Synopsis
Proteomics is fulfilling its potential and beginning to impact the diagnosis and therapy of cardiovascular disease. The field continues to develop, taking on new roles in both de novo discovery and targeted approaches. As de novo discovery – using mass spectrometry alone, or in combination with peptide or protein separation techniques – becomes a reality, more and more attention is being directed toward the field of cardiovascular serum/plasma biomarker discovery. With the advent of quantitative mass spectrometry, this focus is shifting from the basic accumulation of protein identifications within a sample to the elucidation of complex protein interactions. Despite technical advances, however, the absolute number of biomarkers thus far discovered by proteomics’ systems biology approach is small. Although several factors contribute to this lack, one step we must take is to build “translation teams” involving a close collaboration between researchers and clinicians.
Proteomics provides a snapshot of the proteome of a sample (or a subfraction/subproteome) at any given point in time. Any change in function is preceded by a change on the protein level. As this can be induced by the slightest alteration in the microenvironment of the protein (e.g. fluctuation in pH), the strength of proteomics to detect these changes is at the same time the weakness of the method in the unstable context of a clinical setting. In order to take cardiovascular proteomics from bench to bedside, great care must be taken to achieve reproducible results.
Introduction
Cardiovascular proteomics is a rapidly evolving field within life sciences. During the last decade, proteomics has been driven by the advent of new techniques in mass spectrometry (MS), and advances in classic techniques (such as 1- and 2-dimensional protein separation gel electrophoresis and liquid chromatography).
Proteomics aims to identify and characterize the set of proteins present at a certain point in time. Unlike the mere identification of a gene, proteomics is extremely complex, requiring many technologies and strategies. A majority of genes, once they are expressed, undergo extensive posttranslational modification (PTM); this results in different functional gene products arising from the same gene. So far, there are 560 types of PTMs that can be detected by MS alone, with oxidation and phosphorylation being the most common ones. The number of possible protein variants from a single gene further increases when all splice variants are taken into account. A functional change is typically preceded by a change on the protein level, and this can be induced by the slightest fluctuation in the microenvironment of the protein (e.g. change in pH). The strength of proteomics to detect these sometimes minute changes, is at the same time the weakness of the method when translated to the “real-world” context of a clinical setting. This review addresses the safeguards and pitfalls involved in taking proteomics from bench to bedside.
Manuscript
Most clinicians tend to underestimate the efforts that are necessary to conduct a proteomics study. Proteomics is often presented as, per se, a high-throughput single method, which (although it employs high-throughput techniques) it is not. In fact, the goal of proteomics is not speed but rather breadth of observation. Proteomics can simultaneously investigate tens, hundreds, even thousands of proteins. Because proteomics techniques are able to detect even the slightest changes in protein conformation (e.g. posttranslational modifications), the methods themselves are sensitive to very slight modifications, and extreme care must be taken to achieve reproducible results. Over the last several years, plasma and serum have become the most common sample types for routine clinical use, because they are quite easy to obtain and provide a good reference for systemic processes.
One challenge with serum and plasma is the sheer depth of the proteome. The human genome project has identified only ~30,000 genes; but human plasma contains about 106 different molecules, and the concentrations of proteins in the plasma span 9 magnitudes (from < 1 picogram/ml up to 50 mg/ml). Currently, there is no proteomics technique with a dynamic range that even comes close to covering these differences in protein abundance. Even after removing the 10 most abundant proteins (which account for 90% of the total protein content in the blood), analysis of the mid- and low-abundance proteins is feasible but challenging (1). While this makes proteomics seem, on first view, like a less-than-ideal candidate for clinical applications, this strategy has been incorporated into many biomarker studies based on the assumption that potentially interesting biomarkers are still hidden from standard proteomic techniques.
Biomarker discovery is conducted along two lines: one is the systems biology approach, driven by identifying all protein differences between disease and control; the other is a more targeted approach. While several authors argue for one or the other, most researchers combine the two approaches (2). The known biology of the disease informs choices as to the sample type, the collection method, and the proteomic techniques to be used. After the initial phase of discovery work, candidate markers are evaluated on the basis of the underlying biology, insofar as it is known. Proteins with preexisting availability of antibodies and enzyme-linked immunosorbent assay (ELISA) kits are often preferentially selected for validation. In fact, the need for highly specific antibodies presents a considerable and unfortunate bias in biomarker discovery, since this should not be the default selection criterion. Tools that can develop robust ELISAs in a timely fashion, as well as MS-based methods that allow for absolute quantification of proteins, are required. Indeed, there is considerable ongoing effort to overcome some of the difficulties in this area.
De novo discovery
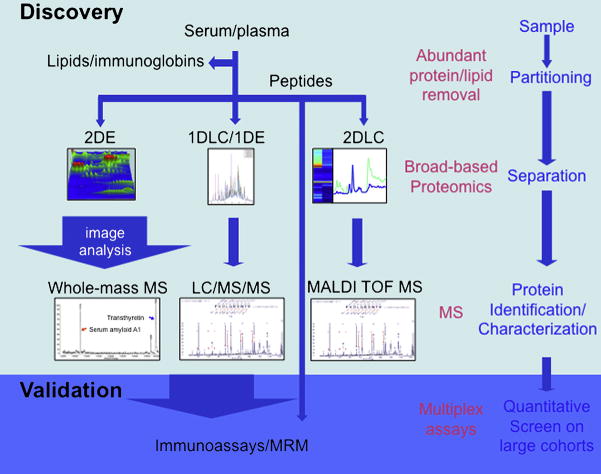
Biomarker discovery (3, 4, 5, 6) uses one or more proteomics techniques to detect as many changes as possible between the proteomes of the disease and the control (7, 8). In order to achieve this, the essence of biomarker discovery methodology is to investigate two samples that differ only in one characteristic – the disease. Sample processing (Fig. 1) differs according to the nature of the sample (serum, plasma, cells, or tissue) and other case-specific factors, such as the biology of the disease (9). In general, samples will undergo analysis employing a gel-based or gel-free approach either using proteins or peptides methods.
Fig. 1.
Workflow in biomarker discovery and validation using proteomics techniques.
In a gel-based approach, the proteins are separated in the first dimension by their isoelectric point (IEF), and then in the second dimension according to their mass within a polyacrylamide gel (SDS-PAGE). (Today, most research groups use strips with fixed pH-gradients (IPG); these have been a tremendous improvement over ampholyte-based gradients, which are prone to instability over time. IPG strips are available in several pH-ranges, so that a very high resolution can be achieved if the isoelectric point of the proteins in question is known in advance.) At this point, the spots on the gel (each containing several proteins) represent all of the proteins in the sample. Depending on their intended utilization, the gels are now stained with silver, Coomassie Blue, or more elaborate techniques like fluorescent dyes. Computer-assisted analysis of the gels identifies spots containing proteins of interest; these can then be excised, digested using trypsin, and subjected to further analysis by MS. Although there are limits to the dynamic range of the method, its ability to detect posttranslational modifications makes it worthwhile. A gel-free approach could accomplish separation of the sample’s proteins by liquid chromatography coupled with tandem mass spectrometry.
Once an individual combination of peptides is identified, the result is cross-referenced with protein databases to identify the protein in question. Because the overlap between gel-based and gel-free methods is only ~40%, combining protein separation methods enhances the breadth of protein coverage.
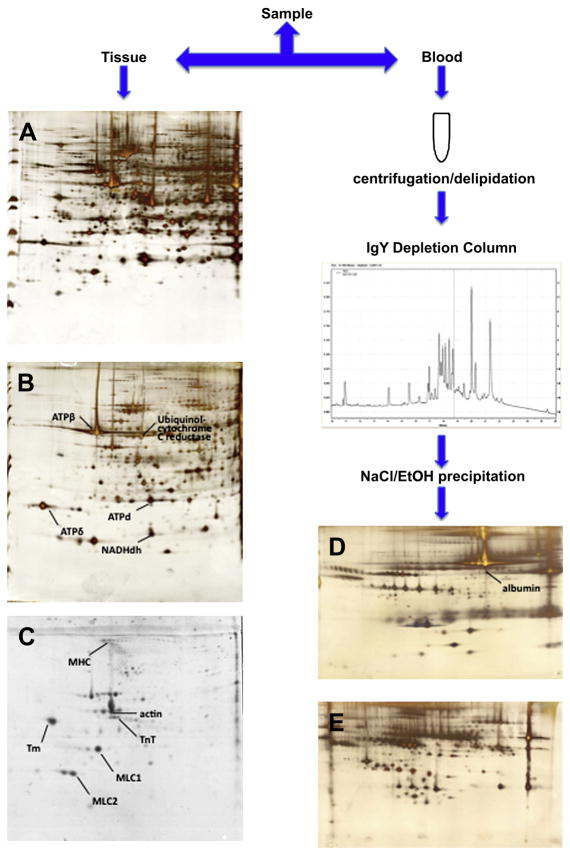
Regardless of the peptide or protein approach, and in order to enhance the depth of coverage in proteomes isolated from cells, samples are fractionated into subproteomes (10), by a process such as enrichment of mitochondrial or myofilament fractions. Depletion of samples from the most abundant proteins can increase the resolution of the subsequent proteomics technique. Several protocols have been developed to deplete serum and plasma samples from albumin and immunoglobulins, by either chemical means (using sodium chloride and ethanol) or affinity-based methods (Fig. 2). But recent research on the “albuminome” raises questions about this methodology, showing that several important proteins are actually bound to albumin and will be removed with it from the sample. Furthermore, the “albuminome” has been linked with cardiovascular disease, suggesting that investigation of albumin samples may be fruitful in and of itself.
Fig. 2.
Removing the top abundant proteins from the plasma sample is an essential step in enhancing sensitivity and depth of coverage of the downstream techniques. In tissue samples, this can be achieved by enriching the sample for certain subproteomes. Panel (A) shows a silver stained 2D-PAGE image of homogenized cardiac tissue; (B) depicts a silver stained 2D-PAGE image of mitochondria enriched fractions; and (C) shows myofilament-enriched fractions in blue-silver staining (Courtesy of G. Agnetti, Johns Hopkins University). Panel (D) shows a 2D-PAGE image of albumin-associated retenate after depletion of human plasma; whereas (E) shows the protein separation of the depleted plasma itself.
The importance of pre-analytical issues in proteomics is often underestimated; such issues in fact play a major role in successful proteomics studies, especially when dealing with complex matrices such as plasma or serum. The two keys to reducing pre-analytical variation are consistency and thorough knowledge of the underlying biology. Samples must be harvested under precisely the same conditions, controlling for: time point (circadian rhythm); pH; temperature; posture (supine vs. upright); and equipment (e.g. syringes, needles, and tubes). Processing must follow a similarly strict protocol. The purpose of the analysis, and the characteristics of the potential analytes, should be kept in mind when selecting the sample type. For example, serum is easy to obtain, but due to ex vivo processes (e.g. formation of additional peptides, or binding of proteins to the blood clot) it might not be ideal for proteomics-based biomarker discovery; in such cases, plasma should be preferred (11). The main anticoagulants used in the preparation of plasma are citrate, EDTA, and heparin, but each of these additives limits the usability of the sample. Citrate binds Ca2+, and has been shown to interfere with immunoassays (12); and EDTA can preclude the analysis of enzymes such as matrix metalloproteinases (MMPs), because the chelating properties inhibit the co-factors of the enzyme. In 2005, the HUPO Plasma Proteome Project published a comprehensive study (13) on sample protocols and varying experimental conditions. An overview of the pre-analytical factors found to influence proteomics-based biomarker discovery is given in Table 1. As stated, the underlying biology (as exemplified by the variance in TGF-beta levels when using polystyrene instead of polypropylene plates, due to the interaction of the protein with different surfaces) also plays a major role in analysis and technique.
Table 1.
Pre-analytical factors influencing proteomics-based techniques. Adapted from Rai 2005.
Patient information
|
For use in a clinical setting, it is very important to plan and implement a protocol that will be feasible in the field, and yet will yield scientifically meaningful results. Table 2 depicts a protocol (for collecting samples for proteomics analysis) that has been successfully implemented in the cardiac surgery operating rooms of our institution. Although there is no broad evidence regarding the optimal storage temperatures for proteomic samples, we strongly recommend storage at −80°C. Because repeated freeze/thaw cycles have a detectable and deleterious effect on the protein content of the samples, appropriate aliquoting of the initial sample should be one of the first steps in consistent processing of samples. Access to liquid nitrogen and sophisticated, cooled centrifuges may constitute a particular problem in routine clinical workflows. Therefore, initial storage of the centrifuged sample at −80°C, followed by aliquoting and snap-freezing in liquid nitrogen in a lab environment, presents a compromise that requires only minimal time in the operating room while effectively preserving sample quality for further analysis. It should be noted that even approved, standard laboratory operating procedures (National Committee for Clinical Laboratory Standards (NCCLS), for instance) are sometimes not sufficient for proteomics analysis. A popular example is the family of the transforming growth factors (TGF-beta), which have recently received attention for their involvement in several key biological processes (14). TGF-beta is released ex vivo from platelets, and samples must therefore be platelet-free to ensure that TGF-beta levels are not overestimated. One could argue that if a biomarker is not robust enough to “survive” small deviations from a protocol, it is inherently not useful for the clinical routine. While this might hold true for the strict validation part of a proteomics study, it would seriously hamper any attempts at biomarker discovery using a proteomics approach.
Table 2.
Sample protocol successfully implemented in a clinical environment.
|
Validation/verification or targeted discovery
After identification of candidate markers during the discovery stage, the proteins are forwarded to validation or verification. Validation is performed to confirm that protein identification is consistent with the way the protein is actually represented in the sample. Validation of MS-based protein identifications can be misleading, because antibody-based validation techniques sometimes exhibit difficulty in correctly recognizing isoforms of PTMs. When there are no antibodies or recombinant proteins available, the researcher has to judge for himself if the candidate marker justifies the cost and labor required to develop new and specific high-affinity antibodies. For this reason, the future of candidate marker validation might be in techniques that allow for antibody-independent quantitation.
Once a marker has been validated in the animal model or in pooled samples of the initial patient population, the marker has to be evaluated in a larger population with known and well-characterized phenotypes. This evaluation stage also collects information regarding the potential marker’s stability, detectability and release/clearance kinetics. Although many people are on the hunt for the “one and only” biomarker, and proteomics certainly has the potential to discover this marker, there are few examples of important, widely applied biomarkers that are the result of proteomic discovery. Most biomarkers that are broadly applied in clinical routines – like cardiac Troponin I (cTnI) or T (cTnT) (demonstrating diagnostic, therapeutic, and prognostic implications in cardiovascular disease) – have matured over years, and are the product of a specific biological hypothesis.
The classic literature differentiates among three standard types of biomarker: type 0 (natural history marker), type 1 (drug activity), and type 2 (surrogate marker). Recent authors tend to use more functional designations: biomarkers that describe the risk of getting the disease (antecedent markers); screening biomarkers; biomarkers used to diagnose the disease; biomarkers that describe the levels of disease (staging); and prognostic markers. Prognostic markers include those that make a prediction regarding the natural course of the disease, but they may also predict response to therapy, recurrence rate, and efficacy of a therapy (15,16,17).
Once a candidate marker has undergone validation within the initial patient population from which it was generated (and, ideally, including the same samples), it must then be evaluated across a broader patient population. A common error is to move instead to an immediate comparison with a group of healthy controls. This is likely to yield highly significant results, but there is a substantial risk that identifications will be for very general markers of disease, such as an inflammatory process. The next step is to define normal and abnormal values; the most common method is to measure average values in the population, and then use the 95th, 97.5th or 99th percentile as a cut-off. Care has to be taken, though, as these discrimination limits will in many cases define decision thresholds. Because many cases demonstrate a significant overlap between the general population and the patient population, it may be useful to establish longitudinal control values, meaning that each patient serves as his own control.
An important factor when judging biomarkers is considering, beyond sensitivity and specificity, the positive (or negative) predictive value of a marker. For example, while C-reactive protein (CRP) might not be a very specific marker, unable to differentiate between pneumonia and rheumatoid arthritis, it does, if the patient is already diagnosed with pneumonia, provide valuable information about the progress of the disease and the response to therapy.
Whereas most clinicians tend to favor single biomarkers, and design their trials accordingly, researchers in the proteomics field largely believe that the future of biomarkers will be in biomarker panels. Strictly speaking, the panel philosophy is not new. Every clinician would accept the fact that measuring creatinine in an oliguric patient does not yield sufficient information for an accurate diagnosis. Nevertheless, no clinician would exchange this marker for another one; he would rather order additional tests (and in so doing, effectively create his own biomarker panel). Predefined multi-marker panels can enhance sensitivity and specificity compared to each marker alone. As an example, CRP and LDL are both markers for cardiovascular risk; but when combined, they consistently demonstrate higher predictive power than either by itself.
Multiplex assays represent a promising new strategy in cardiovascular diagnostics. As de novo proteomics analysis gets more and more streamlined, and robust high-throughput methods are developed, validation becomes the bottleneck for biomarker development. Multiplex assays allow the simultaneous assessment of several biomarkers within the same sample, greatly enhancing both the standardization and the accuracy of the validation. A successful multiplex assay requires less sample quantity, and is far more cost-effective, than a monoplex assay. But this approach only gains ground slowly in the clinical lab setting, because there are still obstacles to putting a set of clinically important biomarkers in a single multiplex assay. Recently, however, an alternative way of performing multiplex assays has arisen. Multiple reactions monitoring (MRM) is a long-standing MS-based technique that has been used in the pharmaceutical industry for drug development and other small-molecule applications. MRM utilization was recently introduced for peptides in complex mixtures (such as plasma), with the possibility to perform quantitative, antibody-independent multiplexing where the abundance of peptides derived from a parent peptide is compared to a known amount of spiked-in peptide (18). Although this is a very promising technique, antibody-based methods, such as sandwich assays, are currently the most common.
Creation of a meaningful assay, of course, requires a common theme – whether it is related to a certain pathological condition (e.g. heart failure) or a particular class of proteins (e.g. cytokines). Because the plasma levels of secreted proteins span up to 9 magnitudes, analytes must be selected based on their concentration. Most multiplex assays, for instance, cannot deal with the >100-fold average difference expected between the analytes of healthy individuals and patients. After a set of potential analytes for multiplexing has been identified, each analyte must be optimized within a monoplex to establish a baseline. A crucial role is played by the diluent used to dilute the samples and antibodies. While assays might perform very well with bovine serum albumin as a standard diluent, they might produce completely different results when a diluent resembling the more complex nature of the human serum is used. Contemporary multiplexes generally employ one of two different strategies: a sandwich immunoassay, where the analyte is bound to a capture antibody printed onto a surface; or a single-antibody, direct-labeling assay. Multiplex platforms such as Bio-Plex (Bio-Rad Laboratories, Hercules, CA) use a detection antibody-labeled bead population, analyzed with a modified flow cytometer. Although this is an established platform, bead-based assays seem to require more hardware maintenance compared to other systems, and may be somewhat less ideal in a non-clinical setting, where the hardware is not used on a daily basis and quality checks are not rigorously performed. We prefer a platform with robust hardware that requires minimal maintenance, such as the Ruthenium-based multiplex platform Sector Imager (Mesoscale Discovery, Gaithersburg, MD). It is important to note that research and clinical applications may place different demands on assay platforms.
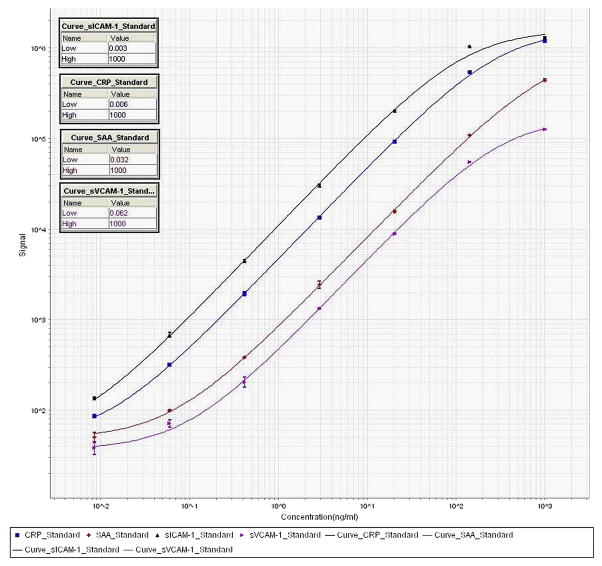
Reproducibility is critically important when deploying multiplex platforms in a clinical setting, even if they are not for direct diagnostic use. In our hands, we consider results valid when: 1) the recovery of the standards/calibrators is 100±20% (recovery = expected concentration divided by calculated concentration multiplied by 100); 2) the coefficient of variation is <20% (CV = standard deviation divided by the average of duplicates multiplied by 100); 3) intra-assay CV is <10%; and 4) interassay CV is <20%. A full run is considered valid when >85% of samples lie within these result specifications. At least for research use, transparency in any data analysis performed according to manufacturers’ algorithms (e.g. as a part of instrument operation) is imperative (Fig. 3). Otherwise, parameters like LLOD (lowest level of detection) and LLOQ (lowest level of quantification) cannot be judged objectively. When results from different multiplex platforms have to be compared, we recommend exporting the raw data and analyzing it using commercially available software; this removes any differential between manufacturers’ calculation parameters, and ensures comparable results across the board.
Fig. 3.
Standard curves of a 4-plex vascular injury kit (Mesoscale Discovery, Gaithersburg, MD), showing the sigmoidal dose-response curve typical of immunoassays.
The selection criteria used for candidate biomarkers can include such factors as the diagnostic window requirements, the extent of maximum change between control and disease population, overlap with other diseases, or lack of change due to age or drugs. Some of the most promising biomarkers are present exclusively or extensively in diseased patients: Cardiac Troponin I and T, used for diagnosis of myocardial infarction, are great examples of such markers. These isoforms are highly abundant proteins in cardiac muscle, but are only present in minuscule quantities in the blood of patients without myocardial necrosis (19,20,21). This situation led researchers to screen whole tissue samples (e.g. heart) for candidate marker identification, and then move to easily-accessible sample types (like serum/plasma or urine) for detection. This approach is appropriate for diseases where necrosis or cell leakage is a major factor. In other cases, however, protein biomarkers must arise from cellular secreted proteins or from proteins present on the cell membrane. B-natriuretic peptide (as a biomarker for heart failure) is an example of a secreted protein: it is present in healthy individuals, but the change in biomarker plasma levels over time, within the individual patient, is more important than the absolute value at any given time.
Cardiovascular disease is a systemic condition, and most patients do have other associated diseases, such as diabetes. Thus, multiple disease markers, which are specific to different diseases, may be present in blood, confounding de novo discovery. If, in the search for biomarkers, researchers simply compare samples (e.g. between patients with cardiovascular disease and healthy controls), any discovered marker could easily be for diabetes and not for cardiovascular disease. This is further complicated by the large numbers of initially promising biomarkers that turn out to be very general markers of disease, demonstrating only systemic, instead of disease-specific, involvement. Recent research estimates that only a small percentage of all proteins, about 4%, are truly cell type specific (22). Therefore it might not be the mere presence of a protein that makes the cell unique, but rather the amount of the protein in question. There is, at the same time, a strong case to be made that isoforms from the same protein can be products either of different genes (e.g. fast skeletal TnI vs. cardiac TnI) or of splice variants of the same gene. Thus, disease-induced modifications of a protein, such as degradation products of cardiac TnI, may have the ability to increase tissue specificity (23,24,25).
Proteomics and clinical physician partnerships
Successful proteomics studies require a team effort, a partnership. It is only after careful, joint review of the study protocol – with both a clinician and the proteomics scientist – that a proteomics study can be successfully launched. The clinician has to understand the demands and limits of the proteomics approach, not only in terms of sample quality but also regarding resource requirements (i.e. time and money). The proteomics scientist must learn what is feasible within the clinical setting, and what disease-specific circumstances will influence the analysis. For example, a proteomics scientist engaged in a promising study in renal failure should be eager to learn that the definition of the nephrotic syndrome is a protein loss through the kidneys of >3g/day: accurate representation will require that he normalize his results to total protein content, and this will greatly influence his analysis compared to a control population. The same holds true with the analysis of blood from patients undergoing open heart surgery using cardiopulmonary bypass: the surgeon has to tell the scientist that he stopped the heart using high molar potassium solutions; that he primed the heart lung machine with exogenously supplied albumin; that he circulated the blood for two hours through plastic tubing, and counteracted the activation of the coagulation cascade with the use of aprotinin or tranexamic acid (table 3).
Table 3.
Non-gaseous emboli present in cardiopulmonary bypass circuit, potentially interfering with analysis using proteomics techniques.
|
We encourage the basic scientist, when working in the cardiovascular field, to spend a day with a cardiologist in the chest-pain unit or with a cardiac surgeon in the operating room, in order to get a feeling for how samples are won and what is really being done – physically and functionally – in the particular study. On the other side, we encourage clinicians to listen carefully to the scientist when he describes the variety of factors influencing subsequent sample analysis. All of this collaborative preparation should happen before the first sample is taken. Additionally, most researchers seek statistical advice when planning a new study. And newcomers to the complex field of biomarker discovery should similarly consult with an established scientist in order to plan and carry out research that will achieve meaningful results.
Summary
Proteomics holds tremendous promise for the diagnosis, prognosis, and treatment of cardiovascular disease, and is already changing the way we look at biomarkers of all kinds. But the field and techniques are complex and rigorous, and even in the most capable proteomics facilities in the world, more significant biomarkers are passed over than discovered every day.
Acknowledgments
Jennifer Van Eyk’s work is supported by the National Heart Lung Blood Institute Proteomic Initiative (contract N01-HV-28120), the SCCOR program (Specialized Centers of Clinically Oriented Research, grant 1 P50 HL 084946-01), and by the Daniel P. Amos Foundation. Florian Schoenhoff is supported by the Novartis Research Foundation. We thank Shandev Rai for improving the manuscript with editorial comments and suggestions.
Footnotes
No competing interests to disclose.
References
- 1.Gerszten RE, Accurso F, Bernard GR, Caprioli RM, Klee EW, Klee GG, Kullo I, Laguna TA, Roth FP, Sabatine M, Srinivas P, Wang TJ, Ware LB. Challenges in translating plasma proteomics from bench to bedside: update from the NHLBI Clinical Proteomics Programs. Am J Physiol Lung Cell Mol Physiol. 2008;295:16–22. doi: 10.1152/ajplung.00044.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malmstrom J, Lee H, Aebersold R. Advances in proteomic workflows for systems biology. Curr Opin Biotechnol. 2007;18:378–84. doi: 10.1016/j.copbio.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arrell DK, Neverova I, Van Eyk JE. Cardiovascular proteomics: evolution and potential. Circ Res. 2001;88:763–73. doi: 10.1161/hh0801.090193. [DOI] [PubMed] [Google Scholar]
- 4.Jaffe AS, Babuin L, Apple FS. Biomarkers in acute cardiac disease: the present and the future. J Am Coll Cardiol. 2006;48:1–11. doi: 10.1016/j.jacc.2006.02.056. [DOI] [PubMed] [Google Scholar]
- 5.Vivanco F, Martin-Ventura JL, Duran MC, Barderas MG, Blanco-Colio L, Darde VM, Mas S, Meilhac O, Michel JB, Tunon J, Egido J. Quest for novel cardiovascular biomarkers by proteomic analysis. J Proteome Res. 2005;4:1181–91. doi: 10.1021/pr0500197. [DOI] [PubMed] [Google Scholar]
- 6.Stanley BA, Gundry RL, Cotter RJ, Van Eyk JE. Heart disease, clinical proteomics and mass spectrometry. Dis Markers. 2004;20:167–78. doi: 10.1155/2004/965261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arab S, Gramolini AO, Ping P, Kislinger T, Stanley B, van Eyk J, Ouzounian M, MacLennan DH, Emili A, Liu PP. Cardiovascular proteomics: tools to develop novel biomarkers and potential applications. J Am Coll Cardiol. 2006;48:1733–41. doi: 10.1016/j.jacc.2006.06.063. [DOI] [PubMed] [Google Scholar]
- 8.Parikh SV, de Lemos JA. Biomarkers in cardiovascular disease: integrating pathophysiology into clinical practice. Am J Med Sci. 2006;332:186–97. doi: 10.1097/00000441-200610000-00006. [DOI] [PubMed] [Google Scholar]
- 9.Matt P, Fu Z, Fu Q, Van Eyk JE. Biomarker discovery: proteome fractionation and separation in biological samples. Physiol Genomics. 2008;33:12–7. doi: 10.1152/physiolgenomics.00282.2007. [DOI] [PubMed] [Google Scholar]
- 10.McDonald T, Sheng S, Stanley B, Chen D, Ko Y, Cole RN, Pedersen P, Van Eyk JE. Expanding the subproteome of the inner mitochondria using protein separation technologies: one- and two-dimensional liquid chromatography and two-dimensional gel electrophoresis. Mol Cell Proteomics. 2006;5:2392–411. doi: 10.1074/mcp.T500036-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Tammen H, Schulte I, Hess R, Menzel C, Kellmann M, Mohring T, Schulz-Knappe P. Peptidomic analysis of human blood specimens: comparison between plasma specimens and serum by differential peptide display. Proteomics. 2005;5:3414–22. doi: 10.1002/pmic.200401219. [DOI] [PubMed] [Google Scholar]
- 12.Tubes and Additives for Venous Blood Specimen Collection; Approved Standard. 4. NCCLS; Wayne PA: 2003. [Google Scholar]
- 13.Rai AJ, Gelfand CA, Haywood BC, Warunek DJ, Yi J, Schuchard MD, et al. HUPO Plasma Proteome Project specimen collection and handling: towards the standardization of parameters for plasma proteome samples. Proteomics. 2005;5:3262–3277. doi: 10.1002/pmic.200401245. [DOI] [PubMed] [Google Scholar]
- 14.Matt P, Habashi J, Carrel T, Cameron DE, Van Eyk JE, Dietz HC. Recent advances in understanding Marfan syndrome: should we now treat surgical patients with losartan? J Thorac Cardiovasc Surg. 2007;135:389–94. doi: 10.1016/j.jtcvs.2007.08.047. [DOI] [PubMed] [Google Scholar]
- 15.Jaffe AS, Babuin L, Apple FS. Biomarkers in acute cardiac disease: the present and the future. J Am Coll Cardiol. 2006;48:1–11. doi: 10.1016/j.jacc.2006.02.056. [DOI] [PubMed] [Google Scholar]
- 16.Vasan RS. Biomarkers of cardiovascular disease: molecular basis and practical considerations. Circulation. 2006;113:2335–62. doi: 10.1161/CIRCULATIONAHA.104.482570. [DOI] [PubMed] [Google Scholar]
- 17.Anderson L. Candidate-based proteomics in the search for biomarkers of cardiovascular disease. J Physiol. 2005;563:23–60. doi: 10.1113/jphysiol.2004.080473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5:573–88. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Apple FS, Murakami MM. Serum and plasma cardiac troponin I 99th percentile reference values for 3 2nd-generation assays. Clin Chem. 2007;53:1558–60. doi: 10.1373/clinchem.2007.087718. [DOI] [PubMed] [Google Scholar]
- 20.Apple FS, Pearce LA, Doyle PJ, Otto AP, Murakami MM. Cardiac troponin risk stratification based on 99th percentile reference cutoffs in patients with ischemic symptoms suggestive of acute coronary syndrome: influence of estimated glomerular filtration rates. Am J Clin Pathol. 2007;127:598–603. doi: 10.1309/17220CY5MK5UCQRP. [DOI] [PubMed] [Google Scholar]
- 21.Apple FS. Cardiac troponin monitoring for detection of myocardial infarction: newer generation assays are here to stay. Clin Chim Acta. 2007;380:1–3. doi: 10.1016/j.cca.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 22.Berglund L, Bjorling E, Oksvold P, Fagerberg L, Asplund A, Szigyarto CA, Persson A, Ottosson J, Wernerus H, Nilsson P, Lundberg E, Sivertsson A, Navani S, Wester K, Kampf C, Hober S, Ponten F, Uhlen M. A genecentric human protein atlas for expression profiles based on antibodies. Mol Cell Proteomics. 2008;10:2019–27. doi: 10.1074/mcp.R800013-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Bovenkamp DE, Stanley BA, Van Eyk JE. Optimization of cardiac troponin I pull-down by IDM affinity beads and SELDI. Methods Mol Biol. 2007;357:91–102. doi: 10.1385/1-59745-214-9:91. [DOI] [PubMed] [Google Scholar]
- 24.Madsen LH, Christensen G, Lund T, Serebruany VL, Granger CB, Hoen I, Grieg Z, Alexander JH, Jaffe AS, Van Eyk JE, Atar D. Time course of degradation of cardiac troponin I in patients with acute ST-elevation myocardial infarction: the ASSENT-2 troponin substudy. Circ Res. 2006;99:1141–7. doi: 10.1161/01.RES.0000249531.23654.e1. [DOI] [PubMed] [Google Scholar]
- 25.McDonough JL, Van Eyk JE. Developing the next generation of cardiac markers: disease-induced modifications of troponin I. Prog Cardiovasc Dis. 2004;47:207–16. doi: 10.1016/j.pcad.2004.07.001. [DOI] [PubMed] [Google Scholar]