1033) was prepared by dissolving the puromycin dihydrochloride powder in sterile water to a concentration of 50 mM and added to growth media to achieve the final concentrations listed. Puromycin was added to YPD agar plates prior to pouring. O-propargyl-puromycin (OP-puromycin) was custom synthesized (Medchem Source LLP, Federal Way, WA) following the synthesis scheme established by Liu et al. [Liu et al., 2012]. OP-puromycin was dissolved in dimethyl sulfoxide (DMSO) to a working concentration of 150 mM and subsequently diluted to achieve the final concentrations listed.
Antibodies
The 12D10 monoclonal antibody against puromycin-incorporated proteins and peptides (gift from Philippe Pierre, now commercially available, Millipore, MABE343) was used at a 1:5000 dilution for western blot analyses. Mouse anti-HA antibody clone 12CA5 (Santa Cruz Biotechnology, sc-57592) was used at a 1:100 dilution and the pan-actin antibody mAbGEa (Novus Biologicals, NB100-74340) at a 1:1000 dilution was used for loading control. All antibodies were detected using the goat anti-mouse IRDye 800CW (LiCor, 926-32210) at a 1:10000 dilution and detected using a LiCor Odyssey Infrared Imaging System.
Growth assays
Strains were grown overnight in liquid YPD and normalized by cell count to the lowest cell concentration. Eight 5-fold serial dilutions were prepared and 5 μl of each dilution was plated onto YPD agar plates with or without puromycin. Plates were incubated at 30° C and imaged on an Epson Perfection 2480 Photo scanner at 300 dpi resolution every 24 hours for 2 days. Two replicate assays were performed.
For liquid growth assays, strains were grown overnight and counted the following day. Cells were diluted in YPD and 5×103 cells (in 135 μl total volume) were added to each well of a tissue culture treated, flat bottomed, 96-well plate. Then, 15 μl of a 10x working drug stock was added to each well to achieve the final drug concentration. Each strain and drug concentration pair was assayed in triplicate for every experiment. Plates were sealed with sterile, gas-permeable, optical adhesive sealing film for microplates and incubated in a Sunrise 96-well optical plate reader (Tecan Austria GmbH, Austria) controlled by the Tecan Magellan v6.55 software. Cells were grown at 30° C with continuous shaking. Prior to reading, the plate was subjected to 15 seconds of high intensity shaking, and the OD600 was read using the Tecan accuracy method. Growth was measured for 2–3 days or until the OD of the wells exhibiting growth had plateaued. For puromycin spike-in assays, cells and 96-well plates were prepared as above and grown to log phase (OD600 ~ 0.4) at which point the Tecan was stopped, the plate was removed, and 15 μl of a 10x working drug stock was added to each well. The plates were then re-sealed and returned to the Tecan for continued growth monitoring.
Growth rates were calculated from liquid growth assays by applying a linear fit in 90-minute sliding windows to the log-transformed OD600 values. Once the growth rates in the sliding windows had been estimated, the maximum growth rate achieved was reported for each dose and strain combination. For comparisons across strain backgrounds, the maximum growth rate for each strain at each drug concentration was normalized to its maximum growth rate in YPD. For analysis of the puromycin spike-in experiments, growth rates following the puromycin spike-in were normalized to growth rates in YPD and the maximum decrease in the relative rate for each dose is reported. The number of minutes required to achieve a 25% reduction in growth rate at various doses was also calculated using the normalized growth rate parameter.
Microscopy and image analysis
For microscopic evaluation of Edc3-GFP aggregation following puromycin treatment, cells were grown to log phase (OD600 ~ 0.8) and incubated with puromycin or vehicle for 30 minutes or 120 minutes. Following puromycin treatment, cells were fixed in 3.7% formaldehyde in PBS for 2 minutes at room temperature, followed by a 10-minute incubation in PBS on ice. Fixed cells were imaged using a DeltaVision deconvolution imaging system (Applied Precision, Issaquah, WA) outfitted with an Olympus IX-71 wide field microscope. Cells were imaged with a PlanApo 60x oil objective (N.A. 1.42) using differential interference contrast (DIC) bright field illumination and a 250W xenon LED transillumination light source. For GFP fluorescence imaging a polychroic beam splitter was used along with excitation 490/20 and emission 528/38 bandpass filters. A set of up to 30, 0.2 micron z-sections were captured for each image, deconvolved, and exported as tiff files. Tiff files were imported into ImageJ to generate image stacks and image levels were adjusted to be identical across all images. Tiff stacks were then batch analyzed by the Cell3 MATLAB program for segmentation of the bright field and GFP images [Selinummi]. We implemented the local comparison and selection (LOC-CS) algorithm within Cell3 for fluorescent segmenation [Ruusuvuori et al., 2010]. The LOC-CS parameters were r=5 and alpha=0.6. For bright field segmentation, the minimum was set to 120 pixels, the maximum to 2040 pixels, and any item smaller than 40 pixels or larger than 3000 pixels was excluded [Niemisto et al., 2007]. Statistical significance was determined using the Mann Whitney Rank-Sum U test implemented in the R statistical computing language.
Puromycin Incorporation, HA-protein induction, and Western Blot Analysis
To test the in vivo incorporation of puromycin into yeast proteins, overnight cultures were diluted to an OD600 of 0.2 and allowed to grow to log phase (OD600 ~ 0.8). Puromycin was added directly to the culture at the concentrations indicated and allowed to grow for the amount of time indicated, at which time cells were rapidly pelleted (5 min at 3000g), and cell pellets were frozen in liquid nitrogen and stored at −80°C for further analysis. Lysates were prepared by alkaline lysis [Kushnirov, 2000] and volumes corresponding to equivalent cell numbers were used for western blot analysis with the anti-puromycin antibody and Coomassie stain (Imperial Stain, Thermo Pierce) loading control.
For galactose induction of the HA-tagged constructs, overnight cultures in SC medium with 2% raffinose were diluted to an OD600 of ~0.2 into a fresh aliquot of the same medium. The culture was grown to log phase (OD600 ~ 0.7) and galactose was added to a final concentration of 2%. After 15 minutes of galactose induction, 0 mM-2 mM puromycin was added to the cultures. The cultures were allowed to grow in galactose and puromycin for an additional 15 to 30 minutes before collection and analysis as above. The induced proteins were visualized by anti-HA western blot.
CuAAC Click Reaction & Mass Spectrometry
To test OP-puromycin-peptide fragmentation by tandem mass spectrometry (MS), a synthetic peptide was ordered with the sequence NH2-ISHVSTGGGASLELLEG[K(N3]-COOH containing an azide-modified lysine terminal residue (Thermo Fisher Scientific). This peptide was resuspended in PBS (pH 7.4) and mixed with OP-puromycin at a molar ratio of 1:2 peptide:OP-puromycin, 250 μM CuSO4, 1.25 mM Tris(3-hydroxyropyltiazolmethyl)amine (THPTA) ligand (gift from M.G. Finn, Scripps), 5 mM aminioguanidine, and 5 mM ascorbic acid. The mixture was incubated at 30 °C for 1 hour. The reaction was then desalted on C18 resin, evaporated on a speedvac and resuspended in 5% acetonitrile and 0.1% formic acid to prepare for mass spectrometry.
The experiments were carried out on the LTQ ion trap MS of an LTQ-FT MS instrument (Thermo Scientific, San Jose, CA) the front end of which, including the ion source, was replaced by a homebuilt electrodynamic ion funnel [Shaffer et al., 1997; Page et al., 2007]. Samples were dissolved into a solution mixture of H2O/MeOH (1:1 v:v). A direct infusion with 2kV of spray voltage was used for ionization. A sample flow rate of 3 μl/min flow rate was applied and an atmospheric pressure ionization (API) inlet capillary temperature of 300 °C was used. Collision induced dissociation was used for the structural identification. Normalized collision energies of 20% for OP-puromycin at 495 m/z, 24% for Pgk1 azido peptide at 1781 m/z, 35% for the CuAAC product at 1135 m/z, and 20% or 27% for the CuAAC product at 755 m/z were used. To mitigate the effect of instantaneous signal fluctuations, the analyzed data were produced from mass spectra averaged over one minute.
Results and discussion
Identification and characterization of a puromycin-sensitive S. cerevisiae strain
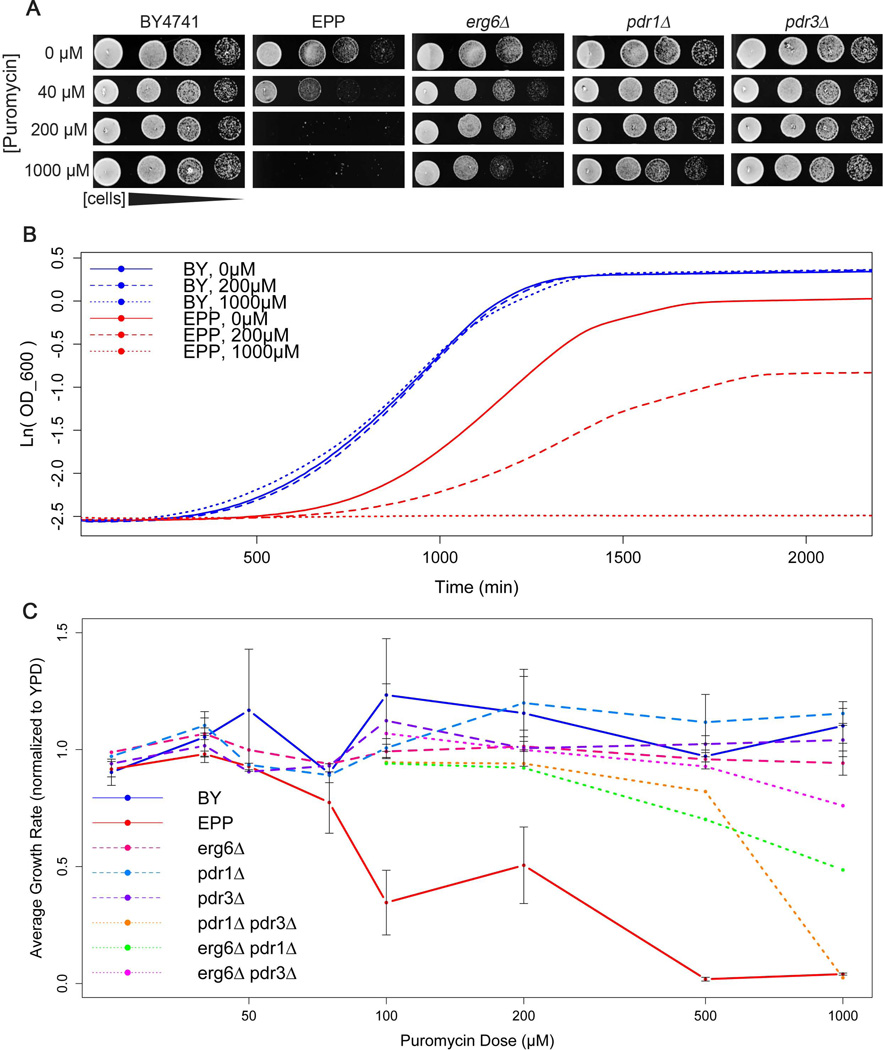
Despite the fact that puromycin is able to disassemble yeast ribosomes in vitro [Barbacid et al., 1975]; [Pfund et al., 1998], live, intact cells of the commonly used laboratory strain of S. cerevisiae are insensitive to the antibiotic effects of puromycin [Schindler & Davies, 1975]; [Melcher, 1971] and deriving sensitivity has required generating spheroplasts [van Wijk, 1968];[Schindler & Davies, 1975]. To identify a strain with the potential to incorporate puromycin into proteins in vivo, we first set out to identify strains that were sensitive to the antibiotic properties of the drug. Yeast strains harboring mutations in the genes encoding the Pdr1p and Pdr3p pleiotropic drug resistance transcription factors and the Erg6p methyltransferase involved in the biosynthesis of the sterol component of yeast membranes (ergosterol) have previously been shown to sensitize strains to a variety of drugs [Dunstan et al., 2002]; [Sipos & Kuchler, 2006]; [Simon & Bedalov, 2004]. To test whether an erg6Δ, pdr1Δ, pdr3Δ triple mutant (EPP) could sensitize yeast to puromycin, we compared the growth of the EPP strain to wild type cells on YPD agar plates supplemented with puromycin at a range of concentrations (Figure 1A). While puromycin concentrations as high as 1 mM had no effect on the growth of wild-type cells, puromycin at concentrations of 40 μM slowed the growth of the EPP strain, with complete growth inhibition at 200 μM. Puromycin showed a similar effect on EPP growth in liquid culture, although the drug concentrations required were higher than those required on solid media (Figure 1B). Thus, the triple mutant EPP strain is sensitive to puromycin, although less sensitive by an order of magnitude than metazoan cell culture systems that typically use between 2 μM and 20 μM puromycin [Medina et al., 2000]; [Iwaki & Castellino, 2008]; [Iwaki & Umemura, 2011].
Figure 1.
Characterization of a puromycin-sensitive yeast strain. (A) Growth of 5-fold serial dilutions of wild-type (BY4741), EPP (erg6Δ pdr1Δ pdr3Δ) triple deletion mutant, and single deletion mutants cells spotted on YPD agar containing 0 μM, 40 μM, 200 μM and 1000μM puromycin. (B) Growth curves for BY4741 (blue) and the EPP triple mutant (red) in liquid media containing 0 μM (solid), 200 μM (dash), and 1000 μM (dot) puromycin. Curves are the average of 6 biological replicates. Growth curves were measure using a Tecan Sunrise plate reader sampling OD600 at 15 min intervals at a constant temperature of 30°C. (C) Average maximum growth rate (maximum change in log(OD)/minute) calculated for each strain grown at each concentration of puromycin, normalized to average maximum growth rate for each strain in YPD without puromycin. Error bars +/− SEM. Wild type (blue, solid line), EPP (red, solid line), erg6Δ (magenta, dash line), pdr1Δ (light blue, dash line), pdr3Δ (purple, dash line), pdr1Δ pdr3Δ (orange, dotted line), erg6Δ pdr1Δ (green, dotted line), and erg6Δ pdr3Δ (pink, dotted line).
To characterize the contribution of the individual mutations on the sensitivity of the EPP triple mutant, we examined the puromycin sensitivity of strains harboring each single and all combinations of double erg6Δ, pdr1Δ, and pdr3Δ mutations. With the exception of a modest effect on an erg6Δ mutant on solid medium, there were no measurable effects of puromycin on the growth of any of the single mutants on solid or liquid media. All double mutant strains showed some level of sensitivity to puromycin in liquid culture (Figure 1C), with the pdr1Δ pdr3Δ mutant being the most sensitive of the panel. However, none of the strains were as sensitive to puromycin as the triple mutant. Thus, the strong sensitivity of the EPP strain requires the deletion of all three genes.
Studies using puromycin as a probe for of translational dynamics would be likely to pulse the drug into the growth medium at a specific stage of growth or following an environmental perturbation. To assess the response of the EPP strain to a puromycin pulse, we added puromycin to cultures of logarithmically growing EPP cells and measured the subsequent effects on growth. While the addition of puromycin to EPP cells in mid-log phase growth strongly inhibited their growth, higher doses of the drug were required to achieve similar levels of growth inhibition compared to continuous culture with the drug (Supplemental Figure S1A). The concentration of puromycin applied also affected the rate of growth inhibition; 2 mM puromycin slowed cells to 75% of their maximal growth rate after 105 minutes, while 1 mM puromycin took 120 minutes and 0.5 mM took 225 minutes to slow to the same degree (Supplemental Figure S2B). These data provide an initial framework for dose selection with the EPP strain.
Puromycin exposure induces aggregation of the p-body protein Edc3-GFP
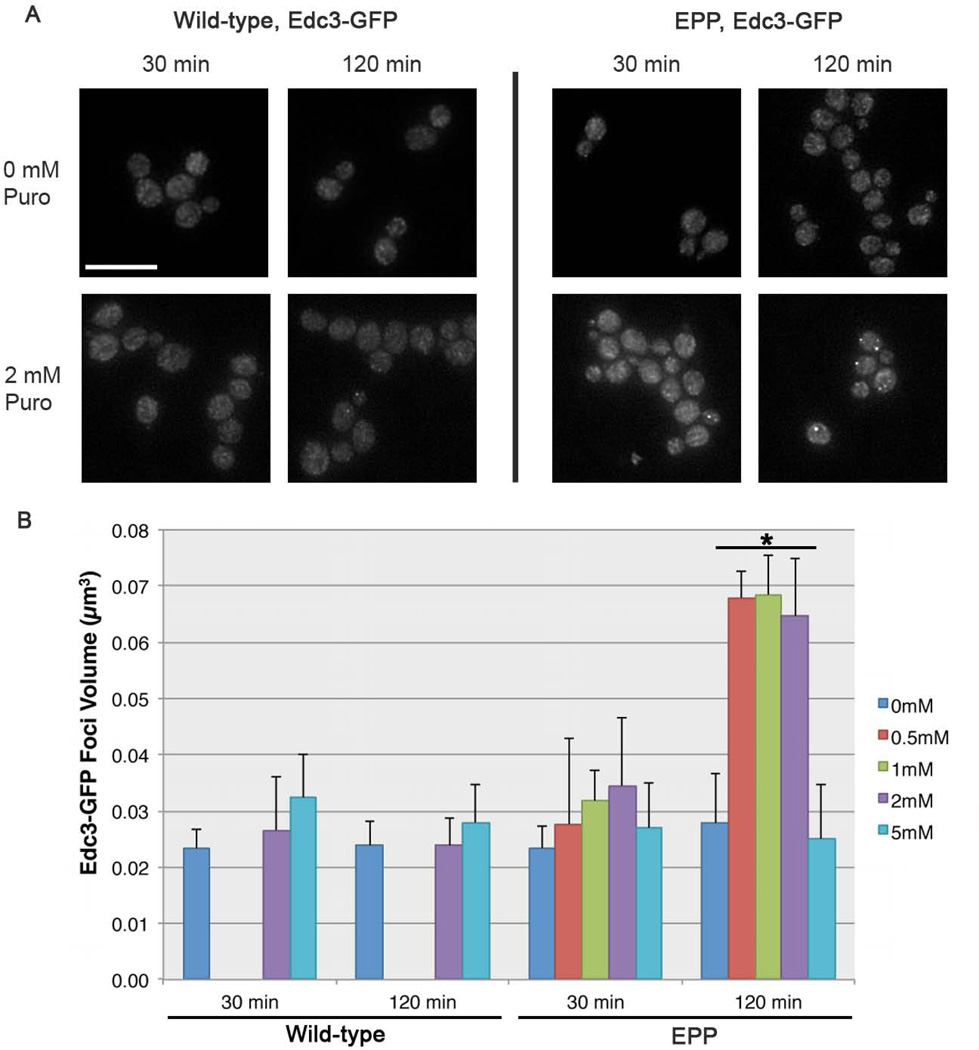
Processing bodies (p-bodies) are cytoplasmic RNP complexes that are conserved across eukaryotes and are composed of RNA decay enzymes, translational repressors and non-translating mRNPs [Parker & Sheth, 2007]. P-bodies are sensitive to environmental perturbations and typically increase in size and number when cells are exposed to conditions that inhibit translation [Brengues et al., 2005]. Puromycin treatment is known to induce the aggregation of p-body proteins into cytoplasmic foci in metazoan systems [Eulalio et al., 2007]; [Aizer et al., 2008]; [Swetloff et al., 2009], a response that likely coincides with ribosomal subunit dissociation and translational arrest. To test whether puromycin exposure could also induce p-bodies in yeast, we introduced a florescent p-body reporter (the p-body component Edc3p fused to GFP) into the EPP strain background. Puromycin was added to cells growing in log phase, and cells were fixed and imaged following 30 minutes and 120 minutes of drug exposure. While no change was observed in the localization of Edc3-GFP in the wild-type strain background, in the EPP strain Edc3-GFP accumulated in distinct cytoplasmic foci (p-bodies) after 120 minutes of exposure to 2 mM puromycin (Figure 2A). To provide a more objective assessment of Edc3 aggregation, we used automated image analysis to measure p-body volume (Materials and Methods). In the wild-type strain background we observed no significant (p < 0.01) increases in p-body volume at any of the doses tested. However, after 2 hours of incubation with puromycin, all EPP mutants treated with concentrations of drug below 5 mM showed significant increases in the volume of Edc3-GFP foci (p < 2.2×10−16) (Figure 2B). Interestingly, at 5 mM puromycin there is no increase in Edc3-GFP accumulation, perhaps representing extreme toxicity of a high dose. These results demonstrate that upon sensitization to the drug via the introduction of the EPP deletion mutations, the effects of puromycin on yeast cellular physiology are similar to those that have been observed in other eukaryotes.
Figure 2.
Exposure to puromycin induces Edc3-GFP accumulation into cytoplasmic foci. (A) Images of Edc3-GFP in wild-type and EPP cells grown in 0 mM or 2 mM puromycin for 30 min and 120 min. Scale bar = 10 μm. (B) Average Edc3-GFP foci volume (in μm3) in wild type and EPP cells after 30 min and 120 min of exposure to a range of concentrations from 0 mM to 5 mM puromycin. Error bars +/− SD. The conditions in which a statistically significant difference in Edc3-GFP foci volume relative to 0 mM puromycin treatment are marked (*; p < 2.2×10−16).
Puromycin is rapidly incorporated into proteins in the sensitized strain
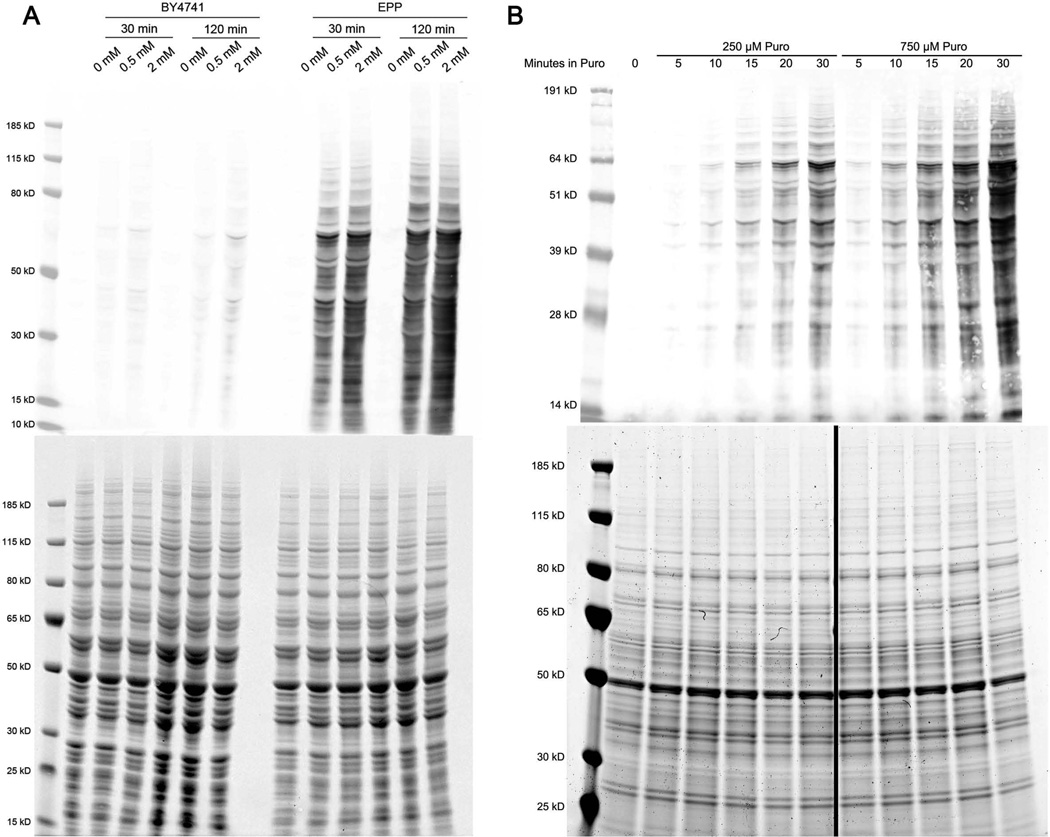
To test whether puromycin could be incorporated into yeast proteins in vivo, thereby enabling a number of puromycin based techniques, we used the anti-puromycin monoclonal antibody 12D10 [Schmidt et al., 2009], which has been used to detect puromycin incorporation into human proteins by western blot and flow cytometry. We observed a dose- and time-dependent increase in 12D10 immunoreactivity in yeast lysates prepared from EPP strains exposed to puromycin for 30 and 120 minutes (Figure 3A), with the highest levels detected in lysates from cells treated for 120 minutes with 2 mM puromycin. We also observed low levels of 12D10 immunoreactivity in wild-type cells treated with puromycin (Figure 3A), suggesting that wild-type cells are at least minimally permeable to the drug. To assess how rapidly puromycin is detectable in yeast proteins, we prepared yeast lysates of the EPP strain harvested at different times following the addition of puromycin. Here we observed puromycin immunoreactivity by western blot with the 12D10 antibody as early as 5–10 minutes after the drug was administered (Figure 3B). Taken together, these results demonstrate that that puromycin is rapidly taken up by the mutant strain, available to ribosomes in vivo, and able to be incorporated into nascent proteins in yeast.
Figure 3.
Puromycin is rapidly incorporated into proteins in the EPP strain. (A) Anti-puromycin (12D10) western blot of lysates from wild type and EPP strains following 30 min and 120 min of exposure to puromycin and a Coomassie stain demonstrating loading equivalency. (B) Time-course of the EPP strain grown in 250 μM and 750 μM puromycin and a Coomassie stain demonstrating loading equivalency.
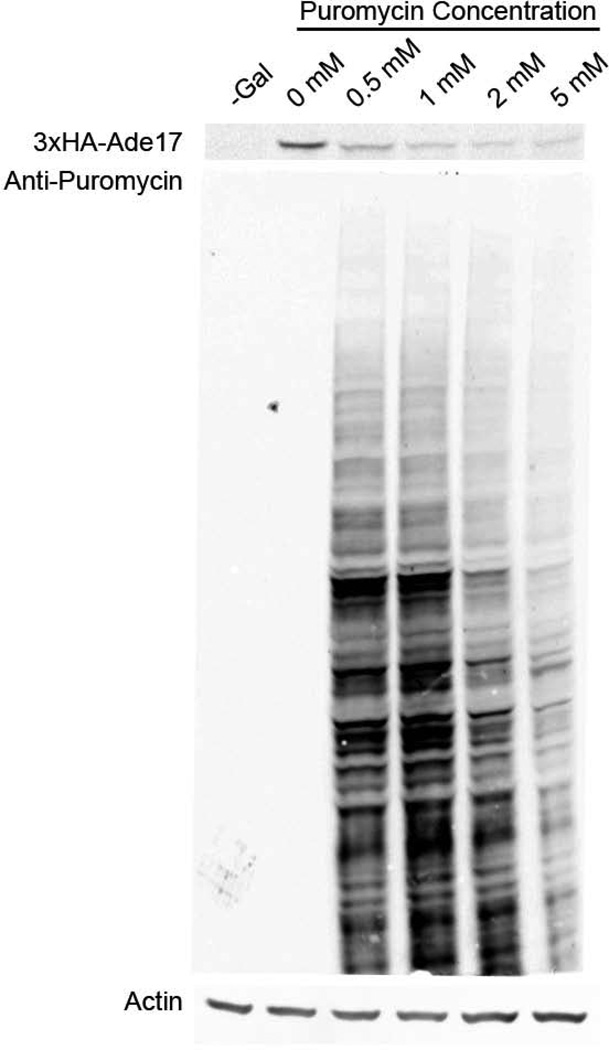
To examine the incorporation of puromycin into individual proteins in vivo, we constructed an EPP strain in which ADE17 is N-terminally 3xHA tagged and expressed under the transcriptional control of the GAL1 promoter. The GAL1 promoter is tightly repressed in the presence of glucose, highly expressed in the presence of galactose, and in the presence of neutral carbon sources, such as raffinose, poised to rapidly initiate transcription upon addition of galactose [Longtine et al., 1998]. To facilitate the detection of puromycin incorporation into newly synthesized proteins, the EPP strain containing the galactose-inducible 3xHA-ADE17 was grown in raffinose, its transcription was induced by the addition of galactose for 15 minutes, and different concentrations of puromycin were added to label proteins synthesized after this timepoint. The results show that while steady state levels of protein do not change (Figure 4, Actin, lanes 1–6), there is a galactose-dependent induction of the 3xHA-ADE17 protein (Figure 4, 3xHA-ADE17, lanes 1 and 2). As described above (Figure 3), we observed drug-dependent puromycin immunoreactivity in total proteins (Figure 4, anti-puromycin, lanes 1–2 versus lanes 3–5) that appeared to decrease at the highest concentration of puromycin (Figure 4, anti-puromycin, lane 6). These results support the hypothesis that puromycin inhibits translation in vivo in a dose-dependent manner and provide further evidence that 5 mM puromycin represents an extreme dose with distinct physiological and translational responses. We observe an increase in low-molecular weight relative to high-molecular weight anti-puromycin immunoreactivity at elevated doses of the drug (Figure 3A, last lane, and Figure 4). This observation could reflects an increased rate of incorporation into smaller proteins or and increased chance of protein truncation at these doses. The work presented here does not distinguish between these two possibilities, and we highlight it as an avenue for further experimentation and optimization. Finally, in the presence of various concentrations of puromycin, we were able to detect nascent molecules of a specific protein, 3xHA-ADE17 (Figure 4, 3xHA-ADE17, lanes 3–6), although at a significantly reduced level relative to the no-drug control (Figure 4, 3xHA-ADE17, lane 2).
Figure 4.
Dose-response effect of puromycin treatment on protein translation in vivo. Anti-HA (12CA5) and anti-puromycin (12D10) western blots of a galactose-induced 3xHA-Ade17p strain grown in 0mM-2mM puromycin for 15 minutes. No galactose induction was present for the sample marked –Gal, while the other samples were grown in 2% galacotse for 15 minutes prior to puromycin addition. Anti-actin is shown to demonstrate loading equivalency.
In addition to dose-dependent effects of puromycin treatment, we also examined time-dependent translational kinetics using three galactose inducible N-terminally 3xHA-tagged proteins of increasing length: Ade17p (592 amino acids), Hsp104p (908 amino acids) and New1p (1196 amino acids). Cells were collected in a time course following puromycin or vehicle addition. In cultures not treated with puromycin, anti-HA immunoreactivity increases starting at 15 minutes of growth in galactose (Supplemental Figure S2). The larger proteins exhibited reduced signal compared with HA-Ade17p, which presumably reflects the extra time involved in translating the additional 300–600 residues per protein and also the decreased efficiency for western blot transfer of large proteins [Bolt & Mahoney, 1997]. When the same strains were treated with puromycin, the production of HA signal was reduced for all three proteins relative to the untreated cells (Supplemental Figure S2). This signifies that proteins of a range of sizes are affected by puromycin incorporation and there does not appear to be a drastic bias toward longer or shorter proteins for puromycin incorporation.
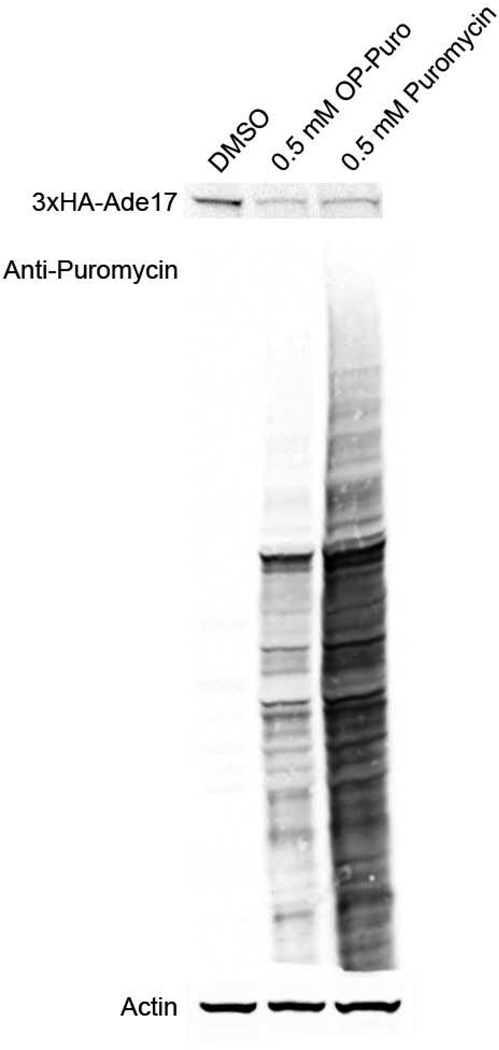
Puromycin analogs, such as O-propargyl-puromycin (OP-puromycin), enable more sophisticated inquiries into translational activity including visualization of translational activity and capture of nascent proteins. We tested whether the EPP mutant strain is also able to incorporate OP-puromycin. Growing the EPP strain in 500μM OP-puromycin was sufficient to inhibit the expression of HA-Ade17p and incorporate into proteins, generating 12D10 immunoreactivity (Figure 5). While the anti-puromycin immunoreactivity of the OP-puromycin treated lysates was reduced compared to the cells treated with an equal dose of puromycin, inhibition of the production of the HA-tagged ORF is equivalent. This suggests there may be different sensitivities of the 12D10 antibody to puromycin and OP-puromycin labeled proteins.
Figure 5.
Anti-puromycin (12D10) western blot of cells grown in either 0.5mM OP-puromycin, puromycin or DMSO and anti-HA (12CA5) western blot of 3xHA-Ade17p expression in each condition.
We also tested the effects of OP-puromycin on peptide fragmentation by collision-induced dissociation (CID) in an ion trap mass spectrometer (MS). To do so OP-puromycin was conjugated to a synthetic peptide with an azido-lysine terminal residue using CuAAC chemistry. However, the puromycin orientation with respect to the peptide is not as would be expected for in vivo incorporated puromycin peptides, through a peptide linkage, but rather conjugated to the terminal lysine side chain (Supplemental Figure S3A and S3B). Subjecting OP-puromycin to CID, the base peak of the tandem mass spectrum corresponds to loss of the dimethylaminopurine ring (Supplemental Figure S3C). This reflects what is known concerning the mass spectrometric fragmentation of puromycin and related compounds, where a common fragment ion forms due to loss of the purine ring from the sugar [Eggers et al., 1966]. Tandem MS of the pre-conjugated synthetic peptide resulted in observation of the b- and y-series fragment ions common to peptides subjected to CID, and which correspond well to the consensus fragmentation pattern reported in the PeptideAtlas for the native peptide (Supplemental Figure S3D). When we subjected the product of the peptide-OP-puromycin conjugation to CID, the base peak in the tandem mass spectra corresponded to loss of dimethylaminopurine (Supplemental Figure S3E) with no detectable b- and y- fragment ions. These data indicate that the loss of the purine ring from a puromycin-peptide represents cleavage of the most labile bond producing a tandem mass spectrum devoid of peptide backbone fragment ions. This suggests that efficient identification of puromycin labeled proteins and peptides of unknown sequence by mass spectrometry will require either enrichment by puromycin capture or the development of MS3 based tandem MS approaches popular in the phosphoproteomic field for similar reasons; i.e. facile loss of phosphate during CID resulting in few detectable b- or y-ions. In such an MS3 based strategy after MS2 detection of the loss of dimethylaminopurine would trigger MS2 on the subsequent base peak.
This work demonstrates in vivo puromycin sensitivity in S. cerevisiae. The increase in sensitivity is attributable to an increase in cell permeability by disruption of the multi-drug efflux system and sterol synthesis. Both the physiological effect of p-body induction and the increase in puromycin immunoreactivity indicate that puromycin is acting in vivo in a similar manner to its actions in other systems. Because puromycin incorporation acts as a label of translational activity, the characterization of the EPP strain enables yeast researchers to utilize the tools of puromycin-based assays including visualizing the location of protein translation across conditions or cell states, the ability to capture puromycin labeled proteins and peptides for proteomic analysis, and the assessment of co-translational versus post-translational protein modifications. Furthermore, development of these and other puromycin-based assays will be facilitated by the many advantages of the yeast model. Yeast researchers have been on the forefront of developing and implementing assays to explore translation, and the ability to incorporate puromycin in vivo enables the development of new and complementary techniques to enrich the understanding post-transcriptional regulatory mechanisms.
Supplementary Material
Figure S1. Characterization of puromycin spike-in kinetics. (A) Minimum pre-saturation average growth rate following puromycin spike-in at several doses normalized to growth rate prior to puromycin addition. Error bars +/− SEM. (B) Length of time to reduce growth rate to 25% of rate in YPD by spike-in of puromycin.
Figure S2. Time course of strains expressing galactose-induced N-terminally 3xHA-tagged ORFs grown in 0 μM or 250 μM puromycin. The ORFs tested are 3xHA-Ade17p (592 amino acids); 3xHA-Hsp104p (908 amino acids); 3xHA-New1p (1196 amino acids). Anti-actin is shown to demonstrate loading equivalency.
Figure S3. Mass spectrometric analysis of OP-puromycin and OP-puromycin-peptide adducts. (A) Structures of the peptide CuAAC scheme. In our experiments R corresponds to the synthetic peptide ISHVVSTGGGASLELLEG with the terminal azido-lysine shown. The expected masses of m1 and p1 from OP-puromycin are 162m/z and 333m/z respectively whereas the expected masses of m1 and p1 from the CuAAC product are 162m/z and 2108m/z respectively. (B) Hypothetical in vivo ribosome mediated puromycinylation. Tandem mass spectra of (C) OP-Puromycin, (D) ISHVVSTGGGASLELLEG-KazideE) and the CuAAC product, OP-puromycin conjugated peptide in +2 charge states, p1 (+1) corresponds to the singly charged fragment ion and p1 (+2) corresponds to the doubly charged fragment ion.
Acknowledgments
SPONSORS: University of Luxembourg - ISB Strategic Partnership, National Institutes of Health P50 GM076547/Center for Systems Biology
References
- 1.Aizer A, Brody Y, Ler LW, Sonenberg N, Singer RH, Shav-Tal Y. The dynamics of mammalian P body transport, assembly, and disassembly in vivo. Mol Biol Cell. 2008;19:4154–4166. doi: 10.1091/mbc.E08-05-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson P. Post-transcriptional regulons coordinate the initiation and resolution of inflammation. Nat Rev Immunol. 2010;10:24–35. doi: 10.1038/nri2685. [DOI] [PubMed] [Google Scholar]
- 3.Aviner R, Geiger T, Elroy-Stein O. Novel proteomic approach (PUNCH-P) reveals cell cycle-specific fluctuations in mRNA translation. Genes Dev. 2013;27:1834–1844. doi: 10.1101/gad.219105.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbacid M, Fresno M, Vazquez D. Inhibitors of polypeptide elongation on yeast polysomes. J Antibiot (Tokyo) 1975;28:453–462. doi: 10.7164/antibiotics.28.453. [DOI] [PubMed] [Google Scholar]
- 5.Bolt MW, Mahoney PA. High-efficiency blotting of proteins of diverse sizes following sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Anal Biochem. 1997;247:185–192. doi: 10.1006/abio.1997.2061. [DOI] [PubMed] [Google Scholar]
- 6.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 7.Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunstan HM, Ludlow C, Goehle S, et al. Cell-based assays for identification of novel double-strand break-inducing agents. J Natl Cancer Inst. 2002;94:88–94. doi: 10.1093/jnci/94.2.88. [DOI] [PubMed] [Google Scholar]
- 9.Eggers SH, Biedron SI, Hawtrey AO. Mass spectra of puromycin and some derivatives. Tetrahedron Lett. 1966;28:3271–3280. doi: 10.1016/s0040-4039(01)82777-5. [DOI] [PubMed] [Google Scholar]
- 10.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freeman JA, Espinosa JM. The impact of post-transcriptional regulation in the p53 network. Brief Funct Genomics. 2013;12:46–57. doi: 10.1093/bfgp/els058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giaever G, Chu AM, Ni L, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 13.Halbeisen RE, Scherrer T, Gerber AP. Affinity purification of ribosomes to access the translatome. Methods. 2009;48:306–310. doi: 10.1016/j.ymeth.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Halegoua S, Hirashima A, Inouye M. Puromycin-resistant biosynthesis of a specific outer-membrane lipoprotein of Escherichia coli. J Bacteriol. 1976;126:183–191. doi: 10.1128/jb.126.1.183-191.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen WJ, Lingappa VR, Welch WJ. Complex environment of nascent polypeptide chains. J Biol Chem. 1994;269:26610–26613. [PubMed] [Google Scholar]
- 16.Hinz FI, Dieterich DC, Tirrell DA, Schuman EM. Non-canonical amino acid labeling in vivo to visualize and affinity purify newly synthesized proteins in larval zebrafish. ACS Chem Neurosci. 2012;3:40–49. doi: 10.1021/cn2000876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 18.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwaki T, Castellino FJ. A single plasmid transfection that offers a significant advantage associated with puromycin selection in Drosophila Schneider S2 cells expressing heterologous proteins. Cytotechnology. 2008;57:45–49. doi: 10.1007/s10616-008-9129-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iwaki T, Umemura K. A single plasmid transfection that offers a significant advantage associated with puromycin selection, fluorescence-assisted cell sorting, and doxycycline-inducible protein expression in mammalian cells. Cytotechnology. 2011;63:337–343. doi: 10.1007/s10616-011-9357-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim W, Kyung Lee E. Post-transcriptional regulation in metabolic diseases. RNA Biol. 2012;9:772–780. doi: 10.4161/rna.20091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kushnirov VV. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 23.Liu J, Xu Y, Stoleru D, Salic A. Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc Natl Acad Sci U S A. 2012;109:413–418. doi: 10.1073/pnas.1111561108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Longtine MS, McKenzie A, 3rd, Demarini DJ, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 25.McCarthy JE. Posttranscriptional control of gene expression in yeast. Microbiol Mol Biol Rev. 1998;62:1492–1553. doi: 10.1128/mmbr.62.4.1492-1553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medina D, Moskowitz N, Khan S, Christopher S, Germino J. Rapid purification of protein complexes from mammalian cells. Nucleic Acids Res. 2000;28:E61. doi: 10.1093/nar/28.12.e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melcher U. Metabolism of puromycin by yeast cells. Biochim Biophys Acta. 1971;246:216–224. doi: 10.1016/0005-2787(71)90130-4. [DOI] [PubMed] [Google Scholar]
- 28.Miyamoto-Sato E, Nemoto N, Kobayashi K, Yanagawa H. Specific bonding of puromycin to full-length protein at the C-terminus. Nucleic Acids Res. 2000;28:1176–1182. doi: 10.1093/nar/28.5.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niemisto A, Korpelainen T, Saleem R, Yli-Harja O, Aitchison J, Shmulevich I. A K-means segmentation method for finding 2-D object areas based on 3-D image stacks obtained by confocal microscopy. Conf Proc IEEE Eng Med Biol Soc. 2007;2007:5559–5562. doi: 10.1109/IEMBS.2007.4353606. [DOI] [PubMed] [Google Scholar]
- 30.Nousch M, Eckmann CR. Translational control in the Caenorhabditis elegans germ line. Adv Exp Med Biol. 2013;757:205–247. doi: 10.1007/978-1-4614-4015-4_8. [DOI] [PubMed] [Google Scholar]
- 31.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 32.Page JS, Tang K, Smith RD. An electrodynamic ion funnel interface for greater sensitivity and higher throughput with linear ion trap mass spectrometers. International Journal of Mass Spectrometry. 2007;265:244–250. [Google Scholar]
- 33.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 34.Pfund C, Lopez-Hoyo N, Ziegelhoffer T, et al. The molecular chaperone Ssb from Saccharomyces cerevisiae is a component of the ribosome-nascent chain complex. EMBO J. 1998;17:3981–3989. doi: 10.1093/emboj/17.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pospisek M, Valasek L. Polysome profile analysis--yeast. Methods Enzymol. 2013;530:173–181. doi: 10.1016/B978-0-12-420037-1.00009-9. [DOI] [PubMed] [Google Scholar]
- 36.Roberts RW, Szostak JW. RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc Natl Acad Sci U S A. 1997;94:12297–12302. doi: 10.1073/pnas.94.23.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rose MDWF, Hieter P. Laboratory Course Manual for Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 38.Ruusuvuori P, Aijo T, Chowdhury S, et al. Evaluation of methods for detection of fluorescence labeled subcellular objects in microscope images. BMC Bioinformatics. 2010;11:248. doi: 10.1186/1471-2105-11-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schindler D, Davies J. Inhibitors of macromolecular synthesis in yeast. Methods Cell Biol. 1975;12:17–38. doi: 10.1016/s0091-679x(08)60949-8. [DOI] [PubMed] [Google Scholar]
- 40.Schmeing TM, Seila AC, Hansen JL, et al. A pre-translocational intermediate in protein synthesis observed in crystals of enzymatically active 50S subunits. Nat Struct Biol. 2002;9:225–230. doi: 10.1038/nsb758. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods. 2009;6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 42.Schwanhausser B, Gossen M, Dittmar G, Selbach M. Global analysis of cellular protein translation by pulsed SILAC. Proteomics. 2009;9:205–209. doi: 10.1002/pmic.200800275. [DOI] [PubMed] [Google Scholar]
- 43.Selinummi J. Supplement Site for Cell3 Software https://sites.google.com/site/cell3supplement/ 2013 Nov 13; [Google Scholar]
- 44.Shaffer SA, Tang K, Anderson GA, Prior DC, Udseth HR, Smith RD. A novel ion funnel for focusing ions at elevated pressure using electrospray ionization mass spectrometry. Rapid Communications in Mass Spectrometry. 1997;11:1813–1817. [Google Scholar]
- 45.Simon JA, Bedalov A. Yeast as a model system for anticancer drug discovery. Nat Rev Cancer. 2004;4:481–492. doi: 10.1038/nrc1372. [DOI] [PubMed] [Google Scholar]
- 46.Sipos G, Kuchler K. Fungal ATP-binding cassette (ABC) transporters in drug resistance & detoxification. Curr Drug Targets. 2006;7:471–481. doi: 10.2174/138945006776359403. [DOI] [PubMed] [Google Scholar]
- 47.Starck SR, Green HM, Alberola-Ila J, Roberts RW. A general approach to detect protein expression in vivo using fluorescent puromycin conjugates. Chem Biol. 2004;11:999–1008. doi: 10.1016/j.chembiol.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 48.Swetloff A, Conne B, Huarte J, Pitetti JL, Nef S, Vassalli JD. Dcp1-bodies in mouse oocytes. Mol Biol Cell. 2009;20:4951–4961. doi: 10.1091/mbc.E09-02-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tabuchi I. Next-generation protein-handling method: puromycin analogue technology. Biochem Biophys Res Commun. 2003;305:1–5. doi: 10.1016/s0006-291x(03)00686-7. [DOI] [PubMed] [Google Scholar]
- 50.van Wijk R. Alpha-glucosidase synthesis, respiratory enzymes and catabolite repression in yeast. IV. B. Studies on the level of protein synthesis at 21 which repression and induction of alpha-glucosidase synthesis occur. Proc K Ned Akad Wet C. 1968;71:300–313. [PubMed] [Google Scholar]
- 51.Whitney ML, Hurto RL, Shaheen HH, Hopper AK. Rapid and reversible nuclear accumulation of cytoplasmic tRNA in response to nutrient availability. Mol Biol Cell. 2007;18:2678–2686. doi: 10.1091/mbc.E07-01-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Characterization of puromycin spike-in kinetics. (A) Minimum pre-saturation average growth rate following puromycin spike-in at several doses normalized to growth rate prior to puromycin addition. Error bars +/− SEM. (B) Length of time to reduce growth rate to 25% of rate in YPD by spike-in of puromycin.
Figure S2. Time course of strains expressing galactose-induced N-terminally 3xHA-tagged ORFs grown in 0 μM or 250 μM puromycin. The ORFs tested are 3xHA-Ade17p (592 amino acids); 3xHA-Hsp104p (908 amino acids); 3xHA-New1p (1196 amino acids). Anti-actin is shown to demonstrate loading equivalency.
Figure S3. Mass spectrometric analysis of OP-puromycin and OP-puromycin-peptide adducts. (A) Structures of the peptide CuAAC scheme. In our experiments R corresponds to the synthetic peptide ISHVVSTGGGASLELLEG with the terminal azido-lysine shown. The expected masses of m1 and p1 from OP-puromycin are 162m/z and 333m/z respectively whereas the expected masses of m1 and p1 from the CuAAC product are 162m/z and 2108m/z respectively. (B) Hypothetical in vivo ribosome mediated puromycinylation. Tandem mass spectra of (C) OP-Puromycin, (D) ISHVVSTGGGASLELLEG-KazideE) and the CuAAC product, OP-puromycin conjugated peptide in +2 charge states, p1 (+1) corresponds to the singly charged fragment ion and p1 (+2) corresponds to the doubly charged fragment ion.