Abstract
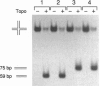
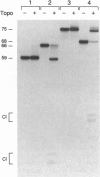
The Holliday junction, a key intermediate in both homologous and site-specific recombination, is generated by the reciprocal exchange of single strands between two DNA duplexes. Resolution of the junctions can occur in two directions with respect to flanking markers, either restoring the parental DNA configuration or generating a genetic crossover. Recombination can be regulated, in principle, by factors that influence the directionality of the resolution step. We demonstrate that the vaccinia virus DNA topoisomerase, a eukaryotic type I enzyme, catalyzes resolution of synthetic Holliday junctions in vitro. The mechanism entails concerted transesterifications at two recognition sites, 5'-CCCTT decreases, that are opposed within a partially mobile four-way junction. Cruciforms are resolved unidirectionally and with high efficiency into two linear duplexes. These findings suggest a model whereby type I topoisomerases may either promote or suppress genetic recombination in vivo.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ariyoshi M., Vassylyev D. G., Iwasaki H., Nakamura H., Shinagawa H., Morikawa K. Atomic structure of the RuvC resolvase: a holliday junction-specific endonuclease from E. coli. Cell. 1994 Sep 23;78(6):1063–1072. doi: 10.1016/0092-8674(94)90280-1. [DOI] [PubMed] [Google Scholar]
- Been M. D., Champoux J. J. DNA breakage and closure by rat liver type 1 topoisomerase: separation of the half-reactions by using a single-stranded DNA substrate. Proc Natl Acad Sci U S A. 1981 May;78(5):2883–2887. doi: 10.1073/pnas.78.5.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonven B. J., Gocke E., Westergaard O. A high affinity topoisomerase I binding sequence is clustered at DNAase I hypersensitive sites in Tetrahymena R-chromatin. Cell. 1985 Jun;41(2):541–551. doi: 10.1016/s0092-8674(85)80027-1. [DOI] [PubMed] [Google Scholar]
- Christiansen K., Bonven B. J., Westergaard O. Mapping of sequence-specific chromatin proteins by a novel method: topoisomerase I on Tetrahymena ribosomal chromatin. J Mol Biol. 1987 Feb 5;193(3):517–525. doi: 10.1016/0022-2836(87)90264-6. [DOI] [PubMed] [Google Scholar]
- Christiansen K., Westergaard O. Characterization of intra- and intermolecular DNA ligation mediated by eukaryotic topoisomerase I. Role of bipartite DNA interaction in the ligation process. J Biol Chem. 1994 Jan 7;269(1):721–729. [PubMed] [Google Scholar]
- Christman M. F., Dietrich F. S., Fink G. R. Mitotic recombination in the rDNA of S. cerevisiae is suppressed by the combined action of DNA topoisomerases I and II. Cell. 1988 Nov 4;55(3):413–425. doi: 10.1016/0092-8674(88)90027-x. [DOI] [PubMed] [Google Scholar]
- Dixon J. E., Sadowski P. D. Resolution of immobile chi structures by the FLP recombinase of 2 microns plasmid. J Mol Biol. 1994 Oct 21;243(2):199–207. doi: 10.1006/jmbi.1994.1647. [DOI] [PubMed] [Google Scholar]
- Dixon J. E., Sadowski P. D. Resolution of synthetic chi structures by the FLP site-specific recombinase. J Mol Biol. 1993 Dec 5;234(3):522–533. doi: 10.1006/jmbi.1993.1608. [DOI] [PubMed] [Google Scholar]
- Duckett D. R., Murchie A. I., Diekmann S., von Kitzing E., Kemper B., Lilley D. M. The structure of the Holliday junction, and its resolution. Cell. 1988 Oct 7;55(1):79–89. doi: 10.1016/0092-8674(88)90011-6. [DOI] [PubMed] [Google Scholar]
- Dunderdale H. J., Benson F. E., Parsons C. A., Sharples G. J., Lloyd R. G., West S. C. Formation and resolution of recombination intermediates by E. coli RecA and RuvC proteins. Nature. 1991 Dec 19;354(6354):506–510. doi: 10.1038/354506a0. [DOI] [PubMed] [Google Scholar]
- Fisher C., Parks R. J., Lauzon M. L., Evans D. H. Heteroduplex DNA formation is associated with replication and recombination in poxvirus-infected cells. Genetics. 1991 Sep;129(1):7–18. doi: 10.1093/genetics/129.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz B., Landy A. Interactions between lambda Int molecules bound to sites in the region of strand exchange are required for efficient Holliday junction resolution. J Mol Biol. 1990 Oct 20;215(4):523–535. doi: 10.1016/s0022-2836(05)80165-2. [DOI] [PubMed] [Google Scholar]
- Gangloff S., McDonald J. P., Bendixen C., Arthur L., Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol Cell Biol. 1994 Dec;14(12):8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel S. J., Johnson G. P., Perkus M. E., Davis S. W., Winslow J. P., Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990 Nov;179(1):247-66, 517-63. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- Halligan B. D., Davis J. L., Edwards K. A., Liu L. F. Intra- and intermolecular strand transfer by HeLa DNA topoisomerase I. J Biol Chem. 1982 Apr 10;257(7):3995–4000. [PubMed] [Google Scholar]
- Hanai R., Wang J. C. Protein footprinting by the combined use of reversible and irreversible lysine modifications. Proc Natl Acad Sci U S A. 1994 Dec 6;91(25):11904–11908. doi: 10.1073/pnas.91.25.11904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiom K., West S. C. Branch migration during homologous recombination: assembly of a RuvAB-Holliday junction complex in vitro. Cell. 1995 Mar 10;80(5):787–793. doi: 10.1016/0092-8674(95)90357-7. [DOI] [PubMed] [Google Scholar]
- Hoess R., Wierzbicki A., Abremski K. Isolation and characterization of intermediates in site-specific recombination. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6840–6844. doi: 10.1073/pnas.84.19.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. L., Landy A. Resolution of synthetic att-site Holliday structures by the integrase protein of bacteriophage lambda. Nature. 1984 Oct 25;311(5988):721–726. doi: 10.1038/311721a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H., Takahagi M., Shiba T., Nakata A., Shinagawa H. Escherichia coli RuvC protein is an endonuclease that resolves the Holliday structure. EMBO J. 1991 Dec;10(13):4381–4389. doi: 10.1002/j.1460-2075.1991.tb05016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kho S. H., Landy A. Dissecting the resolution reaction of lambda integrase using suicide Holliday junction substrates. EMBO J. 1994 Jun 1;13(11):2714–2724. doi: 10.1002/j.1460-2075.1994.tb06562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim R. A., Caron P. R., Wang J. C. Effects of yeast DNA topoisomerase III on telomere structure. Proc Natl Acad Sci U S A. 1995 Mar 28;92(7):2667–2671. doi: 10.1073/pnas.92.7.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim R. A., Wang J. C. A subthreshold level of DNA topoisomerases leads to the excision of yeast rDNA as extrachromosomal rings. Cell. 1989 Jun 16;57(6):975–985. doi: 10.1016/0092-8674(89)90336-x. [DOI] [PubMed] [Google Scholar]
- Kitts P. A., Nash H. A. Homology-dependent interactions in phage lambda site-specific recombination. Nature. 1987 Sep 24;329(6137):346–348. doi: 10.1038/329346a0. [DOI] [PubMed] [Google Scholar]
- Lee J., Lee J., Jayaram M. Junction mobility and resolution of Holliday structures by Flp site-specific recombinase. Testing partner compatibility during recombination. J Biol Chem. 1995 Aug 11;270(32):19086–19092. doi: 10.1074/jbc.270.32.19086. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Clegg R. M. The structure of the four-way junction in DNA. Annu Rev Biophys Biomol Struct. 1993;22:299–328. doi: 10.1146/annurev.bb.22.060193.001503. [DOI] [PubMed] [Google Scholar]
- Merchlinsky M. Resolution of poxvirus telomeres: processing of vaccinia virus concatemer junctions by conservative strand exchange. J Virol. 1990 Jul;64(7):3437–3446. doi: 10.1128/jvi.64.7.3437-3446.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morham S. G., Shuman S. Phenotypic selection and characterization of mutant alleles of a eukaryotic DNA topoisomerase I. Genes Dev. 1990 Apr;4(4):515–524. doi: 10.1101/gad.4.4.515. [DOI] [PubMed] [Google Scholar]
- Mueller J. E., Kemper B., Cunningham R. P., Kallenbach N. R., Seeman N. C. T4 endonuclease VII cleaves the crossover strands of Holliday junction analogs. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9441–9445. doi: 10.1073/pnas.85.24.9441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Düby S. E., Azaro M. A., Landy A. Swapping DNA strands and sensing homology without branch migration in lambda site-specific recombination. Curr Biol. 1995 Feb 1;5(2):139–148. doi: 10.1016/s0960-9822(95)00035-2. [DOI] [PubMed] [Google Scholar]
- Parsons C. A., Stasiak A., Bennett R. J., West S. C. Structure of a multisubunit complex that promotes DNA branch migration. Nature. 1995 Mar 23;374(6520):375–378. doi: 10.1038/374375a0. [DOI] [PubMed] [Google Scholar]
- Picksley S. M., Parsons C. A., Kemper B., West S. C. Cleavage specificity of bacteriophage T4 endonuclease VII and bacteriophage T7 endonuclease I on synthetic branch migratable Holliday junctions. J Mol Biol. 1990 Apr 20;212(4):723–735. doi: 10.1016/0022-2836(90)90233-C. [DOI] [PubMed] [Google Scholar]
- Pottmeyer S., Kemper B. T4 endonuclease VII resolves cruciform DNA with nick and counter-nick and its activity is directed by local nucleotide sequence. J Mol Biol. 1992 Feb 5;223(3):607–615. doi: 10.1016/0022-2836(92)90977-r. [DOI] [PubMed] [Google Scholar]
- Schofield M. A., Agbunag R., Michaels M. L., Miller J. H. Cloning and sequencing of Escherichia coli mutR shows its identity to topB, encoding topoisomerase III. J Bacteriol. 1992 Aug;174(15):5168–5170. doi: 10.1128/jb.174.15.5168-5170.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman N. C. De novo design of sequences for nucleic acid structural engineering. J Biomol Struct Dyn. 1990 Dec;8(3):573–581. doi: 10.1080/07391102.1990.10507829. [DOI] [PubMed] [Google Scholar]
- Seeman N. C., Kallenbach N. R. DNA branched junctions. Annu Rev Biophys Biomol Struct. 1994;23:53–86. doi: 10.1146/annurev.bb.23.060194.000413. [DOI] [PubMed] [Google Scholar]
- Sekiguchi J., Shuman S. Proteolytic footprinting of vaccinia topoisomerase bound to DNA. J Biol Chem. 1995 May 12;270(19):11636–11645. doi: 10.1074/jbc.270.19.11636. [DOI] [PubMed] [Google Scholar]
- Sekiguchi J., Shuman S. Requirements for noncovalent binding of vaccinia topoisomerase I to duplex DNA. Nucleic Acids Res. 1994 Dec 11;22(24):5360–5365. doi: 10.1093/nar/22.24.5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi J., Shuman S. Vaccinia topoisomerase binds circumferentially to DNA. J Biol Chem. 1994 Dec 16;269(50):31731–31734. [PubMed] [Google Scholar]
- Shah R., Bennett R. J., West S. C. Genetic recombination in E. coli: RuvC protein cleaves Holliday junctions at resolution hotspots in vitro. Cell. 1994 Dec 2;79(5):853–864. doi: 10.1016/0092-8674(94)90074-4. [DOI] [PubMed] [Google Scholar]
- Sharma A., Hanai R., Mondragón A. Crystal structure of the amino-terminal fragment of vaccinia virus DNA topoisomerase I at 1.6 A resolution. Structure. 1994 Aug 15;2(8):767–777. doi: 10.1016/s0969-2126(94)00077-8. [DOI] [PubMed] [Google Scholar]
- Sharples G. J., Chan S. N., Mahdi A. A., Whitby M. C., Lloyd R. G. Processing of intermediates in recombination and DNA repair: identification of a new endonuclease that specifically cleaves Holliday junctions. EMBO J. 1994 Dec 15;13(24):6133–6142. doi: 10.1002/j.1460-2075.1994.tb06960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S. DNA strand transfer reactions catalyzed by vaccinia topoisomerase I. J Biol Chem. 1992 Apr 25;267(12):8620–8627. [PubMed] [Google Scholar]
- Shuman S., Prescott J. Specific DNA cleavage and binding by vaccinia virus DNA topoisomerase I. J Biol Chem. 1990 Oct 15;265(29):17826–17836. [PubMed] [Google Scholar]
- Shuman S. Recombination mediated by vaccinia virus DNA topoisomerase I in Escherichia coli is sequence specific. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10104–10108. doi: 10.1073/pnas.88.22.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S. Site-specific interaction of vaccinia virus topoisomerase I with duplex DNA. Minimal DNA substrate for strand cleavage in vitro. J Biol Chem. 1991 Jun 15;266(17):11372–11379. [PubMed] [Google Scholar]
- Shuman S., Turner J. Site-specific interaction of vaccinia virus topoisomerase I with base and sugar moieties in duplex DNA. J Biol Chem. 1993 Sep 5;268(25):18943–18950. [PubMed] [Google Scholar]
- Shuman S. Two classes of DNA end-joining reactions catalyzed by vaccinia topoisomerase I. J Biol Chem. 1992 Aug 25;267(24):16755–16758. [PubMed] [Google Scholar]
- Shuman S. Vaccinia DNA topoisomerase I promotes illegitimate recombination in Escherichia coli. Proc Natl Acad Sci U S A. 1989 May;86(10):3489–3493. doi: 10.1073/pnas.86.10.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse-Dinh Y. C. Uncoupling of the DNA breaking and rejoining steps of Escherichia coli type I DNA topoisomerase. Demonstration of an active covalent protein-DNA complex. J Biol Chem. 1986 Aug 15;261(23):10931–10935. [PubMed] [Google Scholar]
- Voelkel-Meiman K., Keil R. L., Roeder G. S. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell. 1987 Mar 27;48(6):1071–1079. doi: 10.1016/0092-8674(87)90714-8. [DOI] [PubMed] [Google Scholar]
- Wallis J. W., Chrebet G., Brodsky G., Rolfe M., Rothstein R. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell. 1989 Jul 28;58(2):409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Caron P. R., Kim R. A. The role of DNA topoisomerases in recombination and genome stability: a double-edged sword? Cell. 1990 Aug 10;62(3):403–406. doi: 10.1016/0092-8674(90)90002-v. [DOI] [PubMed] [Google Scholar]
- Wittschieben J., Shuman S. Mutational analysis of vaccinia DNA topoisomerase defines amino acid residues essential for covalent catalysis. J Biol Chem. 1994 Nov 25;269(47):29978–29983. [PubMed] [Google Scholar]