Abstract
The discovery of microRNAs (miRNAs) in 1993 followed by developments and discoveries in small RNA biology have redefined the biological landscape by significantly altering the longstanding dogmas that defined gene regulation. These small RNAs play a significant role in modulation of an array of physiological and pathological processes ranging from embryonic development to neoplastic progression. Unique miRNA signatures of various inherited, metabolic, infectious, and neoplastic diseases have added a new dimension to the studies that look at their pathogenesis and highlight their potential to be reliable biomarkers. Also, altering miRNA functionality and the development of novel in vivo delivery systems to achieve targeted modulation of specific miRNA function are being actively pursued as novel approaches for therapeutic intervention in many diseases. Here we review the current body of knowledge on the role of miRNAs in development and disease and discuss future implications.
Keywords: animal microRNA, miR, oncomiR, apoptomiR, circulating miRNA, biomarkers
MicroRNAs: Introduction and a Brief History
MicroRNAs (miRNAs) are a class of small (~19–24 nucleotides in length), endogenous, evolutionarily conserved RNAs that function as posttranscriptional regulators of gene expression. They primarily function by binding to complementary target sequences in messenger RNA (mRNA) and interfering with the translational machinery, thereby preventing or altering the production of the protein product. Follow-up studies also revealed that in addition to repressing translation, miRNA binding to its target mRNA also triggered the recruitment and association of mRNA decay factors, leading to mRNA destabilization, degradation, and resultant decrease in expression levels. miRNAs were discovered in 1993 by Lee and colleagues93 in the nematode Caenorhabditis elegans. In these organisms, the downregulation of LIN-14 protein was found to be essential for the progression from the first larval stage (L1) to L2. Furthermore, the downregulation of LIN-14 was found to be dependent on the transcription of a second gene called lin-4. Interestingly, the transcribed lin-4 was not translated into a biologically active protein. Instead, it gave rise to 2 small RNAs approximately 21 and 61 nucleotides in length. The longer sequence formed a stem-loop structure and served as a precursor for the shorter RNA. Later this group,93 along with Wightman et al,183 found that the smaller RNA had antisense complementarity to multiple sites in the 3′ UTR of lin-14 mRNA. The binding between these complementary regions decreased LIN-14 protein expression without causing any significant change in its mRNA levels. These 2 studies together brought forth a model wherein base pairing occurred between multiple lin-4 small RNAs to the complementary sites in the 3′ UTR of lin-14 mRNA, thereby causing translational repression of lin-14 and subsequent progression from L1 to L2 during C. elegans development.
This novel mode of regulating gene expression was first thought to be a phenomenon exclusive to C. elegans. In 2000, 2 separate groups discovered that a small RNA, let-7, was essential for the development of a later larval stage to adult in C. elegans.141,159 More importantly, homologues of this gene were subsequently discovered in many other organisms, including humans.133 The period that followed was marked by a deluge of information wherein multiple laboratories cloned numerous small RNAs from humans, flies, and worms. These RNAs were noncoding, around 19 to 24 nucleotides in length, and derived from a longer precursor with a stem-loop or fold-back structure.9 Many were found to be evolutionarily conserved across species and exhibited cell-type specificity. The recognition and confirmation of the existence of these small RNAs, now termed microRNAs (mi-RNAs), led to intense research aimed at identifying new members of this family. This resulted in the discovery of multiple miRNAs across different species of plants and animals. An miRNA registry, named miRBase, set up in 2002 serves as the primary online repository for all potential miRNA sequences, annotation, nomenclature, and target prediction information.53,55 The current release (miRBase 20) contains 24 521 entries representing hairpin precursor miRNAs that express 30 424 mature miRNA products in 206 species. The biological significance of a vast majority of annotated miRNAs, however, remains unknown and requires functional validation.
Biogenesis of miRNAs
miRNA genes can vary widely in their location in the genome. Earlier studies had revealed 2 distinct classes of miRNAs: those that originated from overlapping introns of protein coding transcripts and others that are encoded in exons, underscoring the complexities associated with miRNA maturation.143 Clusters of miRNA genes that coex-press polycistronically with the potential to be transcribed as a single unit were also discovered.86,90 It is estimated that approximately 50% of miRNAs are expressed from non– protein coding transcripts.147 The rest are mostly located in the introns of coding genes and are generally cotranscribed with their host genes and processed separately.147 Since this is a rapidly evolving field, there is potential for future developments to significantly overhaul the current understanding of miRNA genesis. Based on current knowledge, it can be stated that genomic regions capable of generating mature functional miRNAs can be present on diverse locations within the genome.
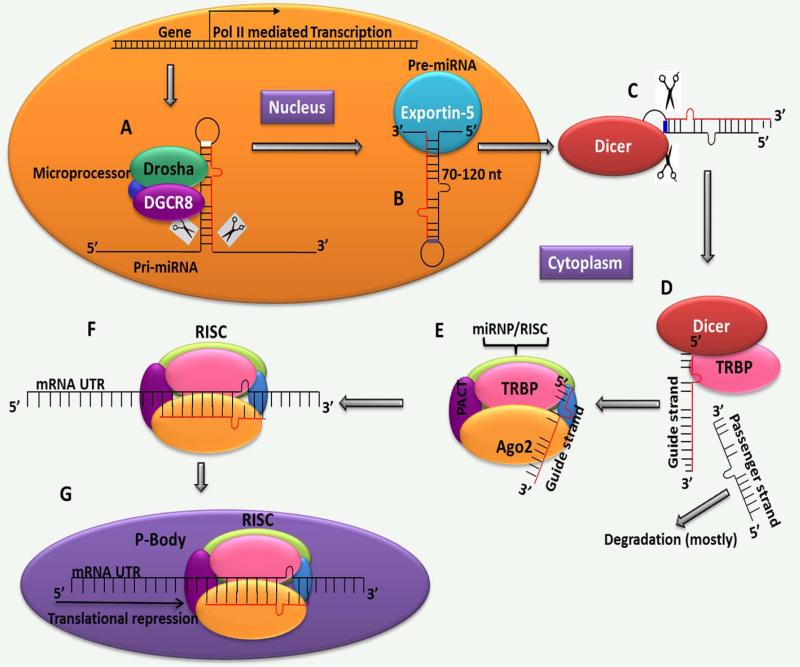
A general overview of the steps involved in miRNA biogenesis is illustrated in Fig. 1. miRNA coding transcripts are initially transcribed by RNA polymerase II as long primary miRNAs (several hundred nucleotides long) with a 5′ guano-sine cap and a 3′ polyadenylated tail. These can be either non-coding or coding (present within the intron of a coding gene). The primary miRNA is then processed into ~70- to 120-nucleotide-long precursor RNA (pre-miRNA) by a multiprotein complex called Microprocessor (Fig. 1A). This complex contains a ~160-kDa nuclear RNase III enzyme called Drosha.94 This enzyme is highly conserved in animals but not in plants.185 Drosha dimerizes with another double-stranded RNA (dsRNA) binding protein, called DiGeorge syndrome critical region gene 8 (DGCR8) or Pasha, to form the functional Microprocessor complex (Fig. 1A).34,52,59,88 The newly transcribed pre-miRNA with a typical 5′ phosphate and ~2-nucleotide 3′ overhang is then exported into the cytoplasm by exportin 5 (Exp-5), a Ran-dependent nuclear transport receptor protein (Fig. 1B).110,195 In the cytoplasm, the pre-miRNAs are finally processed into mature ~18- to 23-nucleotide-long duplexes by another RNase III enzyme, Dicer-1, with help from dsRNA-binding proteins, protein kinase RNA activator and transactivation response RNA binding protein (Fig. 1C,D).28,95,96,195 The 2 miRNA strands are then separated, depending on various factors such as thermodynamic asymmetry of the duplex and stability of base pairing at the 5′ end. One strand, termed the guide strand, along with the aforementioned and other RNA binding proteins that include trinucleotide repeat-containing gene 6A (TNRC6A), associates with catalytic Argonaute (AGO) proteins, forming a microribonuclear protein complex (miRNP) called RNA-induced silencing complex (RISC) (Fig. 1E).154 The miRNA strand with the most unstable base pairing at the 5′ end usually acts as the guide strand, while the strand with stable base pairing at the 5′ end (known as the passenger or miR* strand) is usually degraded or, in rare cases, even associates with AGO proteins, enabling both strands to serve as functional miRNAs.33,129 The guide strand directs the complex to the target mRNA through sequence complementarity and causes its translational repression (Fig. 1F). Ago2 proteins have been localized to cytoplasmic bodies called GW/P-bodies (processing bodies), where miRNAs bound to their mRNA targets are believed to be stored for degradation or translational repression (Fig. 1G).23 However, recent evidence also demonstrates thatmiRNA biogenesiscan be Microprocessor independent. Examples include pre–miRNA-like hairpins named “Mirtrons” formed from spliced and disbranched short hairpin introns, some small nucleolar RNAs (snoRNAs), and endogenous short hairpin RNAs (shRNAs).7,41,128,149
Figure 1.
Canonical microRNA (miRNA) biogenesis pathway: miRNAs are initially transcribed as long, variable-length hairpin RNA substrates called primary miRNAs (pri-miRNAs) directly off the DNA template in the nucleus by RNA polymerase II (A). In the nucleus, these pri-miRNAs are again processed by the Microprocessor, a large complex that includes the RNase type III enzyme called Drosha and the RNA binding protein DGCR8, into ~70- to 120-nucleotide-long premature (pre) hairpin precursor forms called pre-miRNAs (A). The pre-miRNAs are then exported to the cytoplasm by exportin 5 (B), where the loop region of the pre-miRNA is cleaved by the RNAse type III enzyme called Dicer into ~18- to 23-nucleotide-long mature miRNAs (C). A cellular protein called transactivation response RNA binding protein (TRBP) (D) facilitates the entry of the Dicer-miRNA complex into the RNA-induced silencing complex (RISC) that contains Argonaute 2 (Ago2), protein kinase RNA activator (PACT), trinucleotide repeat-containing gene 6A (TNRC6A), and other RNA binding proteins (E). After incorporation of the mature miRNAs into the RNA-induced silencing complex (RISC), the “passenger” strand is degraded, and the “guide” strand is guided to the 3′ UTR of the target messenger RNA (mRNA) to either degrade (in case of perfect base complementarity) or bring about translational inhibition (in case of multiple sequence mismatches between the target mRNA and the miRNA) (F). Mounting evidence suggests that transcripts bound by miRNA-incorporated Ago2 are channeled into structures called P-bodies, resulting in translational repression (G). (Loosely adapted and redrawn from Yeung et al.192)
Various mechanisms have been proposed on the modus operandi of miRNAs that results in posttranscriptional repression of target mRNA. The repression can be the result of either reduced translational efficiency or due to an actual decrease in the mRNA levels. Degree of complementarity between the miRNA and the target regions in the mRNA is thought to have a role wherein sufficient complementarity is believed to result in mRNA degradation while less complementarity leads to translational inhibition.67 Nevertheless, there are exceptions to this rule where even near-perfect complementarity leads to translational suppression but not cleavage.6 An alternative mechanism of action has also been proposed where binding of miRNAs leads to faster deadenylation of mRNAs, thereby decreasing mRNA stability and accelerating their degradation.186 Most recent studies show that mRNA destabilization with a resultant decrease in the mRNA levels is predominantly responsible for the reduced protein output than just inhibition of the translational machinery.57 An independent study also demonstrated that miRNAs can function as “decoys” and interfere with the functioning of regulatory proteins by binding to mRNA without necessitating a seed region.37 The notion that the 3′ UTR is the sole target of miRNA interactions is also changing as some recent studies have demonstrated that miRNA binding sites can be present both in 5′UTR and within the coding sequences.111,168
At least 1 study also identified miRNAs to function as activators of target mRNA transcription that play a role in maintaining quiescence during cell cycle, alluding to the possibility that, in rare instances, they can perform dual roles as activators and repressors.174 Bioinformatic predictions indicate that mammalian miRNAs may regulate up to 30% of all protein coding genes.101 Emerging evidence also suggests that miRNAs not only play a major role in regulating gene expression but also represent a very critical cellular factor with enormous capability to fine-tune biological processes. In this context, it is remarkable to note that these tiny RNA molecules with such important and ubiquitous roles in regulating cellular processes managed to evade the scientific community's radar for most of the 20th century.
Nomenclature of miRNAs
A uniform system of annotation and nomenclature has been adopted to ensure uniformity and ease of cataloging miRNAs.2,53–55 miRNAs are numbered sequentially in the order they are discovered. Those that have been experimentally confirmed are assigned a number that is attached to the prefix “miR” followed by a dash (eg, miR-21). In the identifier hsa-miR-21, the first 3 letters indicate the organism (eg, hsa for Homo sapiens, mml for Macaca mulatta). The mature miRNA is denoted as miR-21 (with a capitalized R), while the uncapitalized mir-21 refers to both the miRNA gene and the predicted stem-loop component of the primary transcript, also known as the precursor miRNA. Identical mature miRNA sequences that originate from discrete precursor sequences and genomic loci are given identifiers that contain a numeric suffix such as hsa-miR-219-1 and hsa-miR-219-2. On the other hand, closely related mature sequences that differ by 1 or 2 nucleotides are named with a lettered suffix. This would mean that hsa-miR-130a and has-miR-130b are derived from precursors hsa-mir-130a and hsa-mir-130b, respectively. Recent deep sequencing studies have revealed that individual pre-miRNAs can give rise to multiple mature miRNA species/sequences that can vary in length or sequence after being subjected to a variety of modifications that include trimming at the 5′/3′ ends, substitutions, insertions, deletions, or additions at the 3′ end.119 Termed isomirs, their biological significance is a subject of intense investigation. Also, efforts are being made to modify the nomenclature system so as to incorporate these variants. Some miRNA precursors give rise to two ~21- to 23-nucleotide-long mature miRNAs. When the predominantly expressed miRNA species can be definitively established, it is named as mentioned above (eg, miR-136) while the one originating from the opposite strand of the precursor is designated the same name but with an asterisk next to the number (miR-136*). However, when determination of the predominantly expressed species is not possible, identifiers such as miR-502-5p (from the 5′ strand) and miR-502-3p (from the 3′ strand) are assigned. miRNAs can also be found as clusters and present in close proximity within the genome. For example, the miR-17 cluster contains 6 precursor miRNAs located within a 1-kilobase region of chromosome 13 that can give rise to 7 mature miRNAs—namely, miR-17, miR-91, miR-18, miR-19, miR-19b, miR-20, and miR-92.96 These clusters have been denoted in the literature as either an miR-17 cluster (designated with only the smallest numbered miRNA) or an miR-17-92 cluster (contains both the lowest and highest numbered miRNA).
MiRNAs in Development and Organogenesis
Animals that do not express miRNAs fail to survive or reproduce normally.12,76,81,182 The universal impairment of the miRNA pathway by knocking down Dicer in fruit flies and mice caused embryonic lethality with abnormal morphology in almost all organs and resulted in significant lack of stem cells.3,12,61,62,81,193 Embryonic stem cell differentiation, a cardinal event in the development of organs and organ systems, is significantly modulated by miRNAs.114 Studies show that miRNAs can either promote or inhibit stem cell renewal depending on the cell type or culture environment. In mice, even though Dicer deficiency did not stop the formation of stem cell colonies, it severely impaired the differentiation capability of embryonic stem cells and caused considerable defects in cellular morphology.75 A subgroup of miRNAs from the miR-290 cluster (cluster implies that these miRNAs are coexpressed from a single transcript) was shown to regulate the embryonic stem cell (ESC) cycle and is now termed the ESCC family of miRNAs (ESC cell cycle–promoting miRNAs).114,178 ESCC miRNAs directly target the cell cycle inhibitors p21 and LATS2, thus facilitating G1-S phase transition.178 Moreover, transcription factors such as Oct3/4, Nanog, and Sox2, which are critical for maintaining pluripotency, have been shown to bind to the promoter of the miR-290 cluster and sustain its expression, thereby promoting self-renewal and maintenance of the pluripotent state.103,112 ESC miRNA knockout (through deletion of Dicer or Dgcr8) in mouse models resulted in an altered cell cycle profile with an extended G1 phase.75,179 As ESCs transition from a self-renewing to a differentiated state, several ESCC miRNAs show a gradual decrease in expression levels. In contrast, miRNA let-7 acts as a suppressor of pluripotency and is known to antagonize the effects of the miR-290 cluster. Unlike the miR-290 cluster, upregulation of let-7 was detected in the differentiated state, suggesting that its antagonistic effect may help stabilize the differentiated state.114,115
Similarly, miRNAs are also important in regulating the proliferation and differentiation of hematopoietic stem cells. miR-125b performs a specialized role not only in regulating hematopoiesis at the stem cell level but also in modulating inflammation and innate immunity by specifically promoting the differentiation and activation of macrophages.25,155 This proinflammatory effect of miR-125b was shown to be mediated predominantly via regulation of the nuclear factor (NF)–κB pathway. Interestingly, the dysregulation of miR-125b has been reported in multiple human cancers, including leukemia, and causes acute myeloid and lymphoid leukemias in mouse models.42,126
miRNAs have been described to play a major role in orchestrating the coordinated development of various organ systems. Although ubiquitously expressed, temporal and spatial expression of distinct sets of tissue-specific miRNAs is important in modeling tissue development and differentiation. miR-273 is required for establishing left-right asymmetry during neuronal development.64 In mouse heart, the fact that even deletion of 1 of the 2 genes coding for miR-1 (miR-1-2) caused severe and irreparable defects in cardiac morphology suggests critical roles for this miRNA in regulating cardiogenesis.198 The highly conserved miR-1 is the most abundant miRNA in adult heart, and its known functions include controlling cardiac morphogenesis, electrical conduction, and the cell cycle. miR-1 has been proposed to regulate cardiogenesis by fine-tuning the expression of Hand2, a transcription factor essential for cardiac development.199 Other validated targets of miR-1 include insulin-like growth factor 1, calmodulin, and myocyte enhancer factor 2A, all of which have been well documented to cumulatively contribute to the development of cardiac hypertrophy.1,39,68 In mice, deletion of miR-208 markedly impaired the ability of the heart to respond to stress stimuli.173 Double gene knockout of yet another muscle-enriched miRNA, miR-133a, in mice resulted in increased proliferation and apoptosis of myocytes, ventricular septal defects, and embryonic lethality. Those that survived developed severe cardiac dilatation and failure.106 In skeletal muscle, upregulation of miR-27 and subsequent downregulation of its target protein, Pax3, were found to be important in reducing myocyte proliferation and facilitating myogenic differentiation.32 Besides cardiac and skeletal muscle, miRNAs also exert specific functions in skin development. miR-203 is induced during differentiation and stratification of mouse skin, which in turn controls the basal to suprabasal transition by repressing p63, a member of the p53 family of transcription factors.99,194 On the contrary, p63 represses miR-34a/c expression to maintain cell cycle progression and thus antagonize the effects of miR-203.5,153 Along similar lines, expression of miR-124 was found to be essential for proper development of the nervous system.36,163
Systematic expression of miR-127 was found to be essential for proper branching of lung in rat fetal lung cultures. Its untimely overexpression caused defective branching and malformation of lung buds.13 Transgenic overexpression of the miR-17-92 cluster induced proliferation and inhibited differentiation in lung epithelial progenitor cells.109 Another important functional role for miRNAs was documented in insulin secretion, where miR-375 expression in pancreatic islets directly altered exocytosis of insulin from pancreatic beta cells.139 In mice, miR-143 regulates adipocyte differentiation and miR-122 regulates lipid metabolism by altering the expression of multiple target genes.43,44 In the proximal convoluted tubules of mouse kidney, miR-192 suppresses Na+/K+-ATPase, the major transporter of solutes and fluids in renal epithelial cells, and contributes to renal handling of fluid balance during increased sodium/water intake.118 There studies, together with the recent developments in high-throughput miRNA profiling that have investigated the spatial and temporal expression patterns in specific organ systems, including whole organisms, have unequivocally established the role and relevance of miRNAs in animal development and organogenesis.
miRNAs in Diseases
miRNAs in cancer
Apoptosis plays a significant role in both animal development and disease, and the dysregulation of this process has been invariably linked to the progression of various neoplastic processes. miRNAs that regulate apoptosis, termed apoptomiRs, can be either pro- or antiapoptotic. The first miRNA described as a regulator of apoptosis was the Drosophila gene bantam, which directly suppressed the proapoptotic factor hid, thus facilitating proliferation.18,152 Understandably, many miRNAs that play a role in modulating apoptosis have also been linked to the initiation and progression of various neoplastic processes. Approximately 50% of miRNAs are located at genomic sites that are disrupted or amplified in various cancers.21 The first evidence of miRNAs playing a role in cancer (termed oncomiRs) development came to light in 2002 in a study20 that attempted to find tumor suppressor genes at chromosome 13q14, which is frequently deleted in chronic lymphocytic leukemia (CLL).69 CLL is characterized by the presence of substantially increased numbers of predominantly nondividing malignant B cells overexpressing the antiapoptotic B-cell lymphoma 2 (Bcl2) protein. In patients with CLL, the tumor suppressor locus on chromosome 13q14 was found to be frequently altered. However, instead of coding for a tumor suppressor protein, this region contained 2 miRNA genes, miR-15a and miR-16-1, which, when overexpressed, were found to negatively regulate antiapoptotic Bcl2 gene at the posttranscriptional level. Later, many miRNAs that have tumor suppressor roles were identified. The miR-34 family, for example, has been shown to exert significant tumor suppressor capabilities. Upregulation of p53 (a potent tumor suppressor/cell cycle regulator) caused increased miR-34 expression that resulted in G1 arrest in a complementary and parallel fashion to mRNAs that are directly activated by p53.17,152 Also, miR-34 was shown to inhibit the silent mating information regulator 1 (SIRT1) gene that resulted in the upregulation of p53, p21, and PUMA (p53-upregulated modulator of apoptosis), thus regulating cell cycle and apoptosis and functioning as a tumor suppressor by modulating the SIRT1-p53 pathway.190 Furthermore, miR-34 has mediated growth arrest via direct regulation of cell cycle regulatory factors, such as cyclin E2 (CCNE2), cyclin-dependent kinase 4 (CDK4), E2F3, and the hepatocyte growth factor receptor (c-Met), ultimately leading to increased caspase-dependent cell death.152,181 In a separate study, miR-34 inhibited the proliferation/growth of human pancreatic tumor–initiating cells, and its overexpression in p53-deficient human pancreatic cancer cells partially restored the tumor-suppressing function of p53.70 MCL-1, a member of the BCL-2 family, was also demonstrated to be posttranscription-ally regulated by miR-29a, b, and c.121,152 Forced expression of miR-29b to induce tumor cell apoptosis by reducing MCL-1 expression may represent a novel intervention for cancer therapy. Along similar lines, let-7a exerts tumor suppressor functions by directly targeting the expression of RAS and HMGA2, 2 widely recognized oncogenes.71,97 Other examples of tumor suppressor miRNAs include miR-7, miR-124, miR-137, miR-146b, miR-15b, miR-128, and miR-326.151 Furthermore, global knockdown of mature miRNAs by selectively targeting Dicer1, RNASEN, and its cofactor DGCR8 increased the oncogenic potential of transformed cell lines, resulting in accelerated tumor formation in mouse models of K-RAS–driven lung cancer and Rb-driven retinoblastoma.47,84,85,87
Apart from functioning as tumor suppressors, miRNAs can also promote tumor development (oncogenes) depending on the functions of the target protein(s) they regulate. These oncogenic miRNAs include miR-155 and the miR-17-92 cluster that accelerated tumor development in B-cell lymphomas.38,63 Ectopic expression of miR-155 in transgenic mice resulted in pre–B-cell expansion, splenomegaly, and lymphopenia that preceded the development of lymphoblastic leukemia and lymphoma.31 mir-155 is now known to play a critical role in the development of lymphomas, although the components of its upstream regulatory pathways and downstream targets remain unclear. It is interesting to note that even before the discovery of miRNAs in mammalian cells, Tam et al166 had reported that the “bic” locus, the common retroviral integration site for the avian leukosis virus, generated a noncoding RNA.151 Later, after the discovery of miRNAs, it was found that this transcript harbored the mature miR-155 coding sequence, thus offering a potential explanation for the function of bic.151 Members of the mir-17-92 cluster are potent activators of cell proliferation and are frequently overexpressed in several neoplasms, including lymphoma, multiple myeloma, medulloblastoma, and cancers of the lung, colon, breast, and prostate.47 miR-21 is another commonly upregulated miRNA in cancers that include glioblastoma, lymphomas, and cancers of the breast, ovary, colon, rectum, pancreas, lung, liver, gallbladder, prostate, stomach, thyroid, and cervix.151 Increased expression of miR-21 was found in glioblastoma tumors and cell lines, and its inhibition resulted in increased cell death, suggesting that miR-21 could play the role of an oncogene that inhibited cell death in these tumors.24 Furthermore, in glioblastoma cells, knockdown of miR-21 induced the activation of caspase-3, transforming growth factor–β, p53, and mitochondrial apoptotic pathways mainly through upregulation of its validated targets, heterogeneous nuclear ribonucleoprotein K, p53-related TAp63, and PDCD4, acting in synergy with the aforementioned proteins.27,132,151
An individual miRNA can also potentially perform dual functions as an oncogene and tumor suppressor if its targets include antiproliferative genes and growth-promoting genes, respectively. miR-26 functions as an oncogene in glioma and glioblastoma multiforme by regulating PTEN, the molecular antagonist of the Akt pathway, resulting in the inhibition of RB1 and MAP3K2/MEKK2 expression and JNK-dependent apoptosis.66,78 miR-26 is also thought to function as a tumor suppressor as its expression was either downregulated or its downregulation resulted in increased anaplasia or metastasis in various neoplasms (eg, hepatocellular carcinoma, breast cancer, squamous cell carcinoma, thyroid cancers, rhabdomyosarcoma, Myc-induced lymphomas) (reviewed in Gao and Liu49). More recently, miRNA expression in cancers was shown to be controlled by epigenetic mechanisms such as DNA methylation. In primary lung tumors, methylation of CpG islands in genes for miR-9-1, miR-9-3, miR-34b/c, and miR-193a resulted in their downregulation with a concurrent increase in the expression of their target genes, RAR-β2 and NKIRAS1.77 Other examples include hypermethylation of miR-91, miR-124a-3, miR-148, miR-152, and miR-663 in human breast cancer and aberrant DNA methylation and downregulation of miR-127 in prostate and bladder cancers.98,148 In the latter example, chromatin-modifying drugs successfully restored miR-127 expression and downregulated its predicted target Bcl6, a proto-oncogene, thereby highlighting the therapeutic potential of chromatin-modifying drugs in modulating miRNA expression.
High-throughput techniques such as miRNA microarrays, which provide unique miRNA expression signatures (miRNomes) of different cancers and their subclassifications, are now being used for both diagnostic and classification purposes. miRNomes have proved to be more reliable than mRNA profiles in some tumor subclassifications wherein arrays of ~200 miRNAs provided a better classification of tumors by type and source than a collection of >15 000 mRNAs.108 Furthermore, miRNAs are more stable than mRNAs in both body fluids and routinely collected formalin-fixed, paraffin-embedded tissues.108 Recent studies have also detected their presence in plasma and serum of patients (discussed later), thus providing a more convenient and noninvasive approach for miRNA profiling. These developments, coupled with the unique properties of miRNAs, have opened up exciting new avenues for the classification, diagnosis, prognosis, and treatment of various cancers.
miRNAs in infectious diseases
miRNAs can influence the manifestation and pathogenesis of infectious diseases in a multitude of ways. These include modulating the (1) pathogenicity of individual pathogens, (2) the efficiency of host innate and adaptive immune response, and (3) the magnitude and resolution of inflammatory responses. Providing an extensive coverage of the broad range of regulatory roles played by miRNAs in immunity and inflammation is beyond the scope of this review, and the readers are referred to a number of excellent reviews† that have dealt with this topic in greater detail. However, it is relevant to note that changes in miRNA expression during inflammation/immune response are controlled not only by altered transcriptional levels but also by interaction of products of inflammatory/immune responses with the structural and functional components of the miRNA biogenesis pathway.
Studies that have focused on viral pathogenesis pathways revealed that viral infections can alter the levels of host miRNAs that specifically regulate antiviral mechanisms, viral latency, and lytic cycles. These miRNAs either directly target the viral RNA or the host cell factors vital for their replication. Several viruses encode their own miRNAs (v-miRNAs) that in turn regulate their production in host cells.136 v-miRNAs target and downregulate specific host genes, thereby creating a cellular environment that is permissive for virus replication. Also, from an evolutionary standpoint, it is advantageous and faster to generate a miRNA complementary to a new target gene than produce a regulatory protein that performs the same function. Unlike eukaryotic miRNAs, v-miRNAs are generally not conserved.151 Pioneering studies done in Epstein-Barr virus (EBV) latently infected B cells have shown that several unique v-miRNAs originated from the Bam H1 fragment H open reading frame 1 (BHRF1) and the Bam H1-A region rightward transcript (BART) present in the viral genome.136 Some of these v-miRNAs are thought to target lytic genes, suppress viral proliferation, and sustain latency. EBV miRNAs have also been detected in various types of lymphomas that tested positive for EBV. Interestingly, EBV is also known to increase its copy numbers in latently infected cells by hijacking the functions of the host cellular miRNAs, particularly miR-155.82,189 Other miRNAs that were reported to be upregulated during latency include miR-21, miR-23a, miR-24, miR-27a, and miR-34a, while miR-96 and miR-128a/b were downregulated in lymph nodes of patients with EBV-positive classic Hodgkin lymphoma.22,123
Viruses such as human immunodeficiency virus 1 (HIV-1) have also been reported to suppress miRNA-mediated silencing during replication in host cells.169 Components of the host miRNA processing machinery—namely, Dicer and Drosha—were found to be essential in inhibiting virus replication both in HIV-1–infected peripheral blood mononuclear cells and latently infected cells. In this study, when Dicer and Drosha were knocked down using small interfering RNAs (siRNAs), virus replication kinetics was enhanced compared with cells transfected with a nonfunctional siRNA. Furthermore, HIV-1 also actively suppressed the expression of the polycistronic miRNA cluster miR-17/92, which enabled efficient viral replication by upregulating its target histone acetyltransferase Tat cofactor, named PCAF. Along similar lines, the HIV-1 Tat protein, a transcriptional activator with a basic RNA binding domain that can inhibit interferon response, actively suppressed miRNA-siRNA processing by interfering with Dicer activity.11 In addition, cellular miRNAs such as miR-28, miR-125b, miR-150, miR-223, and miR-382, can inhibit HIV-1 replication by binding to complementary sites located within the viral genome.65,170 These anti-HIV miRNAs were found to be enriched in resting CD4+ T cells and were thought to contribute to the development of proviral latency. Interestingly, these miRNAs were also found to be differentially expressed between monocytes and macrophages, providing an explanation as to why macrophages and not monocytes are permissive to HIV infection and replication.177
Other examples of cellular miRNAs modulating viral expression include targeting of (1) influenza virus replication by miR-323, miR-491, miR-654, and let-7c; (2) primate foamy virus 1 by miR-32; (3) vesicular stomatitis virus by miR-24 and miR-93; (4) hepatitis B virus by miR-125a-5p, miR-199a-3p, and miR-210; and (5) hepatitis C virus (HCV) by miR-196, miR-296, miR-351, miR-431, and miR-448.‡ A unique and unexpected finding in HCV was that a liver-specific miRNA, miR-122, directly targeted the viral RNA sequence to upregu-late virus replication.72 This finding is very intriguing as it represents a major deviation from the usual norm that host miRNAs generally repress viral replication.80
Given the dependence on miR-122 for HCV replication, blockade of miR-122 function using an experimental DNA-based drug SPC3649 (locked nucleic acid–modified antagomir to miR-122) successfully inhibited HCV replication in chimpanzees.89 SPC3649 administration at the highest dose produced prolonged suppression of HCV viremia in blood and liver without the emergence of escape mutants. Furthermore, the drug successfully alleviated HCV-induced liver pathology and produced no side effects in treated animals. This drug is currently being evaluated in phase II clinical trials in humans and, due to the lack of any detectable side effects, has great potential to become the first miRNA-based therapeutic intervention for HCV infection.
Various bacterial diseases also cause marked alterations in the expression of miRNAs, especially in cells involved with the immune response. Recent studies have characterized changes in host miRNA expression following infection with a wide array of bacterial organisms that include both extracellular (eg, Helicobacter pylori) and intracellular (eg, Salmonella enterica) pathogens. Dysregulated miRNAs have also been demonstrated to affectthe severity of sepsis that follows bacterial infections. Upregulation of miR-155 and downregulation of miR-146a resulted in increased severity of sepsis in murine models of endotoxemia. miR-155 repressed SHIP1 and SOCS1, 2 negative regulators of inflammation, while repression of miR-146a led to upregulation of its targets IRAK1 and TRAF6, 2 proinflammatory signaling proteins associated with the TLR/IL-1R pathways.4,16,79 Upregulation of miR-155 has also been shown to potentiate the immune response against Salmonella typhimurium.144
In Mycobacterium tuberculosis infections, a novel host evasion mechanism mediated by miR-99b has been described wherein miR-99b expression was high in infected dendritic cells and macrophages, and its blockade resulted in significantly reduced bacterial growth.158 This study also found that knockdown of miR-99b resulted in the upregulation of proinflammatory cytokines such as interleukin (IL)–6, IL-12, and IL-1b and augmented tumor necrosis factor–α (TNF-α) and TNFRSF-4 production. A second study showed that miR-21 can be induced after Bacillus Calmette-Guérin (BCG) vaccination via NF-κB activation.188 miR-21 suppressed IL-12 production by targeting IL-12p35, which in turn impaired anti-mycobacterial T-cell responses and promoted dendritic cell apoptosis by targeting Bcl-2. An analysis of differentially expressed miRNAs that might play a role in the preferential development of a lepromatous form of leprosy over the tuberculoid form found significant enrichment of miR-21 in Mycobacterium leprae–infected monocytes in the lepromatous form.107 In this study, miR-21 directly downregulated TLR2/1-induced CYP27B1 and IL-1b expression and indirectly upregulated IL-10. The end result of these changes was the inhibition of expression of genes encoding 2 vitamin D–dependent antimicrobial peptides, CAMP and DEFB4A, thus providing an effective mechanism for the bacteria to escape from the vitamin D–dependent antimicrobial pathway. This ability of M. leprae to upregulate miR-21 is thought to aid in the progression from the self-limiting tuberculoid form to the progressive lepromatous form of leprosy. The miRNA expression profiles of various infectious diseases, including fungal pathogens, are now available, and the functional implications of these profiles, especially in understanding the pathogenic mechanisms and in developing miRNA-based antimicrobial therapeutics, are aggressively pursued areas of research.
miRNAs in other noninfectious diseases
Dysregulation of miRNA expression has been not only associated with the manifestation of developmental defects in various organisms and organ systems as discussed earlier but also implicated in a wide array of other noninfectious diseases, including autoimmune diseases, metabolic disorders, and genetic diseases. miR-155 and miR-326 are overexpressed in human multiple sclerosis (MS).35,73,74,127 In vivo silencing of these miRNAs in a mouse model of MS—namely, experimental autoimmune encephalomyelitis—demonstrated that their functional relevance could be attributed to their ability to enhance Th17 response and modulate T-cell developmental pathways by affecting the expression of a complex array of targets.73 Excess Th17 response is thought to play a key role in the manifestation of various autoimmune diseases. Multiple studies have looked at miRNA expression profiles in various tissues from patients with systemic lupus erythematosus (SLE) or animal models of SLE with considerable variation in the miRNA signatures reported between studies. Most notably, miR-146a was significantly decreased in patients with SLE and was strongly associated with clinical disease activity and activation of the type I interferon (IFN) pathway.134,156 Reduced expression of miR-146a resulted in aberrant accumulation of its target proteins (STAT1, IRF5, TRAF6, and IRAK1), leading to a long-lasting hyperactivation of the IFN pathway and disease manifestation.167 Patients with SLE are known to display aberrant DNA hypomethylation, and elevated expression of 2 miRNAs, miR-21 and miR-148, has been proposed to contribute to the development of SLE through their effects on inhibiting DNA methylation in T cells.156,167
The role of miRNAs as key regulators of metabolism and the therapeutic implications of these miRNAs in treating metabolic disorders are also being investigated. miRNAs are known to play a major role in controlling glucose homeostasis and insulin signaling. Aberrant miRNA expression in cardiometabolic disorders such as obesity, fatty liver disease, insulin resistance, type 2 diabetes, and coronary artery disease highlights their roles in the manifestation of their respective pathologies. miR-122, expressed primarily in the liver, was the first miRNA to be linked to metabolic control.100 Early studies showed that miR-122 played a significant role in cholesterol and lipid regulation. Even partial silencing of miR-122 in mice resulted in a ~25% to 30% reduction in plasma cholesterol levels, decreased hepatic cholesterol and fatty acid biosynthesis, and an increase in fatty acid β-oxidation.43 The silencing of miR-122 also resulted in decreased hepatosteatosis in mice fed a high-fat diet. Similar effects of miR-122 silencing on circulating cholesterol levels were also observed in African green monkeys.40 However, the exact mechanisms underlying cholesterol lowering in response to miR-122 silencing remains unknown as genes in liver that are involved in cholesterol and lipid metabolism do not seem to be direct targets of miR-122. miR-33a is another miRNA known to control cholesterol homeostasis by cooperating with SREBP2, a cholesterogenic transcription factor, to boost intracellular cholesterol levels (reviewed in Rottiers and Naar146). Other miRNAs that play important roles in metabolic disorders include (1) miR-29, which activates insulin secretion, and miR-9 and miR-375, which inhibit insulin secretion via their effect on monocarboxylate transporter 1, one cut homeobox 2, sirtuin 1, phosphoinositide-dependent kinase 1, and myotrophin; (2) modulation of insulin signaling in adipose tissue by miR-103 and miR-107 by targeting caveolin 1 and miR-29; and (3) miR-34a in humans and rodent models of hepatic metabolic diseases, including obesity, type 2 diabetes, nonalcoholic fatty liver disease, and nonalcoholic steatohepatitis, most likely through an intricate regulatory network of miR-34a, SIRT1 (a key sensor and regulator of metabolic states), farnesoid X receptor, and p53 (reviewed in Rottiers and Naar146). Furthermore, miR-375 and miR-223 are also present in circulating high-density lipoproteins, indicating that disease-associated miRNAs can be transported in lipoprotein particles, thereby exerting their regulatory effect in a paracrine fashion on distant target tissues.175
The importance of miRNAs in genetic diseases is evident from their roles in development and organogenesis described earlier, wherein mutation of relevant miRNAs or target genes is associated with manifestation of a defective phenotype. However, only a few studies have established a clear link between miRNAs and specific genetic disorders. An excellent example of point mutations in the mature miRNA sequence playing an etiopathogenic role in a Mendelian disease includes 2 different nucleotide substitutions in the seed region of miR-96 (13G>A and +14C>A) in 2 Spanish families affected by an autosomal dominant form of deafness, DFNA50.116 A single base change (A>T) in the seed region of miR-96 in mice also resulted in a progressive hearing loss phenotype, providing convincing evidence that loss of miR-96 target gene regulation arising from point mutations in its seed region results in hearing loss phenotype in both human and mouse.102 Similarly, mutations and subsequent sequence variations can also occur in the 3′ UTR of mRNAs, thereby altering miRNA recognition/binding sites. Such a mechanism has also been demonstrated in an autosomal dominant form of hereditary spastic paraplegia (SPG31), in which 2 different point mutations in the predicted miR-140 binding site on the 3′ UTR of the REEP1 gene resulted in failure to regulate its putative target gene expression.10,201 At least a couple more studies have looked at human genetic diseases characterized by mutations in genes involved in miRNA processing/biogenesis. Mutations in the DGCR8 gene, a component of the Drosha complex, is present in DiGeorge syndrome, and loss of function of the FRM1 gene that codes for an RNA binding protein occurs in fragile X syndrome.104,157
miRNAs in Veterinary Medicine
Most miRNA studies are conducted in laboratory animals and cell lines in which the primary objective is to understand their roles in development and diseases specific to humans. Although studies have documented organ- and breed-specific miRNA signatures in various animal species through computational analysis and polymerase chain reaction/microarray profiling, studies that have directly addressed diseases/disorders specific to food, companion, avian, and exotic animals are extremely limited. The handful of studies in this direction is mostly limited to screening of miRNAs in specific conditions, tissues, or species and have not explored deeper into their mechanism of action and target validation. At least 2 miRNAs, miR-17-5p and miR-181a, have been described to be significantly upregulated in canine B- and T-cell lymphomas.120 Similarly, in canine osteosarcoma samples, an inverse correlation between decreased expression of miR-134 and miR-544 originating from the 14q32 locus and aggressive tumor growth characteristics was observed.150 These findings are in agreement with those reported in human osteosarcoma. In canine mammary cancers, especially in tubular papillary carcinomas, miR-29b and miR-21 were found to be significantly upregulated.15 miRNA expression profiling in canine oral melanoma tissues found that decreased expression of miR-203 was associated with a shorter survival time, thereby highlighting its potential as a new prognostic marker for this disease.125 The same study also showed marked downregulation of miR-203 and miR-205 in canine and human melanoma cell lines. Over-expression of miR-205 significantly inhibited the growth of canine and human melanoma cells in vitro by targeting erbb3, a member of the epidermal growth factor receptor (EGFR) family of receptor tyrosine kinases. A recent study compared miRNA expression profiles in the serum of Doberman Pinschers with dilated cardiomyopathy to healthy controls.164 The study detected the expression of a total of 404 miRNAs in serum, of which 22 showed differential expression between the 2 groups. Although none of the differentially expressed miRNAs attained statistical significance, the study certainly highlights the potential of miRNAs to serve as disease biomarkers in veterinary medicine. Another study along similar lines performed in cats detected a significant increase in miR-122 (>40-fold) and miR-139b (>14-fold) in the serum of newly diagnosed diabetic cats when compared with healthy lean cats and cats in diabetic remission.48
An interesting study in Texel sheep, renowned for their exceptional meatiness, identified a new class of regulatory mutations that offer some novel insights into the heritability of complex traits.29 In this study, the GDF8 allele that encoded for the myostatin gene was found to contain a G to A transition in its 3′ UTR, which in turn resulted in the creation of a target site for miR-1 and miR-206, 2 highly expressed miRNAs in skeletal muscle. This resulted in translational inhibition of the myostatin gene, thereby contributing to the development of muscular hypertrophy in this breed. Similar studies to investigate the role of miRNAs in economically important traits such as mam-mary gland development and skeletal muscle and adipose tissue development, which defines meat quality, are being pursued in multiple species and breeds of production livestock.56,113
miR-181, a well-known positive regulator of immune response, was shown to directly impair or even inhibit porcine reproductive and respiratory syndrome virus (PRRSV) infection.58 Synthetic miR-181 mimics significantly inhibited PRRSV replication in vitro in a specific and dose-dependent manner by binding to a highly conserved region downstream of open reading frame 4 (ORF4) of the viral genomic RNA. Studies that have profiled miRNAs in lung tissues of pigs infected with Actinobacillus pleuropneumoniae and porcine epithelial cells infected with pseudorabies have identified several miRNAs that might play roles in immune and inflammatory responses specific to both pathogens.137,187
miR-146a was elevated in the blood of ferrets, horses, and in a human cervical carcinoma cell line (HeLa) infected with Hendra virus (paramyxovirus, genus Henipavirus).165 Blockade of miR-146a function significantly reduced Hendra virus replication in vitro, suggesting a role for this miRNA in Hendra virus replication. This effect was mostly mediated through its target, ring finger protein, a member of the A20 ubiquitin editing complex that negatively regulates NF-κB activity. Increased NF-κB activity is thought to aid in activating and sustaining Hendra virus replication. A study that looked at specific miRNA profiles of myopathic horses with polysaccharide storage myopathy or recurrent exertional rhabdomyolysis in 2 different breeds indicated that it might be possible to distinguish one form from the other based on unique miRNA profiles.8 Interestingly, these miRNA profiles were also found to be breed specific.
In birds, several virus-encoded miRNAs were found to be conserved among different field strains of oncogenic Marek's disease virus (MDV1), and their expression has been detected in both lytic and latent infections, including MDV1-derived tumors.19 This study found that even though each avian herpes-virus had unique sequences, all originated from similar locations on the viral genome. Hence, these miRNAs tended to be clustered in the rapidly evolving repeat regions of the viral genomes. MDV1 and herpesvirus of turkeys (HVT) encode homologs of the host miRNA, miR-221, which targets a gene important in cell cycle regulation. MDV1 also encodes another miRNA (mdv1-miR-M4), which, together with Kaposi sarcoma–associated herpesvirus-encoded miR-K12-11, was verified as functional orthologs of miR-155, a well-characterized miRNA previously linked to lymphoid malignancies and modulation of immune responses.200 Novel miRNAs were also found to be encoded by duck enteritis virus, the functions of which remain to be determined.191 In zebrafish, miR-142-3p was found to be essential for hematopoiesis and affected the cardiac cell fate.124
Circulating miRNAs
Recent studies have shown that a significant number of miRNAs are present extracellularly in various body fluids that include serum, plasma, urine, saliva, and other body fluids.60,180,196 The origin and function of these miRNAs are not well understood. There are indications that they might play a role in cell-to-cell communication and can be exported from one cell and recognized, taken up, and used by another cell, thereby drawing parallels with the endocrine system.45,83,175,176 Termed circulating miRNAs, they are highly stable and resistant to RNase activity and extreme pH and temperatures, unlike conventional RNA molecules.26,117 Since body fluids contain ribonucleases, it is suggested that they are protected from degradation by being packaged in lipid vesicles (microvesicles and exosomes), in complexes with RNA binding protein or both.45,51,171 More importantly, they can be readily detected in serum and plasma, and their expression patterns have correlated positively with various diseases conditions.
In a study that looked at miRNAs in the serum of patients with diffuse large B-cell lymphoma (DLBCL), expression levels of 3 tumor-associated miRNAs—miR-155, miR-210, and miR-21—were significantly upregulated in patient sera.91 Also, patients with DLBCL with high miR-21 serum levels showed increased relapse-free survival but not overall survival. A comprehensive analysis of miRNAs in serum to characterize blood miRNA profiles from healthy individuals and patients found that patients with lung cancer, colorectal cancer, and diabetes had unique serum miRNA profiles.26 Patients with oral squamous cell carcinoma had higher levels of miR-31 in their plasma that decreased significantly following tumor removal, a trend that was also observed in patient saliva.105 In patients with rheumatoid arthritis and osteoarthritis, plasma miR-132 levels were significantly higher than those in healthy controls and correlated well with disease activity.122 Other studies have shown significant correlation between expression levels of specific circulating miRNAs and their expression in specific tissues with a few exceptions (reviewed in Zen and Zhang196). These findings also suggest that the levels of circulating miRNAs strongly correlate with the extent of tumor development.196 These properties and the opportunity to detect their expression by noninvasive means have led to numerous investigations geared toward developing miRNAs as reliable bio-markers. However, there are a few stumbling blocks in this direction. For instance, the specificity of biomarkers based on a single miRNA or a very small group of individual miRNAs is relatively poor, which might be due to the complex nature of certain malignancies, thus highlighting the need to discover and validate signature miRNA panels in complex malignances.196 Since all biological fluid samples are essentially cell free, a lot more needs to be done to optimize several key steps such as RNA extraction, normalization, quantitation, and analysis of serum miRNA profiles before they can be used as reliable biomarkers of specific disease conditions.
Future Implications and Concluding Remarks
The discoveries and current developments in miRNA biology have essentially revolutionized the biological landscape and have created renewed interest within the scientific community to reevaluate and significantly modify the conventional perceptions about gene expression, gene regulation, and RNA functionality. miRNA discovery effectively challenges the “central dogma of molecular biology,” in which RNA was assigned the role of a mere intermediary to the flow of information between the genetic material (DNA) and the end product (protein). The discovery and validation of miRNA function also refute the faulty “junk DNA hypothesis,” which stated that more than 80% of our DNA serves no biological purpose and is “junk” that has accumulated over time as evolutionary fossils.142 The implications of the insights coming out of studies in miRNA biology are far reaching. Many knockout or overexpression studies performed in the past to determine the function of individual genes invariably included deletion of introns. Since a significant number of miRNAs are now known to be present in introns, the inadvertent deletion of any miRNA coding sequences contained in these regions might also be a contributing factor to the phenotypic consequences observed in these individual knockout/overexpression studies that were attributed solely to the protein coding gene. Data in this direction indicate that many knockout studies need to be reexamined to determine if loss of a miRNA contributed to the phenotype, warranting careful planning of knockout studies in future.130
Besides regulating gene expression, very recent studies have identified novel functions for miRNAs as ligands capable of signal transduction by binding to cell surface receptors. In this regard, tumor-secreted miR-21 and miR-29a were recently shown to bind TLR7 and TLR8 in immune cells, resulting in the activation of a TLR-mediated prometastatic inflammatory response that enhanced tumor growth and metastasis.46
Since a single miRNA can regulate a number of genes and a single gene can be regulated by multiple miRNAs, loss of function of 1 or more miRNAs may not be evident at a functional level due to the inherent ability of the biological system to substitute 1 molecule with another capable of performing similar functions. Also, there is growing consensus that miRNAs, in many cases, are “fine-tuners” of gene expression, and hence, unlike genes encoding proteins, changes in spatial and temporal expression of miRNAs can be fleeting, can fluctuate rapidly, and might involve minimal changes in their expression levels. Hence, the ability to observe these changes is limited by the sensitivity and sophistication of the detection system. Although a lot of progress has been made in addressing these bottlenecks, more work is needed to better understand their biological function.
Therapeutic implications of miRNAs are enormous. Individual miRNAs important in the development of diseases can be specifically targeted using anti-miRNAs, which are antisense oligonucleotides with specific modifications.145 In cancer therapeutics, a class of cholesterol-conjugated (to facilitate better cellular uptake and serum protein binding) anti-miRNAs, termed antagomirs, was used to block oncomirs.145,172 Other approaches that efficiently inhibit miRNAs in vivo include locked nucleic acid (LNA) oligos or 2′-O-methoxyethyl phosphorothioate (MOE) modification.172 In African green monkeys, mature miR-122 was effectively and stably silenced using intravenously administered LNA–anti-miRNA-122 with no adverse effects.40 Many studies in this direction have yielded promising results. In diseases associated with downregulation of miRNAs (eg, tumor suppressors), a novel approach that involves delivering synthetic RNA duplexes that mimic the relevant miRNA duplexes, which can then be recognized by the RISC complex and processed into mature miRNAs, is being actively pursued. Although achieving stability and efficiency of the in vivo delivery of miRNAs is a major challenge, progress made in this direction indicates the rapid emergence of an entirely new field of small RNA-based therapeutics. These developments, together with their role as potential biomarkers (discussed previously), make them invaluable tools in probing multiple aspects of a disease process that includes diagnosis, classification, and treatment.
In conclusion, the aim of this review was not to comprehensively analyze the role of individual miRNAs in a specific disease or development process but rather to present the reader with the broad spectrum of capabilities that these small RNAs possess using some well-established examples. The discovery of miRNAs adds a previously overlooked and important variable that can substantially influence the development, manifestation, and progression of disease processes. Although most of the present studies in this direction are done in laboratory animals and aim to tackle human diseases, the knowledge and progress made in this field should be translatable into veterinary medicine in the near future.
Acknowledgement
We thank Drs Andrew A. Lackner and Chad J. Roy from Tulane National Primate Research Center (TNPRC) for their constructive criticism and advice on the manuscript.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship and/or publication of this article: This work was supported by National Institutes of Health grants OD011104 (formerly RR00164), R01DK083929 (to MM), and T32-OD011124 (MB).
Footnotes
References
- 1.Abdellatif M. Differential expression of microRNAs in different disease states. Circ Res. 2012;110:638–650. doi: 10.1161/CIRCRESAHA.111.247437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambros V, Bartel B, Bartel DP, et al. A uniform system for micro-RNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andl T, Murchison EP, Liu F, et al. The miRNA-processing enzyme Dicer is essential for the morphogenesis and maintenance of hair follicles. Curr Biol. 2006;16:1041–1049. doi: 10.1016/j.cub.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Androulidaki A, Iliopoulos D, Arranz A, et al. The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity. 2009;31:220–231. doi: 10.1016/j.immuni.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antonini D, Russo MT, De Rosa L, et al. Transcriptional repression of miR-34 family contributes to p63-mediated cell cycle progression in epidermal cells. J Invest Dermatol. 2010;130:1249–1257. doi: 10.1038/jid.2009.438. [DOI] [PubMed] [Google Scholar]
- 6.Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Babiarz JE, Ruby JG, Wang Y, et al. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrey E, Bonnamy B, Barrey EJ, et al. Muscular microRNA expressions in healthy and myopathic horses suffering from polysaccharide storage myopathy or recurrent exertional rhabdomyolysis. Equine Vet J Suppl. 2010;(38):303–310. doi: 10.1111/j.2042-3306.2010.00267.x. [DOI] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Beetz C, Schule R, Deconinck T, et al. REEP1 mutation spectrum and genotype/phenotype correlation in hereditary spastic paraplegia type 31. Brain. 2008;131:1078–1086. doi: 10.1093/brain/awn026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bennasser Y, Jeang KT. HIV-1 Tat interaction with Dicer: requirement for RNA. Retrovirology. 2006;3:95. doi: 10.1186/1742-4690-3-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernstein E, Kim SY, Carmell MA, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 13.Bhaskaran M, Wang Y, Zhang HH, et al. MicroRNA-127 modulates fetal lung development. Physiol Genomics. 2009;37:268–278. doi: 10.1152/physiolgenomics.90268.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhaumik D, Patil CK, Campisi J. MicroRNAs: an important player in maintaining a balance between inflammation and tumor suppression. Cell Cycle. 2009;8:1822–1822. [PubMed] [Google Scholar]
- 15.Boggs RM, Wright ZM, Stickney MJ, et al. MicroRNA expression in canine mammary cancer. Mamm Genome. 2008;19:561–569. doi: 10.1007/s00335-008-9128-7. [DOI] [PubMed] [Google Scholar]
- 16.Boldin MP, Taganov KD, Rao DS, et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med. 2011;208:1189–1201. doi: 10.1084/jem.20101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bommer GT, Gerin I, Feng Y, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 18.Brennecke J, Hipfner DR, Stark A, et al. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 19.Burnside J, Morgan R. Emerging roles of chicken and viral micro-RNAs in avian disease. BMC Proc. 2011;5(suppl 4):S2. doi: 10.1186/1753-6561-5-S4-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calin GA, Sevignani C, Dan Dumitru C, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cameron JE, Fewell C, Yin QY, et al. Epstein-Barr virus growth/ latency III program alters cellular microRNA expression. Virology. 2008;382:257–266. doi: 10.1016/j.virol.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castilla-Llorente V, Spraggon L, Okamura M, et al. Mammalian GW220/TNGW1 is essential for the formation of GW/P bodies containing miRISC. J Cell Biol. 2012;198:529–544. doi: 10.1083/jcb.201201153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 25.Chaudhuri AA, So AY, Sinha N, et al. MicroRNA-125b potentiates macrophage activation. J Immunol. 2011;187:5062–5068. doi: 10.4049/jimmunol.1102001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 27.Chen Y, Liu W, Chao TF, et al. MicroRNA-21 down-regulates the expression of tumor suppressor PDCD4 in human glioblastoma cell T98G. Cancer Lett. 2008;272:197–205. doi: 10.1016/j.canlet.2008.06.034. [DOI] [PubMed] [Google Scholar]
- 28.Chendrimada TP, Gregory RI, Kumaraswamy E, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clop A, Marcq F, Takeda H, et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38:813–818. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- 30.Contreras J, Rao DS. MicroRNAs in inflammation and immune responses. Leukemia. 2012;26:404–413. doi: 10.1038/leu.2011.356. [DOI] [PubMed] [Google Scholar]
- 31.Costinean S, Zanesi N, Pekarsky Y, et al. Pre–B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E mu-miR155 transgenic mice. Proc Natl Acad Sci U S A. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crist CG, Montarras D, Pallafacchina G, et al. Muscle stem cell behavior is modified by microRNA-27 regulation of Pax3 expression. Proc Natl Acad Sci U S A. 2009;106:13383–13387. doi: 10.1073/pnas.0900210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Czech B, Zhou R, Erlich Y, et al. Hierarchical rules for Argonaute loading in Drosophila. Mol Cell. 2009;36:445–456. doi: 10.1016/j.molcel.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denli AM, Tops BB, Plasterk RH, et al. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 35.Du C, Liu C, Kang J, et al. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009;10:1252–1259. doi: 10.1038/ni.1798. [DOI] [PubMed] [Google Scholar]
- 36.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eiring AM, Harb JG, Neviani P, et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140:652–665. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eis PS, Tam W, Sun L, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elia L, Contu R, Quintavalle M, et al. Reciprocal regulation of microRNA-1 and insulin-like growth factor-1 signal transduction cascade in cardiac and skeletal muscle in physiological and pathological conditions. Circulation. 2009;120:2377–2385. doi: 10.1161/CIRCULATIONAHA.109.879429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elmen J, Lindow M, Schutz S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 41.Ender C, Krek A, Friedlander MR, et al. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 42.Enomoto Y, Kitaura J, Hatakeyama K, et al. Emu/miR-125b transgenic mice develop lethal B-cell malignancies. Leukemia. 2011;25:1849–1856. doi: 10.1038/leu.2011.166. [DOI] [PubMed] [Google Scholar]
- 43.Esau C, Davis S, Murray SF, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 44.Esau C, Kang X, Peralta E, et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 45.Etheridge A, Lee I, Hood L, et al. Extracellular microRNA: a new source of biomarkers. Mutat Res. 2011;717:85–90. doi: 10.1016/j.mrfmmm.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fabbri M, Paone A, Calore F, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109:E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Farazi TA, Spitzer JI, Morozov P, et al. miRNAs in human cancer. J Pathol. 2011;223:102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fleischhacker SN, Bauersachs S, Wehner A, et al. Differential expression of circulating microRNAs in diabetic and healthy lean cats. Vet J. doi: 10.1016/j.tvjl.2013.03.027. [published online April 23, 2013] [DOI] [PubMed] [Google Scholar]
- 49.Gao J, Liu QG. The role of miR-26 in tumors and normal tissues. Oncol Lett. 2011;2:1019–1023. doi: 10.3892/ol.2011.413. [review] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gatsiou A, Boeckel JN, Randriamboavonjy V, et al. MicroRNAs in platelet biogenesis and function: implications in vascular homeostasis and inflammation. Curr Vasc Pharmacol. 2012;10:524–531. doi: 10.2174/157016112801784611. [DOI] [PubMed] [Google Scholar]
- 51.Gibbings DJ, Ciaudo C, Erhardt M, et al. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11:1143–1149. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 52.Gregory RI, Yan KP, Amuthan G, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 53.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Griffiths-Jones S, Grocock RJ, van Dongen S, et al. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Griffiths-Jones S, Saini HK, van Dongen S, et al. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gu Z, Eleswarapu S, Jiang H. Identification and characterization of microRNAs from the bovine adipose tissue and mammary gland. FEBS Lett. 2007;581:981–988. doi: 10.1016/j.febslet.2007.01.081. [DOI] [PubMed] [Google Scholar]
- 57.Guo H, Ingolia NT, Weissman JS, et al. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo XK, Zhang Q, Gao L, et al. Increasing expression of micro-RNA 181 inhibits porcine reproductive and respiratory syndrome virus replication and has implications for controlling virus infection. J Virol. 2013;87:1159–1171. doi: 10.1128/JVI.02386-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han J, Lee Y, Yeom KH, et al. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hanson EK, Lubenow H, Ballantyne J. Identification of forensically relevant body fluids using a panel of differentially expressed microRNAs. Anal Biochem. 2009;387:303–314. doi: 10.1016/j.ab.2009.01.037. [DOI] [PubMed] [Google Scholar]
- 61.Harfe BD, McManus MT, Mansfield JH, et al. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci U S A. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harris KS, Zhang Z, McManus MT, et al. Dicer function is essential for lung epithelium morphogenesis. Proc Natl Acad Sci U S A. 2006;103:2208–2213. doi: 10.1073/pnas.0510839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hobert O. Architecture of a microRNA-controlled gene regulatory network that diversifies neuronal cell fates. Cold Spring Harb Symp Quant Biol. 2006;71:181–188. doi: 10.1101/sqb.2006.71.006. [DOI] [PubMed] [Google Scholar]
- 65.Huang JL, Wang FX, Argyris E, et al. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4(+) T lymphocytes. Nat Med. 2007;13:1241–1247. doi: 10.1038/nm1639. [DOI] [PubMed] [Google Scholar]
- 66.Huse J, Brennan C, Hambardzumyan D, et al. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 2009;23:1327–1337. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 68.Ikeda S, He AB, Kong SW, et al. MicroRNA-1 negatively regulates expression of the hypertrophy-associated calmodulin and Mef2a genes. Mol Cell Biol. 2009;29:2193–2204. doi: 10.1128/MCB.01222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ji Q, Hao XB, Zhang M, et al. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PloS One. 2009;4:e6816. doi: 10.1371/journal.pone.0006816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 72.Jopling CL. Liver-specific microRNA-122 biogenesis and function. RNA Biol. 2012;9:137–142. doi: 10.4161/rna.18827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Junker A. Pathophysiology of translational regulation by micro-RNAs in multiple sclerosis. FEBS Lett. 2011;585:3738–3746. doi: 10.1016/j.febslet.2011.03.052. [DOI] [PubMed] [Google Scholar]
- 74.Junker A, Krumbholz M, Eisele S, et al. MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain. 2009;132:3342–3352. doi: 10.1093/brain/awp300. [DOI] [PubMed] [Google Scholar]
- 75.Kanellopoulou C, Muljo SA, Kung AL, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ketting RF, Fischer SEJ, Bernstein E, et al. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C-elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Khodyrev DS, Pronina IV, Rykov SV, et al. Involvement of methylation of group of miRNA genes in regulation of expression of RAR-beta2 and NKIRAS1 target genes in lung cancer. Mol Biol. 2012;46:693–704. [PubMed] [Google Scholar]
- 78.Kim H, Huang W, Jiang X, et al. Integrative genome analysis reveals an oncomir/oncogene cluster regulating glioblastoma survivorship. Proc Natl Acad Sci U S A. 2010;107:2183–2188. doi: 10.1073/pnas.0909896107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kinjyo I, Hanada T, Inagaki-Ohara K, et al. SOCS1/JAB is a negative regulator of LPS-induced macrophage activation. Immunity. 2002;17:583–591. doi: 10.1016/s1074-7613(02)00446-6. [DOI] [PubMed] [Google Scholar]
- 80.Klase Z, Houzet L, Jeang KT. MicroRNAs and HIV-1: complex interactions. J Biol Chem. 2012;287:40884–40890. doi: 10.1074/jbc.R112.415448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kloosterman WP, Plasterk RH. The diverse functions of micro-RNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 82.Kluiver J, Haralambieva E, de Jong D, et al. Lack of BIC and microRNA miR-155 expression in primary cases of Burkitt lymphoma. Genes Chromosomes Cancer. 2006;45:147–153. doi: 10.1002/gcc.20273. [DOI] [PubMed] [Google Scholar]
- 83.Kosaka N, Iguchi H, Yoshioka Y, et al. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kumar MS, Lu J, Mercer KL, et al. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 85.Kumar MS, Pester RE, Chen CY, et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700–2704. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lagos-Quintana M, Rauhut R, Meyer J, et al. New microRNAs from mouse and human. RNA. 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lambertz I, Nittner D, Mestdagh P, et al. Monoallelic but not biallelic loss of Dicer1 promotes tumorigenesis in vivo. Cell Death Differ. 2010;17:633–641. doi: 10.1038/cdd.2009.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 89.Lanford RE, Hildebrandt-Eriksen ES, Petri A, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lau NC, Lim LP, Weinstein EG, et al. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 91.Lawrie CH, Gal S, Dunlop HM, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 92.Lecellier CH, Dunoyer P, Arar K, et al. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 93.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 94.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 95.Lee Y, Hur I, Park SY, et al. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lee Y, Jeon K, Lee JT, et al. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lehmann U, Hasemeier B, Christgen M, et al. Epigenetic inactivation of microRNA gene hsa-mir-9-1 in human breast cancer. J Pathol. 2008;214:17–24. doi: 10.1002/path.2251. [DOI] [PubMed] [Google Scholar]
- 99.Lena AM, Shalom-Feuerstein R, Cervo PRD, et al. miR-203 represses ‘stemness’ by repressing Delta Np63. Cell Death Differ. 2008;15:1187–1195. doi: 10.1038/cdd.2008.69. [DOI] [PubMed] [Google Scholar]
- 100.Lewis AP, Jopling CL. Regulation and biological function of the liver-specific miR-122. Biochem Soc Trans. 2010;38:1553–1557. doi: 10.1042/BST0381553. [DOI] [PubMed] [Google Scholar]
- 101.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 102.Lewis MA, Quint E, Glazier AM, et al. An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat Genet. 2009;41:614–618. doi: 10.1038/ng.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li MA, He L. MicroRNAs as novel regulators of stem cell pluripotency and somatic cell reprogramming. Bioessays. 2012;34:670–680. doi: 10.1002/bies.201200019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li YJ, Lin L, Jin P. The microRNA pathway and fragile X mental retardation protein. Biochim Biophys Acta. 2008;1779:702–705. doi: 10.1016/j.bbagrm.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu CJ, Kao SY, Tu HF, et al. Increase of microRNA miR-31 level in plasma could be a potential marker of oral cancer. Oral Dis. 2010;16:360–364. doi: 10.1111/j.1601-0825.2009.01646.x. [DOI] [PubMed] [Google Scholar]
- 106.Liu N, Bezprozvannaya S, Williams AH, et al. MicroRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu PT, Wheelwright M, Teles R, et al. MicroRNA-21 targets the vitamin D–dependent antimicrobial pathway in leprosy. Nat Med. 2012;18:267–273. doi: 10.1038/nm.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 109.Lu Y, Thomson JM, Wong HY, et al. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007;310:442–453. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lund E, Guttinger S, Calado A, et al. Nuclear export of micro-RNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 111.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci U S A. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Marson A, Levine SS, Cole MF, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryo-nic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.McDaneld TG. MicroRNA: mechanism of gene regulation and application to livestock. J Anim Sci. 2009;87:E21–E28. doi: 10.2527/jas.2008-1303. [DOI] [PubMed] [Google Scholar]
- 114.Melton C, Blelloch R. MicroRNA regulation of embryonic stem cell self-renewal and differentiation. Adv Exp Med Biol. 2010;695:105–117. doi: 10.1007/978-1-4419-7037-4_8. [DOI] [PubMed] [Google Scholar]
- 115.Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mencia A, Modamio-Hoybjor S, Redshaw N, et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet. 2009;41:609–613. doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- 117.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mladinov D, Liu Y, Mattson DL, et al. MicroRNAs contribute to the maintenance of cell-type-specific physiological characteristics: miR-192 targets Na+/K+-ATPase beta1. Nucleic Acids Res. 2013;41:1273–1283. doi: 10.1093/nar/gks1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Morin RD, O'Connor MD, Griffith M, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mortarino M, Gioia G, Gelain ME, et al. Identification of suitable endogenous controls and differentially expressed micro-RNAs in canine fresh-frozen and FFPE lymphoma samples. Leukemia Res. 2010;34:1070–1077. doi: 10.1016/j.leukres.2009.10.023. [DOI] [PubMed] [Google Scholar]
- 121.Mott JL, Kobayashi S, Bronk SF, et al. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133–6140. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Murata K, Yoshitomi H, Tanida S, et al. Plasma and synovial fluid microRNAs as potential biomarkers of rheumatoid arthritis and osteoarthritis. Arthritis Res Ther. 2010;12:R86. doi: 10.1186/ar3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Navarro A, Gaya A, Martinez A, et al. MicroRNA expression profiling in classic Hodgkin lymphoma. Blood. 2008;111:2825–2832. doi: 10.1182/blood-2007-06-096784. [DOI] [PubMed] [Google Scholar]
- 124.Nishiyama T, Kaneda R, Ono T, et al. miR-142-3p is essential for hematopoiesis and affects cardiac cell fate in zebrafish. Biochem Biophys Res Commun. 2012;425:755–761. doi: 10.1016/j.bbrc.2012.07.148. [DOI] [PubMed] [Google Scholar]
- 125.Noguchi S, Mori T, Hoshino Y, et al. MicroRNAs as tumour suppressors in canine and human melanoma cells and as a prognostic factor in canine melanomas. Vet Comp Oncol. doi: 10.1002/vco.306. [published online December 8, 2011] [DOI] [PubMed] [Google Scholar]
- 126.O'Connell RM, Chaudhuri AA, Rao DS, et al. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc Natl Acad Sci U S A. 2010;107:14235–14240. doi: 10.1073/pnas.1009798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.O'Connell RM, Kahn D, Gibson WS, et al. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–619. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Okamura K, Hagen JW, Duan H, et al. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Okamura K, Liu N, Lai EC. Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol Cell. 2009;36:431–444. doi: 10.1016/j.molcel.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Osokine I, Hsu R, Loeb GB, et al. Unintentional miRNA ablation is a risk factor in gene knockout studies: a short report. PLoS Genet. 2008;4:e34. doi: 10.1371/journal.pgen.0040034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Otsuka M, Jing Q, Georgel P, et al. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity. 2007;27:123–134. doi: 10.1016/j.immuni.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 132.Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68:8164–8172. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- 133.Pasquinelli AE, Reinhart BJ, Slack F, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 134.Pauley KM, Satoh M, Chan AL, et al. Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Res Ther. 2008;10:R101. doi: 10.1186/ar2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pedersen IM, Cheng G, Wieland S, et al. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pfeffer S, Zavolan M, Grasser FA, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 137.Podolska A, Anthon C, Bak M, et al. Profiling microRNAs in lung tissue from pigs infected with Actinobacillus pleuropneumoniae. BMC Genomics. 2012;13:459. doi: 10.1186/1471-2164-13-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Potenza N, Papa U, Mosca N, et al. Human microRNA hsa-miR-125a-5p interferes with expression of hepatitis B virus surface antigen. Nucleic Acids Res. 2011;39:5157–5163. doi: 10.1093/nar/gkr067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Poy MN, Eliasson L, Krutzfeldt J, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 140.Recchiuti A, Krishnamoorthy S, Fredman G, et al. MicroRNAs in resolution of acute inflammation: identification of novel resolvin D1-miRNA circuits. FASEB J. 2011;25:544–560. doi: 10.1096/fj.10-169599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 142.Robinson VL. Rethinking the central dogma: noncoding RNAs are biologically relevant. Urol Oncol. 2009;27:304–306. doi: 10.1016/j.urolonc.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 143.Rodriguez A, Griffiths-Jones S, Ashurst JL, et al. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rodriguez A, Vigorito E, Clare S, et al. Requirement of bic/ microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rossbach M. Small non-coding RNAs as novel therapeutics. Curr Mol Med. 2010;10:361–368. doi: 10.2174/156652410791317048. [DOI] [PubMed] [Google Scholar]
- 146.Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13:239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proc Natl Acad Sci U S A. 2007;104:17719–17724. doi: 10.1073/pnas.0703890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Saito Y, Liang G, Egger G, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 149.Saraiya AA, Wang CC. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathogens. 2008;4:e1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sarver AL, Thayanithy V, Scott MC, et al. MicroRNAs at the human 14q32 locus have prognostic significance in osteosarcoma. Orphanet J Rare Dis. 2013;8:7. doi: 10.1186/1750-1172-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91:827–887. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- 152.Schickel R, Boyerinas B, Park SM, et al. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27:5959–5974. doi: 10.1038/onc.2008.274. [DOI] [PubMed] [Google Scholar]
- 153.Schneider MR. MicroRNAs as novel players in skin development, homeostasis and disease. Br J Dermatol. 2012;166:22–28. doi: 10.1111/j.1365-2133.2011.10568.x. [DOI] [PubMed] [Google Scholar]
- 154.Schwarz DS, Hutvagner G, Du T, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 155.Shaham L, Binder V, Gefen N, et al. MiR-125 in normal and malignant hematopoiesis. Leukemia. 2012;26:2011–2018. doi: 10.1038/leu.2012.90. [DOI] [PubMed] [Google Scholar]
- 156.Shen N, Liang D, Tang Y, et al. MicroRNAs—novel regulators of systemic lupus erythematosus pathogenesis. Nat Rev Rheumatol. 2012;8:701–709. doi: 10.1038/nrrheum.2012.142. [DOI] [PubMed] [Google Scholar]
- 157.Shiohama A, Sasaki T, Noda S, et al. Molecular cloning and expression analysis of a novel gene DGCR8 located in the DiGeorge syndrome chromosomal region. Biochem Bioph Res Commun. 2003;304:184–190. doi: 10.1016/s0006-291x(03)00554-0. [DOI] [PubMed] [Google Scholar]
- 158.Singh Y, Kaul V, Mehra A, et al. Mycobacterium tuberculosis controls microRNA-99b (miR-99b) expression in infected murine dendritic cells to modulate host immunity. J Biol Chem. 2013;288:5056–5061. doi: 10.1074/jbc.C112.439778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Slack FJ, Basson M, Liu ZC, et al. The lin-41 RBCC gene acts in the C-elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5:659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- 160.Song LP, Liu H, Gao SJ, et al. Cellular microRNAs inhibit replication of the H1N1 influenza A virus in infected cells. J Virol. 2010;84:8849–8860. doi: 10.1128/JVI.00456-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sonkoly E, Pivarcsi A. MicroRNAs in inflammation. Int Rev Immunol. 2009;28:535–561. doi: 10.3109/08830180903208303. [DOI] [PubMed] [Google Scholar]
- 162.Sonkoly E, Stahle M, Pivarcsi A. MicroRNAs and immunity: novel players in the regulation of normal immune function and inflammation. Semin Cancer Biol. 2008;18:131–140. doi: 10.1016/j.semcancer.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 163.Stark A, Brennecke J, Bushati N, et al. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 164.Steudemann C, Bauersachs S, Weber K, et al. Detection and comparison of microRNA expression in the serum of Doberman Pinschers with dilated cardiomyopathy and healthy controls. BMC Vet Res. 2013;9:12. doi: 10.1186/1746-6148-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Stewart CR, Marsh GA, Jenkins KA, et al. Promotion of Hendra virus replication by microRNA-146a. J Virol. 2013;87:3782–3791. doi: 10.1128/JVI.01342-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Tam W, BenYehuda D, Hayward WS. bic, a novel gene activated by proviral insertions in avian leukosis virus–induced lymphomas, is likely to function through its noncoding RNA. Mol Cell Biol. 1997;17:1490–1502. doi: 10.1128/mcb.17.3.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Tang YJ, Luo XB, Cui HJ, et al. MicroRNA-146a contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009;60:1065–1075. doi: 10.1002/art.24436. [DOI] [PubMed] [Google Scholar]
- 168.Tay Y, Zhang JQ, Thomson AM, et al. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 169.Triboulet R, Mari B, Lin YL, et al. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579–1582. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- 170.Umbach JL, Cullen BR. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 2009;23:1151–1164. doi: 10.1101/gad.1793309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Valadi H, Ekstrom K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 172.van Rooij E, Marshall WS, Olson EN. Toward microRNA-based therapeutics for heart disease: the sense in antisense. Circ Res. 2008;103:919–928. doi: 10.1161/CIRCRESAHA.108.183426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.van Rooij E, Sutherland LB, Qi X, et al. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 174.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 175.Vickers KC, Palmisano BT, Shoucri BM, et al. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wang K, Zhang SL, Weber J, et al. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38:7248–7259. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wang X, Ye L, Hou W, et al. Cellular microRNA expression correlates with susceptibility of monocytes/macrophages to HIV-1 infection. Blood. 2009;113:671–674. doi: 10.1182/blood-2008-09-175000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wang Y, Baskerville S, Shenoy A, et al. Embryonic stem cell–specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wang YM, Medvid R, Melton C, et al. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Weber JA, Baxter DH, Zhang SL, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017–5022. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- 182.Wienholds E, Koudijs MJ, van Eeden FJM, et al. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat Genet. 2003;35:217–218. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- 183.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 184.Wittmann J, Jack HM, Mashreghi MF. MicroRNAs in B-cells and T-cells as regulators of inflammation. Z Rheumatol. 2011;70:507–510. doi: 10.1007/s00393-011-0842-2. [DOI] [PubMed] [Google Scholar]
- 185.Wu H, Xu H, Miraglia LJ, et al. Human RNase III is a 160-kDa protein involved in preribosomal RNA processing. J Biol Chem. 2000;275:36957–36965. doi: 10.1074/jbc.M005494200. [DOI] [PubMed] [Google Scholar]
- 186.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wu YQ, Chen DJ, He HB, et al. Pseudorabies virus infected porcine epithelial cell line generates a diverse set of host micro-RNAs and a special cluster of viral microRNAs. PloS One. 2012;7:e30988. doi: 10.1371/journal.pone.0030988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wu ZW, Lu HF, Sheng JF, et al. Inductive microRNA-21 impairs anti-mycobacterial responses by targeting IL-12 and Bcl-2. FEBS Lett. 2012;586:2459–2467. doi: 10.1016/j.febslet.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 189.Xia T, O'Hara A, Araujo I, et al. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 2008;68:1436–1442. doi: 10.1158/0008-5472.CAN-07-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci U S A. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Yao Y, Smith LP, Petherbridge L, et al. Novel microRNAs encoded by duck enteritis virus. J Gen Virol. 2012;93:1530–1536. doi: 10.1099/vir.0.040634-0. [DOI] [PubMed] [Google Scholar]
- 192.Yeung ML, Bennasser Y, Le SY, et al. siRNA, miRNA and HIV: promises and challenges. Cell Res. 2005;15:935–946. doi: 10.1038/sj.cr.7290371. [DOI] [PubMed] [Google Scholar]
- 193.Yi R, O'Carroll D, Pasolli HA, et al. Morphogenesis in skin is governed by discrete sets of differentially expressed micro-RNAs. Nat Genet. 2006;38:356–362. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]
- 194.Yi R, Poy MN, Stoffel M, et al. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature. 2008;452:225–229. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Yi R, Qin Y, Macara IG, et al. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zen K, Zhang CY. Circulating microRNAs: a novel class of bio-markers to diagnose and monitor human cancers. Med Res Rev. 2012;32:326–348. doi: 10.1002/med.20215. [DOI] [PubMed] [Google Scholar]
- 197.Zhang G, Wang Q, Xu R. Therapeutics based on microRNA: a new approach for liver cancer. Curr Genomics. 2010;11:311–325. doi: 10.2174/138920210791616671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Zhao Y, Ransom JF, Li A, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 199.Zhao Y, Samal E, Srivastava D. Serum, response factor regulates a muscle-specific microRNA that targets Hand2 during cardio-genesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 200.Zhao Y, Yao Y, Xu H, et al. A functional MicroRNA-155 ortholog encoded by the oncogenic Marek's disease virus. J Virol. 2009;83:489–492. doi: 10.1128/JVI.01166-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Zuchner S, Wang GF, Tran-Viet KN, et al. Mutations in the novel mitochondrial protein REEP1 cause hereditary spastic paraplegia type 31. Am J Hum Genet. 2006;79:365–369. doi: 10.1086/505361. [DOI] [PMC free article] [PubMed] [Google Scholar]