Summary
Intellectual disability is a prevalent developmental disorder of cognition that remains incurable. Here, we report that knockdown of the X-linked intellectual disability (XLID) protein polyglutamine binding protein 1 (PQBP1) in neurons profoundly impairs the morphogenesis of the primary cilium, including in the mouse cerebral cortex in vivo. PQBP1 is localized at the base of the neuronal cilium, and targeting its WW effector domain to the cilium stimulates ciliary morphogenesis. We also find that PQBP1 interacts with Dynamin 2 and thereby inhibits its GTPase activity. Accordingly, Dynamin 2 knockdown in neurons stimulates ciliogenesis and suppresses the PQBP1 knockdown-induced ciliary phenotype. Strikingly, a mutation of the PQBP1 WW domain that causes XLID disrupts its ability to interact with and inhibit Dynamin 2 and to induce neuronal ciliogenesis. These findings define PQBP1 and Dynamin 2 as components of a signaling pathway that orchestrates neuronal ciliary morphogenesis in the brain.
Introduction
Intellectual disability (ID) is a common developmental disorder with a prevalence of 1 to 3 percent (Bhasin et al., 2006; Larson et al., 2001). No effective treatments are available, and thus there is an urgent need to better understand the molecular pathogenesis of ID. Human genetic studies have led to the identification of many genes whose mutations cause ID (Chelly et al., 2006; Ropers, 2010). However, the functions of ID proteins remain largely to be elucidated.
Nearly 30 to 50 percent more males than females are affected with ID, presumably reflecting mutations on the X-chromosome. Mutations in more than 80 genes on the X chromosome have been associated with ID (Chiurazzi et al., 2008; Gécz et al., 2009). In many cases, XLID mutations cause syndromes that include constellations of symptoms and signs outside the nervous system. The heterogeneity of genes and associated ID syndromes raise the question of whether common or disparate pathogenic signaling mechanisms underlie ID. To begin to address this fundamental question, a better understanding of the functions of ID-associated proteins is required. Notably, a substantial portion of XLID genes encode proteins predicted to localize in the nucleus (Chiurazzi et al., 2008), providing a starting point for investigation of cell-intrinsic regulation of neuronal development and function by these XLID proteins.
Deregulation of neuronal morphogenesis represents a prominent pathological feature in ID. Impaired development of dendrites and dendritic spines may represent an important aspect of neuronal pathology in brains of ID patients (Kaufmann and Moser, 2000; Purpura, 1974), supporting the concept that deregulation of neuronal connectivity contributes to the pathogenesis of ID. Consistent with these studies, XLID proteins regulate the development of dendrite arbors and the formation of dendritic spines in neurons (Irwin et al., 2000; Iwase et al., 2007). These observations raise the question of whether XLID proteins regulate other aspects of neuronal morphogenesis.
The primary cilium represents an intriguing organelle that has received attention in recent years (Gerdes et al., 2009; Singla and Reiter, 2006). Primary cilia play critical roles in early embryonic development and organogenesis in vertebrates by providing a unique cellular domain that facilitates signal transduction in response to morphogens and growth factors (Goetz and Anderson, 2010; Lancaster and Gleeson, 2009). Disruption of the formation or function of primary cilia causes a heterogeneous group of ciliopathies including Bardet-Biedl, Meckel-Gruber and Joubert syndromes, which are characterized by overlapping clinical presentations of polycystic kidney, retinal degeneration, cerebellar atrophy, and notably intellectual disability (Badano et al., 2006; Hildebrandt et al., 2011). These observations support the hypothesis that impairment of neuronal ciliogenesis might represent an important feature in the pathogenesis of developmental cognitive disorders. Growing evidence suggests that the primary cilium functions as a signaling center in neurons (Lee and Gleeson, 2011; Louvi and Grove, 2011). Several G-protein coupled receptors (GPCRs) are localized within the neuronal cilium including the somatostatin receptor 3 (SSTR3), melanin concentrating hormone receptor 1 (MCHR1), serotonin receptor 6 (5HTR6), and dopamine receptors (Domire et al., 2010; Hamon et al., 1999; Händel et al., 1999; Marley and von Zastrow, 2010). In addition, the twelve transmembrane enzyme adenylyl cyclase 3 (AC3), which generates cyclic AMP in response to activation by GPCRs, is localized in the membrane of the neuronal cilium (Bishop et al., 2007). Mice in which the AC3 or SSTR3 gene is disrupted have defects in object recognition memory, suggesting an important role for cilia in regulating signaling events critical to cognitive function (Einstein et al., 2010; Wang et al., 2011). Thus, the primary cilium may play a key role in neuronal signaling and brain function.
The fundamental architecture of the primary cilium is composed of microtubule bundles extending from the basal body and an encasing ciliary membrane on which the ciliary receptors localize. Components of the primary cilium are sorted and trafficked by the intraflagellar transport (IFT) system and ciliary membrane trafficking, which control the formation and function of the cilium (Nachury et al., 2010; Pedersen et al., 2008). Although the mechanisms that control ciliogenesis in non-neuronal cells have been intensely investigated, the mechanisms that specifically orchestrate the development and morphogenesis of the neuronal cilium remain poorly understood.
In this study, we identify a novel function for the major XLID protein polyglutamine binding protein 1 (PQBP1) in the morphogenesis of the primary cilium in postmitotic neurons. Knockdown of PQBP1 profoundly impairs the formation of the primary cilium in hippocampal neurons and the mouse cerebral cortex in vivo. PQBP1 is localized at the base of the neuronal cilium as well as the nucleus, and targeting the PQBP1 WW effector domain to the cilium induces ciliary development. We also identify the protein Dynamin 2 as a cytoplasmic target of PQBP1 in neurons. PQBP1 interacts with Dynamin 2 and thereby inhibits the GTPase activity of Dynamin 2. Accordingly, Dynamin 2 knockdown stimulates ciliary morphogenesis and suppresses the PQBP1 knockdown-induced loss of cilia in neurons. Importantly, we also find that a mutation of the PQBP1 WW domain that causes XLID deregulates PQBP1-Dynamin 2 signaling and consequent ciliary morphogenesis in neurons. The identification of the PQBP1-Dynamin 2 signaling link as a critical regulator of neuronal ciliary development in the brain bears potential implications for our understanding of the mechanisms underlying ID.
Results
To investigate the functions of XLID proteins in ciliary morphogenesis, we performed a targeted RNAi screen of XLID genes in primary rat hippocampal neurons. We focused our attention on genes encoding proteins predicted to localize in the nucleus and regulate transcription or RNA processing. Knockdown of each XLID protein by cognate short hairpin RNAs (shRNAs) was validated (Figures S1F and 2A).
To visualize the primary cilium in hippocampal neurons, we performed immunocytochemical analyses using an antibody that recognizes adenylyl cyclase 3 (AC3). A single bar-shaped ciliary structure extending from the soma of hippocampal neurons was observed (Figure S1A). The base of the cilium was contiguous with the centrosome, the latter identified by expression of Centrin-GFP (Figure S1A). Although the non-neuronal ciliary marker acetylated tubulin was expressed within the AC3-positive cilia in neurons, it was also detected throughout the cytoplasm in neurons (Figure S1B). The neuronal cilium also displayed expression of somatostatin receptor 3 (SSTR3) and melanin concentration hormone receptor 1 (MCHR1) (Figures S1C and S1D). These data establish that primary hippocampal neurons harbor primary cilia. The percentage of hippocampal neurons bearing a cilium increased as neurons matured between 2 days in vitro (DIV2) and DIV7 (Figure S1E).
To assess the effect of knockdown of nuclear XLID proteins on ciliary morphogenesis, we transfected hippocampal neurons with RNAi plasmids targeting each XLID protein together with a GFP expression plasmid at DIV1, and subjected neurons at DIV5 to immunocytochemical analyses using the AC3 and GFP antibodies. Strikingly, we found that knockdown of the XLID protein, PQBP1, using two shRNAs targeting distinct regions of PQBP1 mRNA, consistently and substantially reduced the percentage of neurons harboring a primary cilium (Figures 1, 2A–C, and S2A). Mutations of the PQBP1 gene comprise one of the more frequent causes of XLID. More than 70 affected individuals from more than 20 families with PQBP1 mutations have been reported in ID (Germanaud et al., 2011; Jensen et al., 2011; Rejeb et al., 2011; Sheen et al., 2010; Stevenson et al., 2005). PQBP1 is a predominantly nuclear protein that regulates transcription and splicing (Tapia et al., 2010; Waragai et al., 1999; Zhang et al., 2000), but the pathophysiologically relevant functions of PQBP1 in XLID have remained unknown. Therefore, we further characterized the role of PQBP1 in the development of the primary cilium in neurons. Besides AC3 staining, expression of the intraflagellar transport protein IFT88 fused to GFP (GFP-IFT88) was used as another ciliary marker to interrogate the function of PQBP1 (Figures 2D and S2B). IFT88 is a component of the kinesin-dependent anterograde ciliary trafficking protein complex IFTB (Gerdes et al., 2009). PQBP1 knockdown reduced the number of neurons bearing a GFP-IFT88-positive cilium (Figures 2D and 2E). Likewise, PQBP1 knockdown reduced the number of neurons bearing a SSTR3 positive cilium (Figure S2C). These observations support the conclusion that PQBP1 is required for ciliogenesis in hippocampal neurons. In contrast, knockdown of PQBP1 had little or no effect on ciliogenesis in NIH3T3 fibroblasts and MDCK epithelial cells in the presence or absence of serum (Figures S2D–G). Consistent with these results, in contrast to PQBP1’s localization at the primary clium in neurons, PQBP1 failed to localize to the cilium in NIH3T3 and MDCK cells in the presence or absence of serum (Figures S2H–K). Taken together, these data suggest that PQBP1 promotes ciliogenesis selectively in neurons.
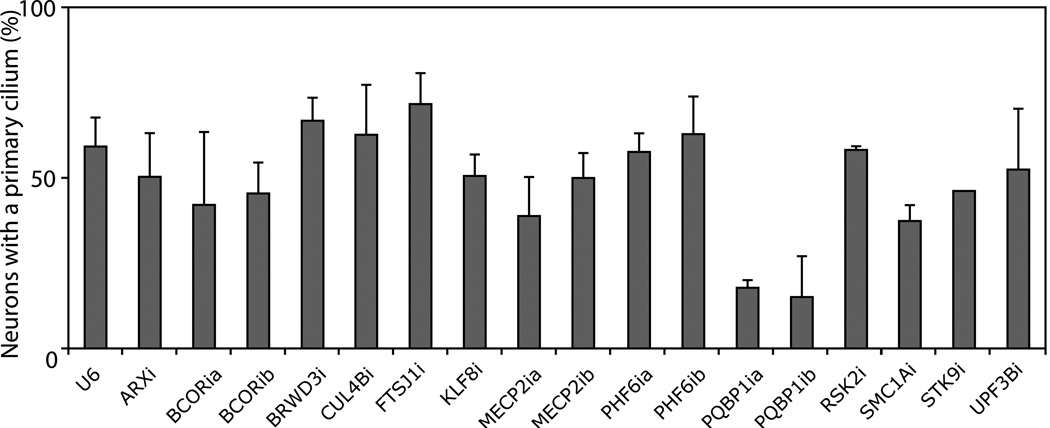
Figure 1. A targeted RNAi screen of XLID genes in ciliary morphogenesis in neurons.
Hippocampal neurons were transfected with an RNAi plasmid encoding shRNAs targeting the indicated XLID gene or the control U6 plasmid together with the GFP expression plasmid and subjected to immunocytochemistry at DIV5 using the AC3 and GFP antibodies. Knockdown of each protein by cognate shRNAs was validated (Figures S1 and 2A). Knockdown of PQBP1 consistently reduced the percentage of neurons harboring a primary cilium (p < 0.05; ANOVA).
Total of 2646 neurons were quantified.
See also Figure S1
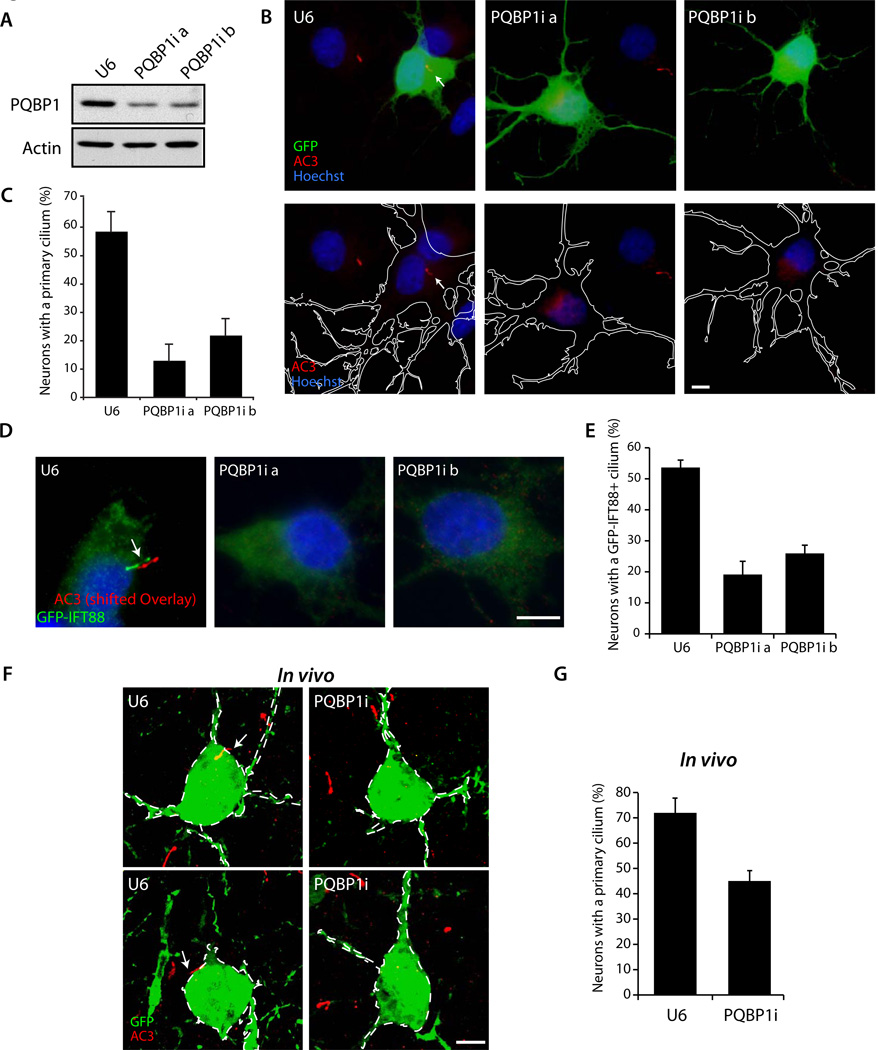
Figure 2. PQBP1 is required for ciliogenesis in primary hippocampal neurons and the cerebral cortex in vivo.
(A) Lysates of 293T cells transfected with the Flag-PQBP1 expression plasmid along with the PQBP1 RNAi or control U6 RNAi plasmid were immunoblotted with the Flag or Actin antibodies, the latter to serve as loading control.
(B) Hippocampal neurons transfected with the PQBP1 RNAi or control U6 RNAi plasmid together with the GFP expression plasmid were subjected to immunocytochemistry using the GFP and AC3 antibodies. Representative neurons are shown. Arrow indicates the primary cilium. The outline of transfected neurons is shown with a white line in the lower panels. Scale bar = 5 µm.
(C) The percentage of neurons harboring a primary cilium was significantly reduced upon PQBP1 knockdown (p < 0.01; ANOVA). Total of 1240 neurons were counted.
(D) Hippocampal neurons transfected with the PQBP1 RNAi or control U6 RNAi plasmid together with a GFP-IFT88 expression plasmid were subjected to immunocytochemistry with the GFP and AC3 antibodies. Representative neurons are shown. Arrow indicates the primary cilium. The AC3 signal is shifted. Scale bar = 5 µm.
(E) The percentage of neurons bearing a GFP-IFT88 positive cilium was significantly reduced upon PQBP1 knockdown (p < 0.05; ANOVA). Total of 244 neurons were counted.
(F) Mouse embryos were electroporated with the PQBP1 RNAi or control U6 RNAi plasmid together with the GFP expression plasmid and subjected to immunohistochemistry at P10 with the GFP and AC3 antibodies. Arrows indicate the primary cilium. PQBP1 RNAi reduced the number of neurons bearing a cilium. Scale bar = 5 µm.
(G) The percentage of neurons harboring a primary cilium was significantly reduced in PQBP1 knockdown animals as compared to the U6 control plasmid (p < 0.01; t-test). Total of 355 neurons were quantified.
See also Figure S2
We next determined the function of PQBP1 in the brain in vivo. We used an in utero electroporation method to transfect embryonic day 15.5 (E15.5) mouse pups with the PQBP1 RNAi or control U6 RNAi plasmid. Animals were sacrificed at postnatal day 10 (P10) and the cerebral cortex was subjected to immunohistochemical analyses using the AC3 antibody. In these experiments, neurons that had migrated from the ventricular zone and reached the cortical plate extended a primary cilium (Figures 2F and S2L). Over 70% of neurons in the cerebral cortex in pups transfected with the control plasmid harbored a primary cilium (Figure 2G). By contrast, only 45% neurons in the cerebral cortex in PQBP1 knockdown animals harbored a primary cilium (Figure 2G). These data suggest that the XLID protein PQBP1 plays a critical role in the morphogenesis of the neuronal cilium in the mouse brain in vivo.
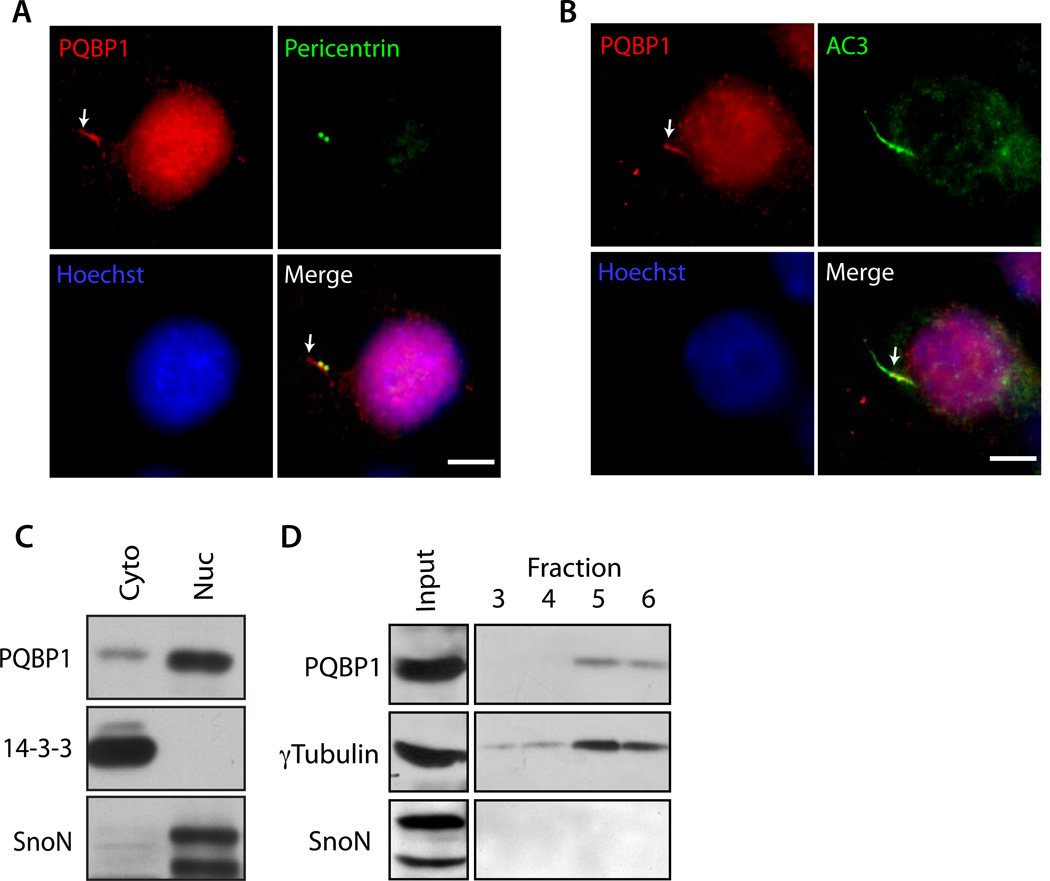
Identification of a critical role for PQBP1 in ciliary morphogenesis in the mammalian brain led us next to investigate the molecular basis of PQBP1’s function. We first characterized the subcellular localization of PQBP1 in neurons. Although overexpressed PQBP1 was predominantly localized in the nucleus (data not shown), in addition to its expression in the nucleus, PQBP1 was localized at the cilium in hippocampal neurons (Figures 3A, B, S3A and D). PQBP1 immunoreactivity was contiguous with the centrosome (Figure 3A), and was often observed at the base of the primary cilium (Figure 3B). Ciliary PQBP1 immunoreactivity was blocked by pre-absorption with PQBP1 protein (Figure S3B), and reduced upon PQBP1 knockdown (Figure S3C), demonstrating the specificity of the PQBP1 immunoreactive signal. Biochemical fractionation of hippocampal neurons revealed that the cytoplasmic fraction contained PQBP1 (Figure 3C). Further fractionation of hippocampal neuron lysates with sucrose gradient demonstrated that PQBP1 co-fractionated with the centrosomal marker γTubulin (Figure 3D). Together, these data reveal that in addition to expression in the nucleus, PQBP1 is localized at the base of the primary cilium in neurons.
Figure 3. PQBP1 localizes at the base of the cilium and centrosome.
(A and B) Hippocampal neurons were subjected to immunocytochemistry with the PQBP1 antibody together with the Pericentrin antibody (A), or AC3 antibody (B). Arrows indicate PQBP1 immunoreactivity at the cilium. Scale bar = 5 µm.
(C) Cytoplasmic and nuclear fractions isolated from rat cortical neurons were immunoblotted using the PQBP1, 14-3-3 and SnoN antibodies. 14-3-3 and SnoN served as the cytoplasmic and nuclear marker, respectively.
(D) Centrosomal fractions prepared from rat cortical neuron lysates were immunoblotted using the PQBP1, γTubulin, and SnoN antibodies. PQBP1 and γTubulin cofractionated, suggesting that PQBP1 is present at centrosome. SnoN served as negative control.
See also Figure S3
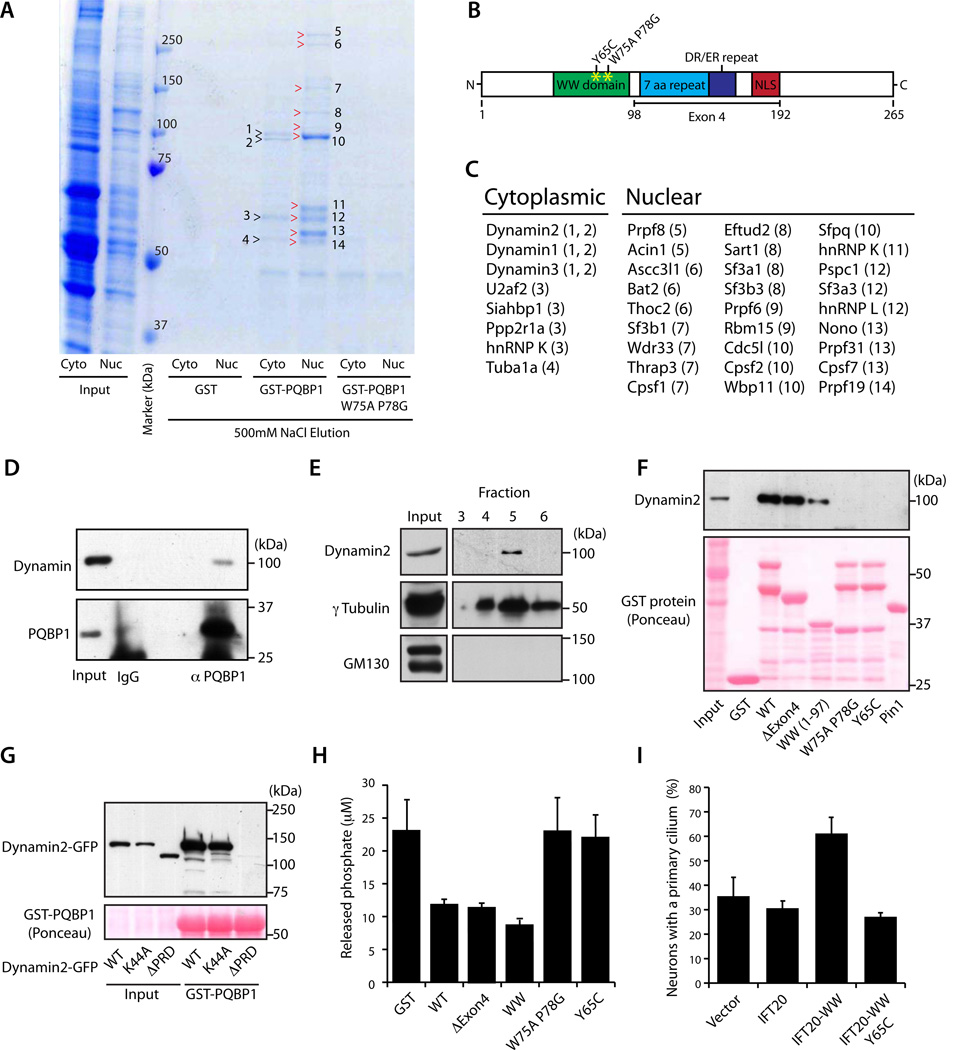
We next characterized the function of PQBP1 outside of the nucleus in the regulation of ciliogenesis. We sought to identify proteins that interact with PQBP1 specifically in the cytoplasm but not nucleus. Therefore, we performed GST pull-down analyses using a GST-PQBP1 fusion protein or the control protein GST as bait and nuclear or cytoplasmic lysates of cortical neurons as the source of protein, followed by high-salt elution, SDS-PAGE and Coomassie staining (Figures 4A and S4A). Notably, the cytoplasmic fraction of cortical neuron lysates contained centrosomal and ciliary proteins (Figure S4B). We found 4 protein bands in the cytoplasmic fraction and 10 protein bands in the nuclear fraction that specifically associated with GST-PQBP1 but not GST (Figure 4A). All of the visible protein bands that coprecipitated with GST-PQBP1 were absent in a pull down assay using a GST-PQBP1 protein containing a mutation at critical amino acids in the WW domain (GST-PQBP1 W75A P78G) (Figures 4A and B), suggesting that the WW domain is required for the interaction. PQBP1-associated proteins were identified by mass spectrometry (Figure 4C). Consistent with PQBP1’s reported function in RNA splicing (Llorian et al., 2005; Tapia et al., 2010; Zhang et al., 2000), pre-mRNA processing factors were among nuclear proteins that interacted with PQBP1, validating the utility of the GST pull down assay for identification of PQBP1-interacting proteins. Importantly, we identified Dynamin 2 in the cytoplasmic fraction as a novel PQBP1-interacting protein (Figure 4C). In coimmunoprecipitation analyses, endogenous PQBP1 formed a complex with endogenous Dynamin 2 in the cytoplasmic fraction of P6 brain lysates (Figure 4D). Taken together, these data suggest that Dynamin 2 represents a new cytoplasmic interaction partner for PQBP1.
Figure 4. PQBP1 forms a complex with Dynamin 2 and thereby inhibits the GTPase activity of Dynamin 2.
(A) Proteins coprecipitated with GST-PQBP1, GST-PQBP1 W75A P78G, or GST were analyzed by SDS-PAGE and Coomassie Brilliant Blue staining. Numbers and arrowheads indicate bands analyzed by mass spectrometry.
(B) Schematic of PQBP1 protein domain structure. The WW domain, 7 aa repeat, DR/ER repeat, and nuclear localization signal (NLS) are indicated.
(C) Identity of PQBP1-associated proteins in cytoplasmic and nuclear fractions of cortical neuron lysates. Numbers in parenthesis indicate protein band in (A).
(D) Cytoplasmic fraction of P6 rat brain lysate was immunoprecipitated with the PQBP1 antibody or rabbit IgG and immunoblotted with the Dynamin or PQBP1 antibody. Asterisk denotes a non-specific band.
(E) Centrosomal fractions isolated from rat cortical neurons were immunoblotted using the Dynamin 2, γTubulin, and GM130 antibodies. Dynamin 2 and γTubulin cofractionated, suggesting Dynamin 2 is present at the centrosome. GM130 served as a negative control.
(F) Lysates of rat cortical neurons subjected to pull-down assay with GST, GST-PQBP1, GST-PQBP1 ΔExon4 (Δ98–192), GST-PQBP1 WW (1-97), GST-PQBP1 W75A P78G, GST-PQBP1 Y65C or GST-Pin1, and immunoblotted with Dynamin 2 antibody or stained with Ponceau S.
(G) Lysates of 293T cells transfected with Dynamin 2-GFP, Dynamin 2 K44A-GFP or Dynamin 2 ΔPRD-GFP were subjected to a pull-down assay with GST-PQBP1 and immunoblotted with the GFP antibody or stained with Ponceau S.
(H) Phosphate released from GTP by Dynamin 2 was measured in the presence of GST, GST-PQBP1, GST-PQBP1 ΔExon4 (Δ98–192), GST-PQBP1 WW (1-97), GST-PQBP1 W75AP78G, or GST-PQBP1 Y65C.
(I) The percentage of hippocampal neurons bearing a primary cilium was significantly higher at DIV3 in neurons transfected with GFP-IFT20-WW compared to control vector, GFP-IFT20 or GFP-IFT20-WW Y65C (p < 0.01; ANOVA). Total of 387 neurons were measured. See also Figure S4
PQBP1 interacted with all three Dynamin isoforms in the cytoplasmic fraction of cortical neurons (Figure 4C). Notably, PQBP1 interacted selectively with Dynamin 1 and 2 in GST-pull down assays (Figure S4C). We focused on Dynamin 2, because Dynamin 2 is reported to localize to the centrosome (Liu et al., 2007; Thompson et al., 2004). Dynamin 2 co-fractionated with the centrosomal marker γTubulin in fractionated lysates of neurons (Figures 4E and S4D), corroborating the conclusion that Dynamin 2 is localized at the centrosome in neurons. In addition, a Dynamin 2-GFP fusion protein partially colocalized with the centrosomal marker Pericentrin (Figure S4E) and PQBP1 in neurons (Figure S4F).
We performed structure-function analyses to identify the structural determinants that mediate PQBP1’s interaction with Dynamin 2. Deletion of PQBP1’s alternatively spliced exon 4, containing amino acids 98–192, did not impair interaction with Dynamin 2 (Figure 4F). Further deletion of the entire C-terminal region (98–265aa) leaving only the WW domain (1–97aa) maintained the ability of PQBP1 to form a complex with Dynamin 2, although the interaction was modestly diminished (Figure 4F). These results suggest that the PQBP1 WW domain is sufficient to interact with Dynamin 2. The WW domain protein Pin1 failed to bind Dynamin 2 (Figure 4F), suggesting that Dynamin 2 specifically interacts with the PQBP1 WW domain.
We next determined the requirement for the WW domain in PQBP1’s interaction with Dynamin 2. Mutation of two conserved amino acids in the WW domain in full length PQBP1 (W75A P78G) abolished PQBP1’s interaction with Dynamin 2 (Figure 4F), suggesting that the WW domain is essential for PQBP1’s association with Dynamin 2. The missense mutation A194G leading to Y65C substitution within the PQBP1 WW domain causes the Golabi–Ito–Hall ID syndrome (Lubs et al., 2006). Importantly, the Y65C mutation blocked the ability of PQBP1 to interact with Dynamin 2 (Figure 4F), suggesting that the interaction of PQBP1 with Dynamin 2 may be deregulated in XLID.
We next identified the Dynamin 2 domain that interacts with PQBP1. A Dynamin 2 protein in which the GTPase active site Lys44 was replaced with alanine (K44A) formed a complex with PQBP1 (Figure 4G), suggesting that the GTPase activity is not required for Dynamin 2’s interaction with PQBP1. In contrast, a Dynamin 2 mutant protein lacking the C-terminal proline/arginine-rich domain (ΔPRD) failed to form a complex with PQBP1, suggesting that the PRD is required for Dynamin 2’s interaction with PQBP1.
Because Dynamin’s GTPase activity is regulated by protein-protein interactions through its PRD (Schmid and Frolov, 2011), we asked if PQBP1 might modulate the GTPase activity of Dynamin 2. Remarkably, the GTPase activity of Dynamin 2 measured in an in vitro assay was substantially attenuated in the presence of PQBP1 (Figure 4H). In structure-function analyses, removal of exon 4 or the entire C-terminal domain had little or no effect on the ability of PQBP1 to inhibit Dynamin 2’s GTPase activity (Figure 4H), suggesting that the PQBP1 WW domain is sufficient to inhibit Dynamin 2 activity. In contrast, mutation of the conserved residues within the WW domain (W75A P78G) impaired the ability of PQBP1 to inhibit the Dynamin 2 GTPase activity (Figure 4H). Importantly, the Golabi-Ito-Hall syndrome mutation Y65C also impaired PQBP1’s ability to inhibit the GTPase activity of Dynamin 2 (Figure 4H). Taken together, our results demonstrate that PQBP1 interacts via its WW domain with Dynamin 2 and thereby inhibits the GTPase activity of Dynamin 2.
Because the WW domain of PQBP1 is sufficient to inhibit Dynamin 2 activity, we employed a gain-of-function approach to target the PQBP1 WW domain to the cilium and interrogate its function in ciliary morphogenesis. The protein IFT20, a component of a protein complex that mediates anterograde ciliary trafficking, localizes to the cilium and basal body as well as the Golgi apparatus (Follit et al., 2006). To localize the PQBP1 WW domain at the cilium, we expressed a protein in which the WW domain was fused to IFT20 (IFT20-WW, Figures S4H and I). Expression of IFT20 alone had little or no effect on ciliary morphogenesis in hippocampal neurons (Figure 4I). By contrast, expression of the IFT20-WW fusion protein substantially increased the percentage of neurons bearing a neuronal cilium (Figure 4I). Remarkably, the Golabi–Ito–Hall ID mutation Y65C abolished the ability of IFT20-WW to promote ciliary morphogenesis (Figure 4I). Expression of the PQBP1 WW domain in the nucleus, achieved by fusing the WW domain to the nuclear localizing signal (NLS-WW), failed to promote ciliogenesis in neurons (Figure S4G), suggesting that PQBP1 operates outside of the nucleus to promote the formation of the primary cilium in neurons. These data suggest that localizing the PQBP1 WW domain to the cilium stimulates ciliary morphogenesis and that impairment of this function may play a pathogenic role in the Golabi–Ito–Hall ID syndrome.
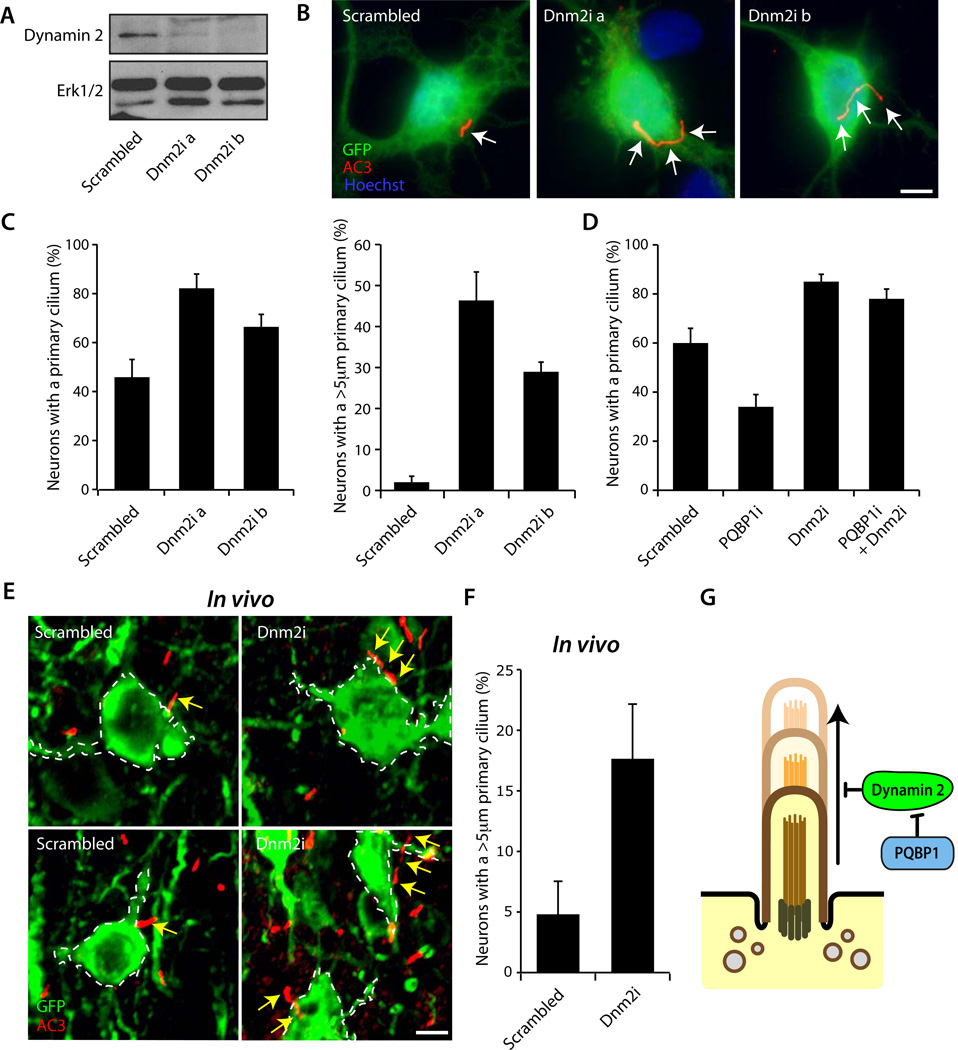
To determine the role of Dynamin 2 in ciliary morphogenesis, we induced Dynamin 2 knockdown in hippocampal neurons using two shRNAs targeting distinct regions of Dynamin 2 mRNA (Figure 5A). Since PQBP1 inhibits Dynamin 2 activity, inhibition of Dynamin 2 would be predicted to induce the formation of the neuronal cilium. Consistent with this prediction, Dynamin 2 knockdown promoted ciliary morphogenesis in hippocampal neurons (Figures 5B, 5C and S5A). The percentage of neurons harboring a primary cilium was increased, and the cilium was substantially longer in Dynamin 2 knockdown neurons (Figure 5C), suggesting that Dynamin 2 inhibits the formation and growth of the primary cilium in neurons. In epistasis analyses, Dynamin 2 knockdown suppressed the PQBP1 knockdown-induced phenotype of loss of cilia (Figure 5D), suggesting that Dynamin 2 operates downstream of PQBP1 in the control of ciliary morphogenesis. Consistently, knockdown of Dynamin 2 had little or no effect on the localization of PQBP1 at the cilium in neurons (Figure S5B).
Figure 5. Dynamin 2 inhibits the formation and elongation of the neuronal cilium.
(A) Lysates of 293T cells transfected with the Dynamin 2-GFP expression plasmid together with the Dynamin 2 RNAi or control Scrambled RNAi plasmid were immunoblotted with the GFP or Erk1/2 antibody, the latter to serve as a loading control. Expression of Dynamin 2 shRNA induced knockdown of Dynamin 2.
(B) Hippocampal neurons transfected with the Dynamin 2 RNAi or control Scrambled RNAi plasmid together with the GFP expression plasmid were subjected to immunocytochemistry with the GFP and AC3 antibodies. Representative neurons are shown. Arrows indicate the primary cilium. Elongated cilia were observed in Dynamin 2 knockdown neurons. Scale bar = 5 µm.
(C) The percentage of hippocampal neurons bearing a primary cilium was significantly higher in Dynamin 2 knockdown neurons as compared to control neurons (Left, p < 0.05; ANOVA). The percentage of hippocampal neurons bearing the primary cilium longer than 5µm was significantly higher in Dynamin 2 knockdown neurons as compared to control neurons (Right, p < 0.01; ANOVA). Total of 389 neurons were measured.
(D) Hippocampal neurons were transfected with the PQBP1 RNAi, Dynamin 2 RNAi, the combination of PQBP1 RNAi and Dynamin 2 RNAi, or control Scrambled RNAi plasmid, and analyzed as in (B). The percentage of neurons bearing a primary cilium was significantly reduced in PQBP1 knockdown neurons as compared to control neurons, and was significantly increased in Dynamin 2 knockdown neurons in the presence or absence of PQBP1 knockdown when compared to control neurons (p < 0.05; ANOVA). Total of 487 neurons were measured.
(E) Cortical slices infected with the Dynamin 2 RNAi or Scrambled RNAi lentivirus with mCitrine expression were subjected to immunohistochemistry with the GFP and AC3 antibodies. Representative neurons are shown. Arrows indicate the primary cilium. Scale bar = 5 µm.
(F) Dynamin 2 knockdown increased the number of neurons with the cilium longer than 5 µm as compared to Scrambled control lentivirus in vivo (p < 0.05; t-test). Total of 152 neurons were measured.
(G) Model depicting the role of PQBP1 and Dynamin 2 in neuronal ciliogenesis. See also Figure S5
We next characterized Dynamin 2 function in neuronal ciliary development in vivo. Dynamin 2 RNAi induced using in utero electroporation led to impaired migration of cortical neurons, suggesting a function for Dynamin 2 in neuronal precursor cells or immature neurons (data not shown). This phenotype precluded the use of in utero electroporation to assess the role of Dynamin 2 in ciliary morphogenesis in mature neurons. To overcome this limitation, we induced Dynamin 2 knockdown in the cerebral cortex in neonatal mice using a lentiviral approach. We found a 3-fold increase in the percentage of neurons in the cerebral cortex in Dynamin 2 knockdown animals that had a cilium longer than 5µm compared to control animals (Figures 5E and 5F), demonstrating that Dynamin 2 inhibits ciliary morphogenesis in vivo. Together, these data reveal a novel function for Dynamin 2 in the control of ciliogenesis in the developing rodent brain. Collectively, our study defines a novel XLID-deregulated PQBP1-Dynamin 2 signaling link that orchestrates ciliary morphogenesis in postmitotic neurons in the brain.
Discussion
In this study, we have discovered a signaling link between the major XLID protein PQBP1 and the GTPase Dynamin 2 that regulates the formation of the primary neuronal cilium in the brain (see model in Figure 5G). PQBP1 is localized at the base of the cilium and plays an important role in ciliogenesis in primary rodent hippocampal neurons and cortical neurons in the mouse brain in vivo. We have also identified the molecular basis by which PQBP1 drives the morphogenesis of the neuronal cilium. PQBP1 forms a complex with the GTPase Dynamin 2 and thereby inhibits its GTPase activity. Knockdown of Dynamin 2 stimulates the formation of the neuronal cilium and suppresses PQBP1 knockdown-induced loss of cilia in neurons. A patient-specific mutation of the PQBP1 WW domain that causes XLID impairs the ability of PQBP1 to interact with and thereby inhibit Dynamin 2 and to promote ciliary morphogenesis. Taken together, our findings define PQBP1-Dynamin 2 signaling as a novel mechanism that orchestrates neuronal ciliary morphogenesis in the brain, with potential implications for the pathogenesis of ID.
The identification of a function for PQBP1 in ciliary morphogenesis in postmitotic neurons may shed light on the pathogenesis of XLID caused by mutations of the PQBP1 gene in syndromic as well as non-syndromic ID (Germanaud et al., 2011; Jensen et al., 2011; Rejeb et al., 2011; Sheen et al., 2010; Stevenson et al., 2005). Mutations of PQBP1 cause recessive XLID, suggesting that these disorders result from loss of PQBP1 function. Accordingly, several XLID-associated PQBP1 mutations cause premature termination and downregulation of PQBP1 protein due to nonsense-mediated RNA decay (Musante et al., 2010). Notably, a missense mutation in the Golabi–Ito–Hall syndrome alters a conserved residue within the WW domain of PQBP1 (Lubs et al., 2006). These observations highlight the importance of the WW domain in the pathogenesis of ID.
We have found that the WW domain is essential for PQBP1’s ability to drive ciliary formation in neurons. PQBP1 forms a complex via its WW domain with the GTPase Dynamin 2, attenuates the GTPase activity of Dynamin 2, and thereby triggers ciliogenesis, potentially through regulation of the ciliary protein/membrane trafficking machinery. Importantly, the Golabi-Ito-Hall mutation in theWW domain disrupts the PQBP1-Dynamin 2 interaction and consequent inhibition of Dynamin 2 activity, and thus impairs the ability of PQBP1 to promote ciliogenesis in neurons. Thus, our findings suggest that deregulation of PQBP1-Dynamin 2 signaling in neuronal ciliogenesis may play a role in the pathogenesis of ID. It will be important to further explore the role of impairment of PQBP1-Dynamin 2 signaling and ciliogenesis in the pathogenesis of ID.
Previous studies and our GST pull down analyses revealed that the WW domain of PQBP1 mediates interaction with nuclear proteins including splicing factors (Figure 4C) (Llorian et al., 2005; Tapia et al., 2010). However, the PQBP1 WW domain targeted to the nucleus failed to promote ciliogenesis, whereas targeting the WW domain to the cilium substantially promoted ciliogenesis (Figure S4G). These data suggest that PQBP1 operates at the cilium, but not in the nucleus, to promote ciliogenesis in neurons. Whether disruption of PQBP1 function at the cilium contributes to the pathological mechanisms of ID remains to be determined, since mutation of the WW domain disrupts the interaction of PQBP1 with Dynamin 2 as well as other interaction partners (Figure 4A) (Tapia et al., 2010).
Interestingly, PQBP1 binds to polyglutamine proteins and may contribute to the pathogenesis of polyglutamine-dependent neurodegenerative disorders (Okazawa et al., 2002; Okuda et al., 2003). Therefore, PQBP1’s novel function in ciliogenesis may also be deregulated in neurodegenerative disorders, raising interesting parallels between developmental disorders of cognition and neurodegeneration. The PQBP1-interacting protein Huntingtin also regulates ciliogenesis (Keryer et al., 2011; Liu and Zeitlin, 2011; Waragai et al., 1999), raising the question of whether PQBP1 and Huntingtin cooperate in the control of neuronal ciliogenesis. The cytoplasmic protein Dynactin forms a complex with PQBP1 (Kunde et al., 2011). Because Dynactin is essential for microtubule-dependent transport and regulates the organization of microtubules at the centrosome (Quintyne et al., 1999), it will be interesting to determine whether Dynactin cooperates with PQBP1 in ciliogenesis in neurons.
Identification of the signaling link between the XLID protein PQBP1 and the GTPase Dynamin 2 illuminates a mechanism that governs the formation of the primary cilium in postmitotic neurons. Notably, inhibition of PQBP1 or Dynamin had little or no effect on the morphology of the primary cilium in non-neuronal cells, suggesting that PQBP1-Dynamin 2 signaling regulates ciliary morphogenesis specifically in neurons.
Our study highlights a new fundamental biological function for Dynamin 2. Members of the Dynamin family of GTPases are the subject of intense interest, because of their functions in endocytosis and synaptic vesicle recycling (Murthy and De Camilli, 2003; Schmid and Frolov, 2011). The GTPase activity of Dynamins triggers membrane fission, which is critical in Dynamin function in endocytosis and synaptic vesicle recycling (Schmid and Frolov, 2011). Identification of the PQBP1-Dynamin 2 signaling link raises the question of whether PQBP1 might also regulate endocytosis and synaptic vesicle recycling.
Recent studies suggest that the primary cilium represents a critical site of pathology in several developmental disorders including the Bardet-Biedl syndrome (BBS) characterized by obesity, kidney abnormalities, and notably intellectual disability (Zaghloul and Katsanis, 2009). Several proteins implicated in this syndrome form a BBSome complex at the basal body, which regulates ciliogenesis in non-neuronal cells (Mykytyn et al., 2004; Nachury et al., 2007). Disruption of BBSome leads to loss of SSTR3 and MHCR1, and enrichment of Dopamine receptor 1 at the neuronal cilium (Berbari et al., 2008; Domire et al., 2010). Our findings raise the question of whether the PQBP1-Dynamin 2 pathway and the BBSome coordinately regulate ciliogenesis and ciliary receptor trafficking in neurons. In addition, it will be interesting to test whether inhibition of Dynamin 2 by small molecules might restore ciliary development and function in XLID syndromes in which PQBP1 is mutated as well as in other ID syndromes such the Bardet-Biedl syndrome that feature abnormalities in ciliary development and function. An improved understanding of the mechanisms that govern the morphogenesis of the primary cilium in neurons should provide novel insights into the pathogenesis of intellectual disability and lay the foundation for the development of therapeutic approaches for these devastating disorders.
Methods
Primary neuron culture
Hippocampal neurons were prepared from embryonic day 18 (E18) Sprague-Dawley rat embryos. Neurons were plated on coverslips coated with poly-L-lysine (Sigma) and cultured in Neurobasal media (Invitrogen) with B27 supplement and penicillin/streptomycin. For ciliary morphology analysis, hippocampal neurons were transfected 16 hours after plating using a modified calcium phosphate method as described (Konishi et al., 2004) with the indicated plasmids together with GFP expression plasmid to visualize transfected neurons.
Immunocytochemistry
For visualization of centrosome-associated proteins including PQBP1, Pericentrin and Centrin-GFP, neurons were fixed in methanol for ten minutes at −20°C and subjected to immunocytochemistry analysis. For other immunocytochemistry experiments, neurons were fixed in 4% paraformaldehyde for 10 minutes at room temperature and analyzed as described (Konishi et al., 2004). For double labeling with two rabbit primary antibodies, the PQBP1 antibody and the Pericentrin or AC3 antibody, sequential staining was performed using goat anti-rabbit Fab fragment (Jackson ImmunoResearch) prior to the second primary antibody to eliminate cross reactivity as described (Berbari et al., 2007).
Fractionation
Nuclear and cytoplasmic fractions were prepared as described (Konishi et al., 2004). Centrosomal fractions from cortical neurons were isolated as described (Kim et al., 2009; Puram et al., 2011).
Dynamin GTPase activity
A colorimetric GTPase assay was performed as described with modifications (Quan and Robinson, 2005). Flag-Dynamin 2 was immunopurified from lysates of 293T cells expressing Flag-Dynamin 2. Flag-Dynamin 2 (20 nM) was incubated with 0.5 mM GTP, 10 mM Tris-HCl pH7.4, 10 mM NaCl, 2 mM MgCl2 and 0.05% Tween 80 in 20 µL at 30°C for 60 minutes in the presence of 75 nM GST-PQBP1 protein. The reaction was stopped by adding 20 µL of solution containing 1 mg/mL Malachite Green oxalate, 10 mg/mL ammonium molybdate tetrahydrate and 1 N HCl. After 5 minutes of incubation, absorbance at wavelength 650 nm was measured. The concentration of phosphate generated from GTP hydrolysis was calculated using an inorganic phosphate standard curve.
In utero electroporation and viral injection
All animal surgical procedures were reviewed and approved by the Harvard Center for Comparative Medicine. In utero electroporation was performed as described (Saito, 2010). Briefly, the PQBP1 RNAi or control U6 RNAi plasmid was injected into the lateral ventricle of E15.5 mouse embryo together with the pCAG-GFP plasmid (Matsuda and Cepko, 2007). Three pulses of electricity with 35 V for 50 msec were applied using 5 mm tweezertrodes with the ECM830 electroporator (BTX). Electroporated mice were sacrificed at postnatal day 10. Brains were fixed, and 20 µm cryosections were prepared for immunohistochemical analyses. To eliminate potential variability in analyses of ciliary morphology in different cortical layers, we focused our analyses on neurons in layer 2/3 of the somatosensory cortex.
Lentivirus injection was performed as described (Cetin et al., 2006). Lentivirus was produced in 293T cells and concentrated by ultracentrifuge. The lentiviral solution was injected into the brain of postnatal day 1 (P1) mouse with a stereotaxic frame (Stoelting) at x (lateral) = 1 mm, y (rostral) = 1.8 mm and z (ventral) = 0.8 mm from Lambda. The viral solution (50 nL) was injected 6 times per animals with 30 seconds intervals using Nanoject II (Drummond). Brains were harvested at P10 and analyzed as described above.
Statistics
All analyses were completed from a minimum of three independent experiments. Statistical analyses were performed with GraphPad Prism 4.0. All histograms are presented as mean + s.e.m. unless otherwise noted. Student’s t-test was used for comparisons in experiments with two sample groups. In experiments with more than two sample groups, ANOVA was performed followed by Bonferroni’s post hoc test for pairwise comparison among three and greater than three groups.
Supplementary Material
Highlights.
The XLID protein PQBP1 plays an essential role in neuronal ciliogenesis.
PQBP1's WW domain interacts with Dynamin 2 and inhibits its GTPase activity.
Dynamin 2 operates downstream of PQBP1 in the control of ciliogenesis in neurons.
XLID mutation in PQBP1's WW domain deregulates PQBP1-Dynamin 2 link and ciliogenesis.
Acknowledgement
We thank Samantha Keough, Dr. Albert Kim and members of the Bonni laboratory for technical assistance and helpful discussions, Zachary Waldon for mass spectrometry analyses, Drs. Jerry Chen and Elly Nedivi for sharing protocols on the viral injection method, Drs. Yu-Zhu Zhang and Erica Golemis for PQBP1 cDNA, Dr. Mark McNiven for Dynamin 2-GFP. This work was supported by NIH grant NS051255 (A.B.), the National Science Foundation and the Albert J. Ryan Foundation (L.T.U.), the Japan Society of the Promotion of Science (Y.I.), and the Human Frontier Science Program (Y.I.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Badano JL, Mitsuma N, Beales PL, Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu Rev Genomics Hum Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- Berbari NF, Bishop GA, Askwith CC, Lewis JS, Mykytyn K. Hippocampal neurons possess primary cilia in culture. J Neurosci Res. 2007;85:1095–1100. doi: 10.1002/jnr.21209. [DOI] [PubMed] [Google Scholar]
- Berbari NF, Lewis JS, Bishop GA, Askwith CC, Mykytyn K. Bardet-Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proc Natl Acad Sci U S A. 2008;105:4242–4246. doi: 10.1073/pnas.0711027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhasin TK, Brocksen S, Avchen RN, Van Naarden Braun K. Prevalence of four developmental disabilities among children aged 8 years--Metropolitan Atlanta Developmental Disabilities Surveillance Program, 1996 and 2006. MMWR Surveill Summ. 2000;55:1–9. [PubMed] [Google Scholar]
- Bishop GA, Berbari NF, Lewis J, Mykytyn K. Type III adenylyl cyclase localizes to primary cilia throughout the adult mouse brain. J Comp Neurol. 2007;505:562–571. doi: 10.1002/cne.21510. [DOI] [PubMed] [Google Scholar]
- Cetin A, Komai S, Eliava M, Seeburg PH, Osten P. Stereotaxic gene delivery in the rodent brain. Nat Protoc. 2006;1:3166–3173. doi: 10.1038/nprot.2006.450. [DOI] [PubMed] [Google Scholar]
- Chelly J, Khelfaoui M, Francis F, Chérif B, Bienvenu T. Genetics and pathophysiology of mental retardation. Eur J Hum Genet. 2006;14:701–713. doi: 10.1038/sj.ejhg.5201595. [DOI] [PubMed] [Google Scholar]
- Chiurazzi P, Schwartz CE, Gecz J, Neri G. XLMR genes: update 2007. Eur J Hum Genet. 2008;16:422–434. doi: 10.1038/sj.ejhg.5201994. [DOI] [PubMed] [Google Scholar]
- Domire JS, Green JA, Lee KG, Johnson AD, Askwith CC, Mykytyn K. Dopamine receptor 1 localizes to neuronal cilia in a dynamic process that requires the Bardet-Biedl syndrome proteins. Cell Mol Life Sci. 2010 doi: 10.1007/s00018-010-0603-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einstein EB, Patterson CA, Hon BJ, Regan KA, Reddi J, Melnikoff DE, Mateer MJ, Schulz S, Johnson BN, Tallent MK. Somatostatin signaling in neuronal cilia is critical for object recognition memory. J Neurosci. 2010;30:4306–4314. doi: 10.1523/JNEUROSCI.5295-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit JA, Tuft RA, Fogarty KE, Pazour GJ. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol Biol Cell. 2006;17:3781–3792. doi: 10.1091/mbc.E06-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes JM, Davis EE, Katsanis N. The vertebrate primary cilium in development, homeostasis, and disease. Cell. 2009;137:32–45. doi: 10.1016/j.cell.2009.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germanaud D, Rossi M, Bussy G, Gérard D, Hertz-Pannier L, Blanchet P, Dollfus H, Giuliano F, Bennouna-Greene V, Sarda P, et al. The Renpenning syndrome spectrum: new clinical insights supported by 13 new PQBP1-mutated males. Clin Genet. 2011;79:225–235. doi: 10.1111/j.1399-0004.2010.01551.x. [DOI] [PubMed] [Google Scholar]
- Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gécz J, Shoubridge C, Corbett M. The genetic landscape of intellectual disability arising from chromosome X. Trends Genet. 2009;25:308–316. doi: 10.1016/j.tig.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Hamon M, Doucet E, Lefèvre K, Miquel MC, Lanfumey L, Insausti R, Frechilla D, Del Rio J, Vergé D. Antibodies and antisense oligonucleotide for probing the distribution and putative functions of central 5-HT6 receptors. Neuropsychopharmacology. 1999;21:68S–76S. doi: 10.1016/S0893-133X(99)00044-5. [DOI] [PubMed] [Google Scholar]
- Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med. 2011;364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Händel M, Schulz S, Stanarius A, Schreff M, Erdtmann-Vourliotis M, Schmidt H, Wolf G, Höllt V. Selective targeting of somatostatin receptor 3 to neuronal cilia. Neuroscience. 1999;89:909–926. doi: 10.1016/s0306-4522(98)00354-6. [DOI] [PubMed] [Google Scholar]
- Irwin SA, Galvez R, Greenough WT. Dendritic spine structural anomalies in fragile-X mental retardation syndrome. Cereb Cortex. 2000;10:1038–1044. doi: 10.1093/cercor/10.10.1038. [DOI] [PubMed] [Google Scholar]
- Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Jensen LR, Chen W, Moser B, Lipkowitz B, Schroeder C, Musante L, Tzschach A, Kalscheuer VM, Meloni I, Raynaud M, et al. Hybridisation-based resequencing of 17 X-linked intellectual disability genes in 135 patients reveals novel mutations in ATRX, SLC6A8 and PQBP1. Eur J Hum Genet. 2011 doi: 10.1038/ejhg.2010.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann WE, Moser HW. Dendritic anomalies in disorders associated with mental retardation. Cereb Cortex. 2000;10:981–991. doi: 10.1093/cercor/10.10.981. [DOI] [PubMed] [Google Scholar]
- Keryer G, Pineda JR, Liot G, Kim J, Dietrich P, Benstaali C, Smith K, Cordelières FP, Spassky N, Ferrante RJ, et al. Ciliogenesis is regulated by a huntingtin-HAP1-PCM1 pathway and is altered in Huntington disease. J Clin Invest. 2011;121:4372–4382. doi: 10.1172/JCI57552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim AH, Puram SV, Bilimoria PM, Ikeuchi Y, Keough S, Wong M, Rowitch D, Bonni A. A centrosomal Cdc20-APC pathway controls dendrite morphogenesis in postmitotic neurons. Cell. 2009;136:322–336. doi: 10.1016/j.cell.2008.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi Y, Stegmüller J, Matsuda T, Bonni S, Bonni A. Cdh1-APC controls axonal growth and patterning in the mammalian brain. Science. 2004;303:1026–1030. doi: 10.1126/science.1093712. [DOI] [PubMed] [Google Scholar]
- Kunde SA, Musante L, Grimme A, Fischer U, Müller E, Wanker EE, Kalscheuer VM. The X-chromosome-linked intellectual disability protein PQBP1 is a component of neuronal RNA granules and regulates the appearance of stress granules. Hum Mol Genet. 2011;20:4916–4931. doi: 10.1093/hmg/ddr430. [DOI] [PubMed] [Google Scholar]
- Lancaster MA, Gleeson JG. The primary cilium as a cellular signaling center: lessons from disease. Curr Opin Genet Dev. 2009;19:220–229. doi: 10.1016/j.gde.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson SA, Lakin KC, Anderson L, Kwak N, Lee JH, Anderson D. Prevalence of mental retardation and developmental disabilities: estimates from the 1994/1995 National Health Interview Survey Disability Supplements. Am J Ment Retard. 2001;106:231–252. [PubMed] [Google Scholar]
- Lee JE, Gleeson JG. Cilia in the nervous system: linking cilia function and neurodevelopmental disorders. Curr Opin Neurol. 2011;24:98–105. doi: 10.1097/WCO.0b013e3283444d05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JP, Zeitlin SO. The long and the short of aberrant ciliogenesis in Huntington disease. J Clin Invest. 2011;121:4237–4241. doi: 10.1172/JCI60243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Tan G, Levenkova N, Li T, Pugh EN, Rux JJ, Speicher DW, Pierce EA. The proteome of the mouse photoreceptor sensory cilium complex. Mol Cell Proteomics. 2007;6:1299–1317. doi: 10.1074/mcp.M700054-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorian M, Beullens M, Lesage B, Nicolaescu E, Beke L, Landuyt W, Ortiz JM, Bollen M. Nucleocytoplasmic shuttling of the splicing factor SIPP1. J Biol Chem. 2005;280:38862–38869. doi: 10.1074/jbc.M509185200. [DOI] [PubMed] [Google Scholar]
- Louvi A, Grove EA. Cilia in the CNS: the quiet organelle claims center stage. Neuron. 2011;69:1046–1060. doi: 10.1016/j.neuron.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubs H, Abidi FE, Echeverri R, Holloway L, Meindl A, Stevenson RE, Schwartz CE. Golabi-Ito-Hall syndrome results from a missense mutation in the WW domain of the PQBP1 gene. J Med Genet . 2006;43:e30. doi: 10.1136/jmg.2005.037556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marley A, von Zastrow M. DISC1 regulates primary cilia that display specific dopamine receptors. PLoS One. 2010;5:e10902. doi: 10.1371/journal.pone.0010902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Cepko CL. Controlled expression of transgenes introduced by in vivo electroporation. Proc Natl Acad Sci U S A. 2007;104:1027–1032. doi: 10.1073/pnas.0610155104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy VN, De Camilli P. Cell biology of the presynaptic terminal. Annu Rev Neurosci. 2003;26:701–728. doi: 10.1146/annurev.neuro.26.041002.131445. [DOI] [PubMed] [Google Scholar]
- Musante L, Kunde SA, Sulistio TO, Fischer U, Grimme A, Frints SG, Schwartz CE, Martínez F, Romano C, Ropers HH, et al. Common pathological mutations in PQBP1 induce nonsense-mediated mRNA decay and enhance exclusion of the mutant exon. Hum Mutat. 2010;31:90–98. doi: 10.1002/humu.21146. [DOI] [PubMed] [Google Scholar]
- Mykytyn K, Mullins RF, Andrews M, Chiang AP, Swiderski RE, Yang B, Braun T, Casavant T, Stone EM, Sheffield VC. Bardet-Biedl syndrome type 4 (BBS4)-null mice implicate Bbs4 in flagella formation but not global cilia assembly. Proc Natl Acad Sci U S A. 2004;101:8664–8669. doi: 10.1073/pnas.0402354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peränen J, Merdes A, Slusarski DC, Scheller RH, Bazan JF, Sheffield VC, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- Nachury MV, Seeley ES, Jin H. Trafficking to the ciliary membrane: how to get across the periciliary diffusion barrier? Annu Rev Cell Dev Biol. 2010;26:59–87. doi: 10.1146/annurev.cellbio.042308.113337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazawa H, Rich T, Chang A, Lin X, Waragai M, Kajikawa M, Enokido Y, Komuro A, Kato S, Shibata M, et al. Interaction between mutant ataxin-1 and PQBP-1 affects transcription and cell death. Neuron. 2002;34:701–713. doi: 10.1016/s0896-6273(02)00697-9. [DOI] [PubMed] [Google Scholar]
- Okuda T, Hattori H, Takeuchi S, Shimizu J, Ueda H, Palvimo JJ, Kanazawa I, Kawano H, Nakagawa M, Okazawa H. PQBP-1 transgenic mice show a late-onset motor neuron disease-like phenotype. Hum Mol Genet. 2003;12:711–725. doi: 10.1093/hmg/ddg084. [DOI] [PubMed] [Google Scholar]
- Pedersen LB, Veland IR, Schrøder JM, Christensen ST. Assembly of primary cilia. Dev Dyn. 2008;237:1993–2006. doi: 10.1002/dvdy.21521. [DOI] [PubMed] [Google Scholar]
- Puram SV, Kim AH, Ikeuchi Y, Wilson-Grady JT, Merdes A, Gygi SP, Bonni A. A CaMKII β signaling pathway at the centrosome regulates dendrite patterning in the brain. Nat Neurosci. 2011;14:973–983. doi: 10.1038/nn.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purpura DP. Dendritic spine "dysgenesis" and mental retardation. Science. 1974;186:1126–1128. doi: 10.1126/science.186.4169.1126. [DOI] [PubMed] [Google Scholar]
- Quan A, Robinson PJ. Rapid purification of native dynamin I and colorimetric GTPase assay. Methods Enzymol. 2005;404:556–569. doi: 10.1016/S0076-6879(05)04049-8. [DOI] [PubMed] [Google Scholar]
- Quintyne NJ, Gill SR, Eckley DM, Crego CL, Compton DA, Schroer TA. Dynactin is required for microtubule anchoring at centrosomes. J Cell Biol. 1999;147:321–334. doi: 10.1083/jcb.147.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejeb I, Ben Jemaa L, Abaied L, Kraoua L, Saillour Y, Maazoul F, Chelly J, Chaabouni H. A novel frame shift mutation in the PQBP1 gene identified in a Tunisian family with X-linked mental retardation. Eur J Med Genet. 2011 doi: 10.1016/j.ejmg.2011.01.010. [DOI] [PubMed] [Google Scholar]
- Ropers HH. Genetics of early onset cognitive impairment. Annu Rev Genomics Hum Genet. 2010;11:161–187. doi: 10.1146/annurev-genom-082509-141640. [DOI] [PubMed] [Google Scholar]
- Saito T. Embryonic in vivo electroporation in the mouse. Methods Enzymol. 2010;477:37–50. doi: 10.1016/S0076-6879(10)77003-8. [DOI] [PubMed] [Google Scholar]
- Schmid SL, Frolov VA. Dynamin: Functional Design of a Membrane Fission Catalyst. Annu Rev Cell Dev Biol. 2011 doi: 10.1146/annurev-cellbio-100109-104016. [DOI] [PubMed] [Google Scholar]
- Sheen VL, Torres AR, Du X, Barry B, Walsh CA, Kimonis VE. Mutation in PQBP1 is associated with periventricular heterotopia. Am J Med Genet A. 2010;152A:2888–2890. doi: 10.1002/ajmg.a.33507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla V, Reiter JF. The primary cilium as the cell's antenna: signaling at a sensory organelle. Science. 2006;313:629–633. doi: 10.1126/science.1124534. [DOI] [PubMed] [Google Scholar]
- Stevenson RE, Bennett CW, Abidi F, Kleefstra T, Porteous M, Simensen RJ, Lubs HA, Hamel BC, Schwartz CE. Renpenning syndrome comes into focus. Am J Med Genet A. 2005;134:415–421. doi: 10.1002/ajmg.a.30664. [DOI] [PubMed] [Google Scholar]
- Tapia VE, Nicolaescu E, McDonald CB, Musi V, Oka T, Inayoshi Y, Satteson AC, Mazack V, Humbert J, Gaffney CJ, et al. Y65C missense mutation in the WW domain of the Golabi-Ito-Hall syndrome protein PQBP1 affects its binding activity and deregulates pre-mRNA splicing. J Biol Chem. 2010;285:19391–19401. doi: 10.1074/jbc.M109.084525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson HM, Cao H, Chen J, Euteneuer U, McNiven MA. Dynamin 2 binds gamma-tubulin and participates in centrosome cohesion. Nat Cell Biol. 2004;6:335–342. doi: 10.1038/ncb1112. [DOI] [PubMed] [Google Scholar]
- Wang Z, Phan T, Storm DR. The type 3 adenylyl cyclase is required for novel object learning and extinction of contextual memory: role of cAMP signaling in primary cilia. J Neurosci. 2011;31:5557–5561. doi: 10.1523/JNEUROSCI.6561-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waragai M, Lammers CH, Takeuchi S, Imafuku I, Udagawa Y, Kanazawa I, Kawabata M, Mouradian MM, Okazawa H. PQBP-1, a novel polyglutamine tract-binding protein, inhibits transcription activation by Brn-2 and affects cell survival. Hum Mol Genet. 1999;8:977–987. doi: 10.1093/hmg/8.6.977. [DOI] [PubMed] [Google Scholar]
- Zaghloul NA, Katsanis N. Mechanistic insights into Bardet-Biedl syndrome, a model ciliopathy. J Clin Invest. 2009;119:428–437. doi: 10.1172/JCI37041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lindblom T, Chang A, Sudol M, Sluder AE, Golemis EA. Evidence that dim1 associates with proteins involved in pre-mRNA splicing, and delineation of residues essential for dim1 interactions with hnRNP F and Npw38/PQBP-1. Gene. 2000;257:33–43. doi: 10.1016/s0378-1119(00)00372-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.