Abstract
DNA methylation in the form of 5-methylcytosine (5mC) is a key epigenetic regulator in mammals, and the dynamic balance between methylation and demethylation impacts various processes from development to disease. The recent discovery of the enzymatic generation and removal of the oxidized derivatives of 5mC, namely 5-hydroxymethylcysotine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) in mammalian cells has led to a paradigm shift in our understanding of the demethylation process. Interestingly, emerging evidence indicates that these DNA demethylation intermediates are dynamic and could themselves carry regulatory functions. Here, we discuss 5hmC, 5fC, and 5caC as new epigenetic DNA modifications that could have distinct regulatory functions in conjunction with potential protein partners.
Keywords: demethylation intermediates, 5-hydroxymethylcytosine, 5-formylcytosine, 5-carboxylcytosine, regulatory roles
New paradigm of active DNA demethylation through 5mC oxidation
DNA methylation in the form of 5mC provides critical regulatory information beyond the simple genomic sequence because it impacts a variety of biological processes from gene regulation to disease pathogenesis [1]. Maintaining dynamic DNA methylation status by balancing methylation and demethylation processes is crucial for mammalian development [2]; disruption of this balance could lead to aberrant methylation patterns seen in human diseases such as cancer [3]. DNA methyltransfereases (DNMTs) catalyze the methylation process (Figure 1). The demethylation process, however, is more complicated.
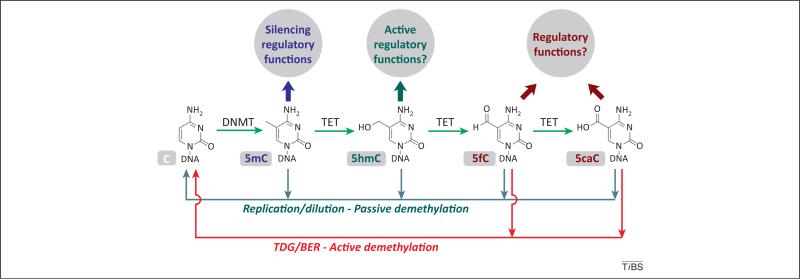
Figure 1.
DNA methylation and demethylation are dynamically balanced. 5-Methylcytosine (5mC) is generated by DNA methyltransfereases (DNMTs), whereas demethylation can be achieved through multiple pathways. 5mC can be passively converted back to cytosine through replication-dependent dilution due to a lack of DNMT activity. 5mC can be actively oxidized by TET proteins to 5-hydroxymethylcysotine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC), which could be followed by passive replication/dilution demethylation. Alternatively, 5fC and 5caC can be removed by thymine DNA glycosylase (TDG) and base excision repair (BER), resulting in an active demethylation of 5mC. Clearly, 5hmC, 5fC, and 5caC all act as demethylation intermediates. However, these oxidized 5mC derivatives may also possess regulatory functions in a complex network.
DNMTs can affect methylation status when they fail to methylate newly synthesized DNA during cell division (Figure 1). However, this passive demethylation process cannot explain all cellular demethylation events during development and differentiation; many of which seem to occur in the absence of DNA replication [4,5]. The finding that 5hmC is in surprisingly high abundance in certain mammalian genomes [6,7] and the discovery that the TET family proteins catalyze the sequential oxidation of 5mC to 5hmC [7], 5fC, and then 5caC [8,9] have changed our view of how the demethylation process could occur (Figure 1). Indeed, many recent studies have shown that genome-wide demethylation events in zygote formation [10–14] and in the germ-cell lineage [15–18] result from multiple parallel and perhaps partially redundant pathways and mechanisms, including the replication-dependent dilution of 5mC, oxidation of 5mC to 5hmC, 5fC, or 5caC, followed by replication-dependent dilution, or the active removal of 5fC and 5caC by thymine DNA glycosylase (TDG) through base excision repair (BER) [9,19] (Figure 1) (for reviews, [2,20,21]). Other pathways based on potential 5mC or 5hmC deamination have also been proposed but still need further experimental support and are not discussed here [22,23].
In this article, we define the TDG/BER-dependent removal of 5fC and 5caC as active demethylation, whereas the replication-dependent dilution of 5hmC, 5fC, and 5caC is still defined as passive demethylation, even though the initial step is a TET-mediated oxidation. The TDG/BER-based active demethylation is less likely to be the major mechanism for genome-wide demethylation events observed in early development, because this would create genome-wide DNA breaks and impose a huge burden on the DNA repair machineries. The replication-dependent passive mechanism, in which the TET oxidation of 5mC facilitates the process, has been proposed as the main pathway of global demethylation [13], although the presence of other pathways and mechanisms should not be completely ruled out. It should be noted that once 5mC is enzymatically converted to 5hmC, 5fC, or 5caC, it may not recruit proteins that selectively bind 5mC and effectively functions as demethylated. TDG/BER-based active demethylation, by contrast, could be a dynamic process occurring to more localized regions of the genome, such as enhancers, during cell differentiation or in response to environmental stimuli. However, this process can still be genome-wide as demonstrated by recent mapping of 5fC and 5caC in mouse embryonic stem cells (ESCs) [24–26].
There are indeed more economical ways to remove actively 5mC than the TET/TDG-based oxidative demethylation (such as a 5mC-specific glycosylase in plants) [27]. In principle, 5hmC could be directly reversed back to cytosine. That nature chooses to use an indirect and more energy-consuming mechanism may suggest that 5hmC, 5fC, and 5caC carry additional functions beyond being just demethylation intermediates. Here, we survey the recent literature and discuss potential regulatory functions of these oxidized 5mC modifications.
5hmC could have regulatory functions
5mC is commonly thought to be a repressive mark, because methylation of CpG islands in promoter regions can recruit methyl-CpG-binding proteins and contribute to gene repression. However, more recent sensitive genome-scale studies have revealed that methylation can also happen at other regions in the genome that have complex functions; for instance, gene body methylation is not associated with repression [28]. Although 5hmC has been well characterized as a demethylation intermediate, increasing data have suggested that 5hmC may also serve as a stable epigenetic mark that possesses unique regulatory functions [29,30]. A recent study divided the mouse ESC DNA methylome into un-, low-, and fully methylated regions (UMRs, LMRs, and FMRs, respectively). LMRs, which have a 10–50% level of methylation, are frequently located in active distal regulatory regions of the genome such as enhancers, and are occupied by DNA-binding factors [31]. Interestingly, 5hmC is strongly enriched in LMRs, reflecting the intermediate demethylation status of these regions [32]. It is possible that 5hmC recruits proteins to engage in demethylation and transcription regulation.
Unlike 5mC, which is evenly distributed across different tissues and cells (~5% of total cytosine), the level of 5hmC varies significantly [33]. For instance, 5hmC abundance is highest in the central nervous system where it can account for up to 1% of total cytosine [8]. The function of the relatively abundant 5hmC in brain tissues is most intriguing. 5hmC also accumulates with age in brain, supporting the idea that it may function as a stable mark [34]. The genomic distribution of 5hmC differs between ESCs and brain tissues. In ESCs, 5hmC is enriched at distal regulatory elements such as enhancers, near but not on transcription factor (TF) binding sites [32]. By contrast, in neurons, 5hmC seems to be enriched at the gene bodies of neuronal function-related genes where 5mC is strongly depleted, and the 5hmC/5mC ratio correlates well with gene expression levels [35]. It seems that gene body 5hmC acts as a stable active mark rather than a demethylation intermediate during neuronal development [34–37]. This observation was further supported by the finding that methyl-CpG-binding protein (MeCP)2 can bind 5hmC in the brain to affect chromatin structure and gene expression [35].
The levels of 5hmC are generally low in cancer cells and cultured somatic cells, but are relatively high in pluripo-tent or multipotent cells such as ESCs or neural stem cells (NSCs) (~0.1% total cytosine) [8,34,38,39]. In line with these observations, recent studies reveal that TET1- and TET2-mediated acquisition of 5hmC are important or essential processes in reprogramming of somatic cells to pluripotent cells by either transduction of reprogramming factors [38,40,41] or fusion with stem cells [42]. 5hmC therefore may serve as a mark during reprogramming, not only as a demethylation intermediate, but may also function in the establishment of pluripotency through potential regulatory roles [43].
Lastly, specific binding proteins have been shown to recognize 5hmC. Besides MeCP2 in the brain, methyl-CpG-binding domain protein (MBD)3 has been suggested to bind 5hmC in ESCs [44]. A recent proteomic study has revealed additional potential 5hmC-specific binders that could exhibit biological function [45]. Overall, current data depict a positive role of 5hmC in regulating neuron function. The function of 5hmC in ECSs is more complicated: although 5hmC in distal regulatory elements may have an overall active regulatory role [32], the role of 5hmC in other regions of the genome such as promoters and gene bodies is unclear and will need further research to clarify.
5fC and 5caC are committed demethylation intermediates derived from 5hmC
5fC and 5caC are further oxidation products of 5hmC and were discovered to be present in the mammalian genome in 2011 [8,9,46]. 5fC and 5caC are 10 to 1,000-fold less abundant than 5hmC, and unlike 5hmC, they are not enriched in brain tissues and their levels are consistently low across all mammalian tissues and cells examined [8]. Given their low abundance, that they are the further oxidized derivatives of 5hmC, and that they are effectively removed by TDG to complete the active demethylation process (Figure 1), 5fC and 5caC are currently considered committed demethylation intermediates [33]. Thus, the presence of 5fC and 5caC could define the portion of 5hmC sites that are demethylation sites. Conversely, those 5hmC sites without concurrent 5fC/5caC are more likely to be stable 5hmC. Indeed, two recent studies mapped the genome-wide distribution of 5fC and 5caC in mouse ESCs and reached similar conclusions [25,26]. In wild type mouse ESCs, 90% of 5fC-marked regions reside in 5hmC-enriched regions; yet, they only mark ~30% of the hydroxymethy-lome [25], consistent with the idea that 5fC/5caC only define part of the hydroxylmethylome. It will be interesting to characterize further the 5fC- and 5hmC-only regions, because these regions may represent functional 5fC and 5hmC sites, respectively.
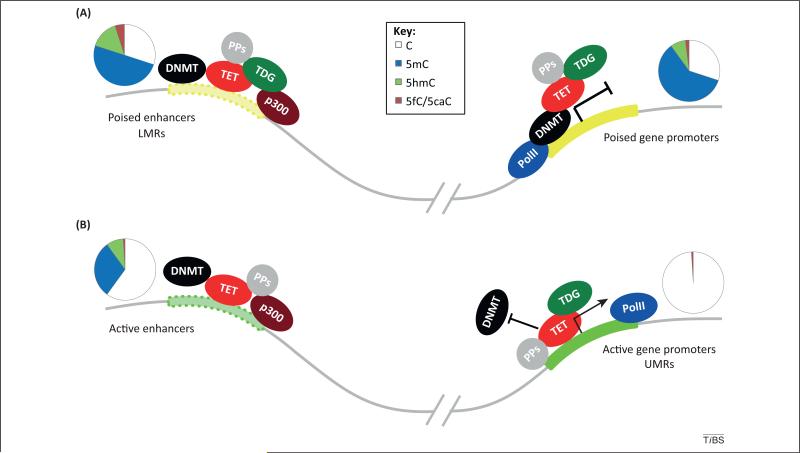
5fC is enriched at distal regulatory elements with a preference for poised enhancers (poised enhancers have an inactive but ‘ready-to-go’ status that can be quickly activated once a particular differentiation signal or other cellular stimuli are given), LMRs, and promoters of ‘poised’ genes (genes with low expression) [25] (Figure 2A). Upon Tdg depletion in mouse ESCs, 5fC and 5caC level increased by 2–10-fold with no apparent changes of the 5mC and 5hmC level [9,25,26]. Although the total 5fC and 5caC levels are still low (10–100-fold less abundant than 5hmC), it is somewhat expected because TDG-mediated active demethylation is unlikely to be a global event as discussed above. The possibility of 5fC or 5caC being converted to other forms independent of TDG cannot be excluded and should be continually investigated. Nevertheless, Tdg depletion leads to an accumulation of 5fC and 5caC at enhancers, multiple TF-binding sites, LMRs, and promoters of genes with low to intermediate expression, all without a change of 5hmC [25,26]. These observations point to a model in which dynamic TET/TDG-mediated 5hmC oxidation and demethylation occur at a portion of the hydroxylmethylome that is marked by 5fC/5caC to maintain those key gene elements in a poised or active state for current and/or subsequent cellular differentiation and development [25,26] (Figure 2A), whereas other 5hmC sites that are less frequently oxidized to 5fC/5caC may play other roles depending on the different protein partners that they interact with (Figure 2B).
Figure 2.
Schematic representation of DNA epigenetic regulation in mouse embryonic stem cells (ESCs). TET/thymine DNA glycosylase (TDG)-mediated active DNA demethylation occurs frequently at distal regulatory elements in ESCs. (A) At poised enhancers and poised gene promoters, where 5-formylcytosine (5fC)/5-carboxylcytosine (5caC) accumulation in the absence of TDG is the most significant, dynamic methylation and demethylation equilibria likely exist to keep these regions at the poised states in order to respond to upcoming development/differentiation events. Accordingly, these 5fC/5caC-marked 5-hydroxymethylcysotine (5hmC) sites are committed demethylation sites. (B) 5hmC and 5fC/5caC are also present at active enhancers where they may have more active regulatory roles. Certain active promoter sites (unmethylated regions; UMRs) are depleted of 5mC and 5hmC but still bound by TET proteins. 5hmC and 5fC may recruit binding proteins to facilitate active demethylation. TET/TDG may interact with diverse protein partners (PPs) that affect their demethylation activities. ESC DNA methylation patterns are represented by pie charts above each gene elements. Abbreviations: DNMT, DNA methyltransferease; LMRs, low-methylated regions; PolII, RNA polymerase II.
A small fraction of 5fC was also detected in the previously defined UMRs [25]. UMRs were originally defined on the basis of conventional bisulfite sequencing, which can only detect 5mC and 5hmC, but not 5fC or 5caC [30,47]. Interestingly, besides LMRs, TET proteins are also highly enriched in UMRs, even though 5mC and 5hmC are not detected at UMRs (primarily active promoters) (Figure 2B) [31,48,49]. These results further indicate that 5fC/5caC could define a portion of UMRs with strong TET binding as active demethylation regions. These regions do get methylated; however, the strong binding and oxidation by TET converts all 5mC and 5hmC to 5fC and perhaps also 5caC for constitutive active demethylation of any 5mC at these UMRs (Figure 2B), whereas at LMRs, TET binding and subsequent 5mC oxidation and BER are dynamic and may reach equilibrium with DNMT-mediated methylation.
Clearly, 5fC and 5caC are more transient intermediates derived from 5hmC as demonstrated by their low levels even after TDG depletion. They define part of the hydro-xymethylome that is destined to demethylation. However, despite the transient nature, they may recruit protein partners to exert potential functional roles as discussed in the next section.
5fC/5caC may exhibit regulatory function
The formyl group of 5fC and carboxyl group of 5caC offer unique chemical anchors for protein recognition, especially the formyl group, which is a relatively active functional group that can react with various cellular components. In vitro, 5fC could form relatively stable Schiff base adducts with amine-containing compounds [50], suggesting 5fC may form transient Schiff bases with proteins in a unique way that differs from traditional TF binding. Thiol groups from either proteins or small molecules can reversibly add to the formyl group; the formed adducts could potentially be stabilized through protein–nucleic acid interactions. A recent proteome-wide analysis identified many more potential binding proteins or readers for 5fC and 5caC than for 5hmC in mouse ESCs, suggesting that 5fC and 5caC may recruit unique proteins for specific functions that may be coupled with active demethylation [45]. Moreover, 5fC and 5caC have been shown to reduce the rate and substrate specificity of RNA polymerase II (PolII) transcription [51]. Future crystallography studies will shed light on the interaction mechanism. Furthermore, 5fC/5caC accumulation in the absence of TDG at distal regulatory elements appears to coordinate with the binding of transcriptional coactivator p300 (Figure 2A) [25]. p300 could serve as a scaffold or bridge for TFs and transcription machinery to facilitate chromatin remodeling and to activate gene transcription [52], therefore, these observations suggest an attractive model in which TET/TDG-mediated iterative oxidation and BER couple with diverse protein partners, such as histone mediators or TFs, through p300 or other 5fC/5caC-binding proteins to dictate active demethylation (Figure 2).
Concluding remarks
DNA methylation is an essential epigenetic modification for mammalian development. Genome-wide studies reveal the complex nature of 5mC rather than simply a silencing mark. Recently, a dynamic DNA epigenetic regulatory network including the oxidized 5mC derivatives, 5hmC, 5fC, and 5caC, as well as the proteins that generate or act on them, the TETs and TDG, has been depicted. It is clear that these oxidized 5mC derivatives serve as DNA demethylation intermediates that are important for programming and reprogramming during development. However, growing evidence also suggests that these new base modifications may possess unique regulatory functions. The relatively high abundance of 5hmC in neurons suggests that it is a stable mark that may directly impact gene expression; both 5hmC and 5fC, and possibility 5caC, could recruit binding proteins to affect chromatin structure and gene expression, in particular at distal regulatory elements. Interweaved with 5mC, histone modifications, and various transcription regulators, these new cytosine modifications add to the complete picture of the epigenome.
Although we are now equipped with many techniques for mapping new 5mC derivatives [25,26,33] to dissect the functional roles of these newly discovered DNA base modifications, many outstanding questions remain (Box 1). Results obtained so far indicate that dynamic methylation–demethylation processes occur at distal gene regulatory elements in mammalian genomes. The scope and specific impacts of this process need to be clarified. During cell development and differentiation, the equilibria established by DNMT-catalyzed methylation and TET-mediated 5mC oxidation with subsequent localized BER-mediated demethylation can shift and re-establish themselves within new dynamic epigenomic states. During this transition 5fC, 5caC, and 5hmC may engage in interactions with different transcription factors, polymerases, potential reader proteins, as well as other cellular epigenetic components, which could affect the demethylation process and the dynamics of the methylation state. Future studies will reveal the exact roles of each base modification and the underlying mechanisms.
Box 1. Outstanding questions and future directions.
Revisit or explore various demethylation events in terminal differentiation processes to confirm TDG-mediated active demethylation indeed happens in vivo and affects specific biological processes.
Detailed characterization of potential binding proteins for 5hmC, 5fC, and 5caC, and how these protein partners contribute to the regulatory functions of 5hmC, 5fC, and 5caC.
New sensitive methods to obtain base-resolution information of 5fC and 5caC.
Correlate the binding of potential reader proteins with the base-resolution maps of 5hmC, 5fC, or 5caC in order to obtain the exact function of reading at specific loci.
Acknowledgments
We thank Drs. P. Jin and K. E. Szulwach for discussion and insight; and S.F. Reichard for editing the manuscript. This study was supported by National Institutes of Health (HG006827 to C.H.).
References
- 1.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 2.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 3.Baylin SB, Jones PA. A decade of exploring the cancer epigenome – biological and translational implications. Nat. Rev. Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat. Rev. Mol. Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhutani N, et al. DNA demethylation dynamics. Cell. 2011;146:866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He YF, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wossidlo M, et al. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat. Commun. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- 11.Iqbal K, et al. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl. Acad. Sci. U.S.A. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu TP, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 13.Inoue A, et al. Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res. 2011;21:1670–1676. doi: 10.1038/cr.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seisenberger S, et al. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell. 2012;48:849–862. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamaguchi S, et al. Tet1 controls meiosis by regulating meiotic gene expression. Nature. 2012;492:443–447. doi: 10.1038/nature11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hackett JA, et al. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2013;339:448–452. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vincent JJ, et al. Stage-specific roles for tet1 and tet2 in DNA demethylation in primordial germ cells. Cell Stem Cell. 2013;12:470–478. doi: 10.1016/j.stem.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J. Biol. Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hackett JA, et al. Parallel mechanisms of epigenetic reprogramming in the germline. Trends Genet. 2012;28:164–174. doi: 10.1016/j.tig.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 21.Guibert S, Weber M. Functions of DNA methylation and hydroxymethylation in mammalian development. Curr. Top. Dev. Biol. 2013;104:47–83. doi: 10.1016/B978-0-12-416027-9.00002-4. [DOI] [PubMed] [Google Scholar]
- 22.Guo JU, et al. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schiesser S, et al. Mechanism and stem-cell activity of 5-carboxycytosine decarboxylation determined by isotope tracing. Angew. Chem. Int. Ed. Engl. 2012;51:6516–6520. doi: 10.1002/anie.201202583. [DOI] [PubMed] [Google Scholar]
- 24.Raiber EA, et al. Genome-wide distribution of 5-formylcytosine in ES cells is associated with transcription and depends on thymine DNA glycosylase. Genome Biol. 2012;13:R69. doi: 10.1186/gb-2012-13-8-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song CX, et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell. 2013;153:678–691. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen L, et al. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013;153:692–706. doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu. Rev. Genet. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 29.Koh KP, Rao A. DNA methylation and methylcytosine oxidation in cell fate decisions. Curr. Opin. Cell Biol. 2013;25:152–161. doi: 10.1016/j.ceb.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen L, Zhang Y. 5-Hydroxymethylcytosine: generation, fate, and genomic distribution. Curr. Opin. Cell Biol. 2013;25:289–296. doi: 10.1016/j.ceb.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stadler MB, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 32.Yu M, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song CX, et al. Mapping recently identified nucleotide variants in the genome and transcriptome. Nat. Biotechnol. 2012;30:1107–1116. doi: 10.1038/nbt.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song CX, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mellen M, et al. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szulwach KE, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci. 2011;14:1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hahn MA, et al. Dynamics of 5-hydroxymethylcytosine and chromatin marks in Mammalian neurogenesis. Cell Rep. 2013;3:291–300. doi: 10.1016/j.celrep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao Y, et al. Replacement of Oct4 by Tet1 during iPSC induction reveals an important role of DNA methylation and hydroxymethylation in reprogramming. Cell Stem Cell. 2013;12:453–469. doi: 10.1016/j.stem.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 39.Jin S-G, et al. 5-Hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res. 2011;71:7360–7365. doi: 10.1158/0008-5472.CAN-11-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doege CA, et al. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature. 2012;488:652–655. doi: 10.1038/nature11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Costa Y, et al. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature. 2013;495:370–374. doi: 10.1038/nature11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piccolo FM, et al. Different roles for Tet1 and Tet2 proteins in reprogramming-mediated erasure of imprints induced by EGC fusion. Mol. Cell. 2013;49:1023–1033. doi: 10.1016/j.molcel.2013.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cantone I, Fisher AG. Epigenetic programming and reprogramming during development. Nat. Struct. Mol. Biol. 2013;20:282–289. doi: 10.1038/nsmb.2489. [DOI] [PubMed] [Google Scholar]
- 44.Yildirim O, et al. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spruijt CG, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 46.Pfaffeneder T, et al. The discovery of 5-formylcytosine in embryonic stem cell DNA. Angew. Chem. Int. Ed. Engl. 2011;50:7008–7012. doi: 10.1002/anie.201103899. [DOI] [PubMed] [Google Scholar]
- 47.Booth MJ, et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- 48.Williams K, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu H, et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu J, et al. Selective chemical labelling of 5-formylcytosine in DNA by fluorescent dyes. Chemistry. 2013;19:5836–5840. doi: 10.1002/chem.201300082. [DOI] [PubMed] [Google Scholar]
- 51.Kellinger MW, et al. 5-formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 2012;19:831–833. doi: 10.1038/nsmb.2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen J, Li Q. Life and death of transcriptional co-activator p300. Epigenetics. 2011;6:957–961. doi: 10.4161/epi.6.8.16065. [DOI] [PubMed] [Google Scholar]