Summary
The large intestine is host to a complex ecological community composed predominantly of obligate anaerobic bacteria belonging to the classes Bacteroidia and Clostridia. This community confers benefit through its metabolic activities and host interactions. However, a microbial imbalance (dysbiosis) characterized by a decreased abundance of Clostridia and a bloom of facultative anaerobic Proteobacteria is commonly observed during inflammation in the large bowel. Here we review recent insights into the principles that favor simultaneous increases in the abundance of closely related species belonging to the Proteobacteria during inflammation, which provides important clues for the rational design of strategies to treat dysbiosis.
Introduction
Our large bowel is host to a complex microbial community (microbiota) composed predominantly of obligate anaerobic bacteria belonging to the phyla Bacteroidetes (class Bacteroidia) and Firmicutes (class Clostridia), while members of the phyla Proteobacteria and Actinobacteria are commonly present in low abundance. Preservation of a balanced microbiota is important for maintaining immune homeostasis, providing nutrients, and conferring resistance against infection (reviewed in (Brestoff and Artis, 2013; Brown et al., 2013; Chu and Mazmanian, 2013; Kamada et al., 2013)). However the general principles that either help conserve a balanced microbial community structure or lead to its disruption during episodes of disease are just beginning to be unraveled.
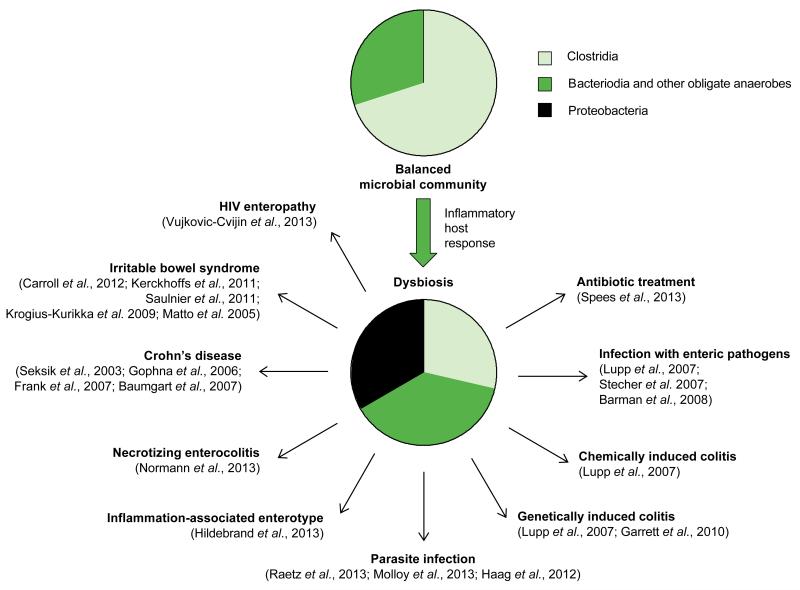
Maintenance of a balanced microbiota in the large bowel has recently been likened to lawn care, in that severe incidents can take the ecosystem back to bare earth where weed-like species can run wild (Lozupone et al., 2012). The resulting microbial imbalance (dysbiosis) is characterized by phylum-level changes in the community structure, which often includes an increased prevalence of facultative anaerobic bacteria belonging to the Proteobacteria and a decreased relative abundance of obligate anaerobic Clostridia (Fig. 1). In other words, conventional wisdom holds that a balanced gut microbiota (i.e. the lawn) occupies an intestinal niche, which can only support a bloom of Proteobacteria (i.e. of weeds) after it has been vacated. While this metaphor has some appeal, a more mechanistic understanding of the underlying processes is desirable for the rational design of potential intervention strategies. Here we review recent mechanistic insights into the processes that lead to phylum-level changes in microbial communities that inhabit the lower gastrointestinal (GI) tract.
Figure 1.
Changes in gut-associated microbial communities commonly associated with inflammation of the lower GI tract.
Pie charts provide a schematic illustration of the balanced microbial community structure of the healthy lower GI tract (top) and the imbalanced microbial community structure (dysbiosis) associated with inflammation of the large bowel (bottom). Black arrows point to recent studies suggesting that this association between the inflammatory host response and dysbiosis is a conserved ecological pattern observed in the lower GI tract.
Similis simili gaudet (like takes pleasure in like)
The principle underlying phylum-level changes in the microbiota composition is that closely related bacterial species bloom simultaneously in the large bowel. This concept was first noted in studies showing that mice harboring microbial communities characterized by a high abundance of commensal Escherichia coli (phylum Proteobacteria) are more susceptible to infection with related enteric pathogens, including Salmonella enterica (Stecher et al., 2010), and Campylobacter jejuni (Haag et al., 2012). The observation that the presence of closely related bacterial species increases the likelihood that a new bacterial species can enter the gut-associated microbial community has become known as the “like will to like” hypothesis (Stecher et al., 2010), a phrase that has its origin in the Latin similis simili gaudet. A possible explanation for this observation is that certain environmental conditions might impose selective forces that confer a fitness advantage upon all members of a phylogenetic group. However the identities of these selective forces are not immediately obvious.
Natural variation in microbial communities inhabiting the lower GI tract of laboratory mice provide clues about possible drivers of phylum-level change in the microbiota composition. Profiling of gut-associated microbial communities from conventional laboratory mice shows that they fall into one of two clusters, termed ‘enterotypes’. One enterotype is characterized by a higher overall diversity and a dominance of Clostridia over Bacteroidia. The second enterotype exhibits lower species diversity, a lower relative abundance of Clostridia and a markedly increased relative abundance of Proteobacteria. Interestingly, the latter enterotype is associated with mice exhibiting low-level intestinal inflammation, as indicated by increased fecal calprotectin levels (Hildebrand et al., 2013)(Fig. 1). Although the concept of human enterotypes has recently been called into question (Koren et al., 2013), the above correlation points to a possible connection between intestinal inflammation, an increased prevalence of Proteobacteria and a decreased abundance of Clostridia within the microbial community.
Inflammation alters microbial communities inhabiting the lower GI tract
Evidence that the inflammatory host response does, in fact, promote an overgrowth of Proteobacteria comes from studies on dysbiosis in mouse models of colitis. Investigation of the mechanism leading to dysbiosis reveals that the inflammatory host response induced by enteric bacterial pathogens, by a chemical trigger, or by genetic predisposition increases the relative luminal abundance of facultative anaerobic bacteria, most commonly members of the family Enterobacteriaceae (phylum Proteobacteria) (Garrett et al., 2010; Lupp et al., 2007) (Fig. 1). Similarly, ileitis induced by the protozoan parasite Toxoplasma gondii results in dysbiosis characterized by an uncontrolled expansion of Enterobacteriaceae within the community (Molloy et al., 2013; Raetz et al., 2013; Haag et al., 2012). Some pathogens within the family Enterobacteriaceae use their virulence factors to cause inflammation, thereby gaining a luminal growth advantage (Kamada et al., 2013; Barman et al., 2008; Lupp et al., 2007; Stecher et al., 2007). Finally, antibiotic treatment has recently been shown to trigger low-level intestinal inflammation (Wlodarska et al., 2011; Atarashi et al., 2008), which is at least in part responsible for the enhanced ability of commensal E. coli to colonize the large bowel of streptomycin-treated mice (Spees et al., 2013).
The principle that conditions of intestinal inflammation are commonly associated with a bloom of Proteobacteria and/or a decreased prevalence of Clostridia also surfaced in a number of studies on a variety of human illnesses (Fig. 1). For instance, microbial communities in patients with Crohn’s disease (CD), a disorder of unknown etiology that can manifest as ileitis or colitis, exhibit an increased prevalence of Enterobacteriaceae and a depletion of Clostridia (Baumgart et al., 2007; Frank et al., 2007; Gophna et al., 2006; Seksik et al., 2003). One of the gravest complications in premature infants is the development of necrotizing enterocolitis (NEC), a disease associated with a bloom of Enterobacteriaceae in the gut (Normann et al., 2013). Human immunodeficiency virus (HIV)-infected individuals can develop chronic diarrhea and an elevated inflammatory tone of the intestinal mucosa without an identified infectious cause, a condition termed HIV enteropathy. A recent study suggests that HIV enteropathy is accompanied by an increased abundance of Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria within gut-associated microbial communities (Vujkovic-Cvijin et al., 2013). Finally, changes in the microbiota composition are observed during irritable bowel syndrome (IBS), a condition, which often follows surgery or repeated courses of antibiotics and is characterized by low-level intestinal inflammation and diarrhea. Changes in the microbiota composition reported from IBS patients include a depletion of Clostridia and a bloom of Proteobacteria belonging to the families Enterobacteriaceae, Pasteurellaceae and Pseudomonadaceae (Carroll et al., 2012; Kerckhoffs et al., 2011; Saulnier et al., 2011; Krogius-Kurikka et al., 2009; Matto et al., 2005).
Collectively, these observations are consistent with the idea that the inflammatory host response imposes selective forces that act on closely related organisms, thereby instigating phylum-level changes in gut-associated microbial communities (Fig. 1). However, the question arises whether the identity of these selective forces are the same for each phylogenetic group. In other words, are the factors responsible for increasing the relative abundance of Proteobacteria during inflammation also responsible for a depletion of Clostridia? As outlined below, the answer is likely to be no.
How the host response feeds Proteobacteria
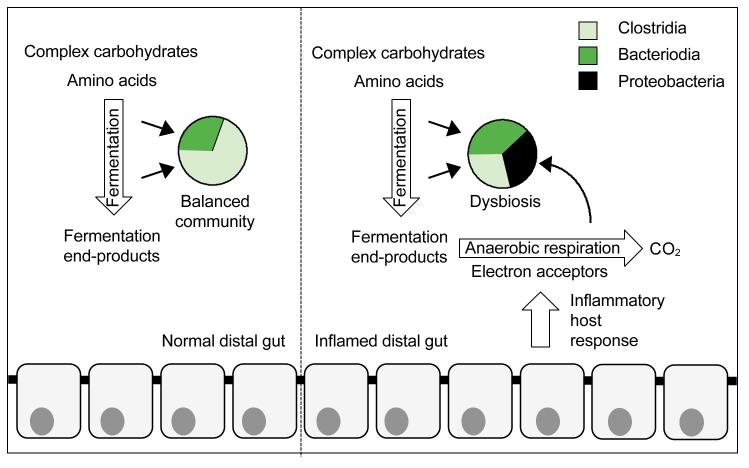
Recent mechanistic insights suggest that the inflammatory host response enhances growth of Proteobacteria by a mechanism that involves production of reactive oxygen species and reactive nitrogen species. As these antimicrobial compounds diffuse away from the epithelium, they react to form respiratory electron acceptors, such as tetrathionate or nitrate, which become available in the lumen during inflammation (Lopez et al., 2012; Winter et al., 2010). Pathogenic S. enterica and commensal E. coli can use electron acceptors produced as a by-product of the inflammatory host response to support their growth by anaerobic respiration, which leads to their uncontrolled expansion in the lumen of the large bowel (Rivera-Chavez et al., 2013; Spees et al., 2013; Winter et al., 2013b; Lopez et al., 2012; Winter et al., 2010). Anaerobic respiration provides a fitness advantage because it enables facultative anaerobic bacteria to use non-fermentable substrates or fermentation end products as carbon sources, which enables them to sidestep the competition over fermentable nutrients with obligate anaerobic Bacteroidia and Clostridia (Thiennimitr et al., 2011)(Fig. 2). These data suggest that by producing electron acceptors as a by-product, the host response selectively feeds facultative anaerobic bacteria, which explains the increased prevalence of Proteobacteria within the microbial community during inflammation (Winter et al., 2013a).
Figure 2.
The host inflammatory response provides a growth advantage for Proteobacteria.
Pie charts provide a schematic illustration of the balanced microbial community structure of the healthy lower GI tract (left) and the dysbiosis associated with inflammation (right). In the absence of inflammation, complex carbohydrates and amino acids support growth of obligate anaerobic bacteria by fermentation (left). The inflammatory host response enables facultative anaerobic bacteria to consume fermentation end products, thereby increasing their prevalence within the community (right).
There are likely additional mechanisms that contribute to changes in the microbial community structure during inflammation. For example, the process expected to be responsible for the reduced prevalence of obligate anaerobic Clostridia during inflammation is likely distinct from anaerobic respiration, because members of this group lack the terminal oxidoreductases needed to use electron acceptors produced by the host response (reviewed in (Fischbach and Sonnenburg, 2011)). The identities of the negative selective forces that explain why the prevalence of Clostridia is sometimes reduced during inflammation remain to be worked out. Furthermore, these selective forces are not likely to act on all members of this class, because Clostridium difficile and some members of the family Lachnospiraceae can increase their prevalence within the community during intestinal inflammation (Chassaing et al., 2013).
Concluding remarks
Although our understanding of factors responsible for the dynamics of gut-associated microbial communities during inflammation is still incomplete, clues gained from studying a bloom of Proteobacteria provides first mechanistic insights into the ‘like will to like’ concept. Our current understanding of the processes responsible for increasing the prevalence of Proteobacteria within the community suggest one of the driving forces is that the host response alters the nutritional environment in the lumen of the lower GI tract. The ‘like will to like’ hypothesis predicts that these changes in the nutritional environment impose similar selective forces on closely related organisms, thereby causing them to bloom simultaneously (Stecher et al., 2010). Consistent with this postulate, the increased availability of exogenous electron acceptors during inflammation is expected to provide an anaerobic respiration-dependent fitness advantage upon facultative anaerobic Proteobacteria, but not upon obligate anaerobic Bacteroidia and Clostridia. The resulting bloom of Proteobacteria during inflammation is arguably one of the most robust ecological patterns observed in the lower GI tract (Fig. 1). Competition between related bacterial species likely arises during this process, especially when they occupy very similar metabolic niches (Deriu et al., 2013; Maltby et al., 2013). Nonetheless, analysis of gut-associated microbial communities suggests that multiple species belonging to the same phylum commonly coexist in this environment.
At a first glance, the above mechanism seems to suggests that clearing the niche might not be necessary, because production of electron acceptors by the inflammatory host response would be expected to support an outgrowth of Protebacteria even in the presence of a balanced community of obligate anaerobic bacteria. However, an additional factor that needs to be considered is the ability of a balanced microbial community to actively contribute to immune homeostasis in the intestine (Atarashi et al., 2011). Clostridia in particular are credited with generating large quantities of short-chain fatty acids (SCFAs) during fermentation of complex carbohydrates in the large bowel. In turn, SCFAs stimulate receptors on regulatory T cells, which results in the resolution of inflammation and maintenance of immune homeostasis (Smith et al., 2013). Thus clearing Clostridia from the niche, for example by antibiotic treatment, might be necessary to first elevate the inflammatory tone of the mucosa (Atarashi et al., 2008) before Protebacteria can increase their abundance through growth by anaerobic respiration (Spees et al., 2013). Even though these skirmishes are still adequately portrayed by the lawn care metaphor (Lozupone et al., 2012), it is becoming increasingly clear that the host immune system – akin to a gardener – plays a critical role in shaping microbial communities in the lower GI tract. Furthermore, elucidation of the mechanisms underlying these complex relationships between the host and its microbiota identifies anaerobic respiration as a potential target for intervention strategies.
Acknowledgements
This work was supported by Public Health Service grants AI107393 and AI096528 to A.J.B. and AI103248 to S.E.W. The authors have no conflict of interest.
References
- Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M, Yagita H, Ishii N, Evans R, Honda K, Takeda K. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455:808–812. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov II, Umesaki Y, Itoh K, Honda K. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barman M, Unold D, Shifley K, Amir E, Hung K, Bos N, Salzman N. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect Immun. 2008;76:907–915. doi: 10.1128/IAI.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart M, Dogan B, Rishniw M, Weitzman G, Bosworth B, Yantiss R, Orsi RH, Wiedmann M, McDonough P, Kim SG, Berg D, Schukken Y, Scherl E, Simpson KW. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. ISME J. 2007;1:403–418. doi: 10.1038/ismej.2007.52. [DOI] [PubMed] [Google Scholar]
- Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol. 2013;14:676–684. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EM, Sadarangani M, Finlay BB. The role of the immune system in governing host-microbe interactions in the intestine. Nat Immunol. 2013;14:660–667. doi: 10.1038/ni.2611. [DOI] [PubMed] [Google Scholar]
- Carroll IM, Ringel-Kulka T, Siddle JP, Ringel Y. Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterology and motility: the official journal of the European Gastrointestinal Motility Society. 2012;24:521–530. doi: 10.1111/j.1365-2982.2012.01891.x. e248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassaing B, Koren O, Carvalho FA, Ley RE, Gewirtz AT. AIEC pathobiont instigates chronic colitis in susceptible hosts by altering microbiota composition. Gut. 2013 doi: 10.1136/gutjnl-2013-304909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H, Mazmanian SK. Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat Immunol. 2013;14:668–675. doi: 10.1038/ni.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriu E, Liu JZ, Pezeshki M, Edwards RA, Ochoa RJ, Contreras H, Libby SJ, Fang FC, Raffatellu M. Probiotic bacteria reduce salmonella typhimurium intestinal colonization by competing for iron. Cell host & microbe. 2013;14:26–37. doi: 10.1016/j.chom.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach MA, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell host & microbe. 2011;10:336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DN, St Amand AL, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, Gordon JI, Onderdonk AB, Glimcher LH. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell host & microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJ. Differences between tissue-associated intestinal microfloras of patients with Crohn’s disease and ulcerative colitis. Journal of clinical microbiology. 2006;44:4136–4141. doi: 10.1128/JCM.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag LM, Fischer A, Otto B, Plickert R, Kuhl AA, Gobel UB, Bereswill S, Heimesaat MM. Intestinal microbiota shifts towards elevated commensal Escherichia coli loads abrogate colonization resistance against Campylobacter jejuni in mice. PLoS One. 2012;7:e35988. doi: 10.1371/journal.pone.0035988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand F, Nguyen TL, Brinkman B, Yunta RG, Cauwe B, Vandenabeele P, Liston A, Raes J. Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice. Genome Biol. 2013;14:R4. doi: 10.1186/gb-2013-14-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Chen GY, Inohara N, Nunez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerckhoffs AP, Ben-Amor K, Samsom M, van der Rest ME, de Vogel J, Knol J, Akkermans LM. Molecular analysis of faecal and duodenal samples reveals significantly higher prevalence and numbers of Pseudomonas aeruginosa in irritable bowel syndrome. Journal of medical microbiology. 2011;60:236–245. doi: 10.1099/jmm.0.022848-0. [DOI] [PubMed] [Google Scholar]
- Koren O, Knights D, Gonzalez A, Waldron L, Segata N, Knight R, Huttenhower C, Ley RE. A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLoS computational biology. 2013;9:e1002863. doi: 10.1371/journal.pcbi.1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogius-Kurikka L, Lyra A, Malinen E, Aarnikunnas J, Tuimala J, Paulin L, Makivuokko H, Kajander K, Palva A. Microbial community analysis reveals high level phylogenetic alterations in the overall gastrointestinal microbiota of diarrhoea-predominant irritable bowel syndrome sufferers. BMC gastroenterology. 2009;9:95. doi: 10.1186/1471-230X-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez CA, Winter SE, Rivera-Chavez F, Xavier MN, Poon V, Nuccio SP, Tsolis RM, Bäumler AJ. Phage-mediated acquisition of a type III secreted effector protein boosts growth of salmonella by nitrate respiration. mBio. 2012;3 doi: 10.1128/mBio.00143-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Maltby R, Leatham-Jensen MP, Gibson T, Cohen PS, Conway T. Nutritional basis for colonization resistance by human commensal Escherichia coli strains HS and Nissle 1917 against E. coli O157:H7 in the mouse intestine. PloS one. 2013;8:e53957. doi: 10.1371/journal.pone.0053957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matto J, Maunuksela L, Kajander K, Palva A, Korpela R, Kassinen A, Saarela M. Composition and temporal stability of gastrointestinal microbiota in irritable bowel syndrome--a longitudinal study in IBS and control subjects. FEMS immunology and medical microbiology. 2005;43:213–222. doi: 10.1016/j.femsim.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Molloy MJ, Grainger JR, Bouladoux N, Hand TW, Koo LY, Naik S, Quinones M, Dzutsev AK, Gao JL, Trinchieri G, Murphy PM, Belkaid Y. Intraluminal Containment of Commensal Outgrowth in the Gut during Infection-Induced Dysbiosis. Cell host & microbe. 2013;14:318–328. doi: 10.1016/j.chom.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normann E, Fahlen A, Engstrand L, Lilja HE. Intestinal microbial profiles in extremely preterm infants with and without necrotizing enterocolitis. Acta paediatrica. 2013;102:129–136. doi: 10.1111/apa.12059. [DOI] [PubMed] [Google Scholar]
- Raetz M, Hwang SH, Wilhelm CL, Kirkland D, Benson A, Sturge CR, Mirpuri J, Vaishnava S, Hou B, Defranco AL, Gilpin CJ, Hooper LV, Yarovinsky F. Parasite-induced TH1 cells and intestinal dysbiosis cooperate in IFN-gamma-dependent elimination of Paneth cells. Nat Immunol. 2013;14:136–142. doi: 10.1038/ni.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Chavez F, Winter SE, Lopez CA, Xavier MN, Winter MG, Nuccio SP, Russell JM, Laughlin RC, Lawhon SD, Sterzenbach T, Bevins CL, Tsolis RM, Harshey R, Adams LG, Bäumler AJ. Salmonella uses energy taxis to benefit from intestinal inflammation. PLoS Pathog. 2013;9:e1003267. doi: 10.1371/journal.ppat.1003267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saulnier DM, Riehle K, Mistretta TA, Diaz MA, Mandal D, Raza S, Weidler EM, Qin X, Coarfa C, Milosavljevic A, Petrosino JF, Highlander S, Gibbs R, Lynch SV, Shulman RJ, Versalovic J. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology. 2011;141:1782–1791. doi: 10.1053/j.gastro.2011.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seksik P, Rigottier-Gois L, Gramet G, Sutren M, Pochart P, Marteau P, Jian R, Dore J. Alterations of the dominant faecal bacterial groups in patients with Crohn’s disease of the colon. Gut. 2003;52:237–242. doi: 10.1136/gut.52.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spees AM, Wangdi T, Lopez CA, Kingsbury DD, Xavier MN, Winter SE, Tsolis RM, Bäumler AJ. Streptomycin-Induced Inflammation Enhances Escherichia coli Gut Colonization Through Nitrate Respiration. mBio. 2013;4 doi: 10.1128/mBio.00430-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B, Chaffron S, Kappeli R, Hapfelmeier S, Freedrich S, Weber TC, Kirundi J, Suar M, McCoy KD, von Mering C, Macpherson AJ, Hardt WD. Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS pathogens. 2010;6:e1000711. doi: 10.1371/journal.ppat.1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, Dougan G, von Mering C, Hardt WD. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Bäumler AJ. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vujkovic-Cvijin I, Dunham RM, Iwai S, Maher MC, Albright RG, Broadhurst MJ, Hernandez RD, Lederman MM, Huang Y, Somsouk M, Deeks SG, Hunt PW, Lynch SV, McCune JM. Dysbiosis of the Gut Microbiota Is Associated with HIV Disease Progression and Tryptophan Catabolism. Science translational medicine. 2013;5:193ra191. doi: 10.1126/scitranslmed.3006438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Lopez CA, Bäumler AJ. The dynamics of gut-associated microbial communities during inflammation. EMBO reports. 2013a;14:319–327. doi: 10.1038/embor.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Bäumler AJ. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, Laughlin RC, Gomez G, Wu J, Lawhon SD, Popova IE, Parikh SJ, Adams LG, Tsolis RM, Stewart VJ, Bäumler AJ. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013b;339:708–711. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarska M, Willing B, Keeney KM, Menendez A, Bergstrom KS, Gill N, Russell SL, Vallance BA, Finlay BB. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infection and immunity. 2011;79:1536–1545. doi: 10.1128/IAI.01104-10. [DOI] [PMC free article] [PubMed] [Google Scholar]