Abstract
Breast cancer is one of the leading causes of cancer death worldwide. This study aimed to analyze the expression of centromere protein H (CENP-H) in breast cancer and to correlate it with clinicopathologic data, including patient survival. Using reverse transcription-polymerase chain reaction and Western blotting to detect the expression of CENP-H in normal mammary epithelial cells, immortalized mammary epithelial cell lines, and breast cancer cell lines, we observed that the mRNA and protein levels of CENP-H were higher in breast cancer cell lines and in immortalized mammary epithelial cells than in normal mammary epithelial cells. We next examined CENP-H expression in 307 paraffin-embedded archived samples of clinicopathologically characterized breast cancer using immunohistochemistry, and detected high CENP-H expression in 134 (43.6%) samples. Statistical analysis showed that CENP-H expression was related with clinical stage (P = 0.001), T classification (P = 0.032), N classification (P = 0.018), and Ki-67 (P < 0.001). Patients with high CENP-H expression had short overall survival. Multivariate analysis showed that CENP-H expression was an independent prognostic indicator for patient survival. Our results suggest that CENP-H protein is a valuable marker of breast cancer progression and prognosis.
Keywords: Centromere protein H (CENP-H), breast cancer, prognosis, biomarker, proliferation
Breast cancer is the most common cancer in females, and its incidence is increasing. The mean 5-year relapse-free survival rate of breast cancer is approximately 60%, but this value differs significantly across individuals[1]. Gene expression profiles have been reported to play an important role in predicting cancer progression or prognosis compared to traditional parameters such as lymph node status and tumor grade, among others[2],[3]. However, few prognostic markers have practical value. Therefore, to find reliable breast cancer molecular markers that can accurately predict outcomes for patients remains a great challenge.
Chromosomal instability, a state of frequent chromosome loss and gain during cell division[4], is a hallmark of human cancers. Like many other malignancies, most breast cancers have significantly abnormal genomic structure, including aberrant numbers of chromosomes. In breast cancer cell lines, both variable and unstable chromosome numbers were observed[5]. Reports have also shown that aneuploidy is a prognostic factor in breast cancer[6]. The gain of chromosome 20 is prevalent in patients with breast carcinoma and may serve as a valuable prognostic marker[7]. Additionally, structural genomic aberrations, including aneuploidy, frequently occur in breast cancer[8]–[10]. Thus, chromosomal instability and aneuploidy may be associated with breast cancer development and progression.
Centromere protein H (CENP-H), a component of the kinetochore that is active in centromeres of stable dicentric chromosomes [11], plays an essential role in appropriate kinetochore assembly as well as accurate chromosome segregation[12]–[14]. CENP-H is considered to be crucial for cell growth and mitotic progression[14]. It is known to be up-regulated in colorectal cancers[15]–[20]. Moreover, ectopic CENP-H expression in a diploid colorectal cancer cell line can induce chromosome missegregation and aneuploidy [15]. CENP-H is also deregulated in oral squamous cell carcinomas. Its expression in oral squamous cell carcinomas is significantly correlated with cell proliferation in malignant conditions[16]. Previously, we found that CENP-H was deregulated in several epithelium-originated carcinomas, such as esophageal carcinoma [17], nasopharyngeal carcinoma[18], non–small cell lung cancer[19], and tongue cancer[17]–[20]. We also provided evidence for a direct role of CENP-H in chromosome instability and Carcinogenesis. CENP-H expression may be a valuable prognostic marker to predict the early stages of epithelium -originated cancers. However, it is unknown whether deregulation of CENP-H is involved in breast cancer development and progression.
In this study, we aimed to investigate CENP-H expression in cell lines simulating different stages of breast cancer development and progression. More specifically, we used immunohistochemistry to investigate CENP-H expression in 307 samples of breast cancer to determine the correlation of CENP-H expression with clinicopathologic characteristics, to evaluate the role of CENP-H in breast cancer progression, and to determine its prognostic value.
Materials and Methods
Patients and tissue specimens
The present study was conducted on 307 samples of paraffin-embedded breast cancer and 30 samples of adjacent non-cancerous tissues, which were histologically and clinically diagnosed at the Sun Yat-sen University Cancer Center between March 1, 1999 and December 25, 2002. The clinical information of these samples is described in detail in Table 1. Conventional clinical features including age, clinical stage, T classification, N classification, distant metastasis, estrogen receptor, C-ErbB-2, and progesterone receptor were all available. All patients were females with an average age of 48.9 years (range, 26 to 80 years). A total of 262 patients were followed up at 6-month intervals for the first 5 years after the diagnosis and then annually in an outpatient clinic. The median follow-up time was 58 months (range, 4 to 78 months). Survival time was calculated from the date of operation to the time of death or the most recent follow-up if the patient was alive. This study was approved by an institutional ethical committee.
Table 1. Clinicopathologic characteristics of patients with breast cancer and expression of centromere protein H (CENP-H) in breast cancer.
| Characteristic | No. of cases | Percentage (%) |
| Age (years) | ||
| ≤45 | 124 | 40.4 |
| >45 | 183 | 59.6 |
| Clinical stage | ||
| I | 35 | 11.4 |
| II | 146 | 47.7 |
| III | 90 | 29.1 |
| IV | 36 | 11.8 |
| T classification | ||
| T1 | 66 | 21.9 |
| T2 | 157 | 52.0 |
| T3 | 59 | 18.0 |
| T4 | 25 | 8.1 |
| N classification | ||
| N0 | 128 | 41.8 |
| N1 | 117 | 38.2 |
| N2 | 49 | 15.7 |
| N3 | 13 | 4.3 |
| Distant metastasis | ||
| Yes | 14 | 4.6 |
| No | 293 | 95.4 |
| Ki-67 expression | ||
| Low | 196 | 63.8 |
| High | 111 | 36.2 |
| CENP-H expression | ||
| Negative (–) | 58 | 18.9 |
| Positive (+∼+++) | 249 | 81.1 |
| Low (–∼+) | 173 | 56.4 |
| High (++∼+++) | 134 | 43.6 |
| Patients with follow-up | 262 | 85.3 |
| Survival status (at follow-up) | ||
| Alive | 171 | 65.3 |
| Death because of breast cancer | 79 | 30.1 |
| Death because of other causes | 12 | 4.6 |
Cell lines
A primary culture of the normal mammary epithelial cell line BN6 was established from a biopsy of non-cancerous mammary epithelium and was cultured in complete Keratinocyte-SFM (Invitrogen, Carlsbad, California, USA). Human immortalized mammary epithelial cell line MCF10A and breast cancer cell lines MCF7, MDA-MB-435S, and MDA231 were purchased from American Type Culture Collection (ATCC). MCF10A was cultured essentially as described[21] and grown in DMEM/F12 plus 5% horse serum. MCF7 and MDA-MB-435S cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (Invitrogen). MDA231 cells were maintained in RPMI-1640 supplemented with 10% FBS and 1% penicillin-streptomycin (Invitrogen). The primary culture of mammary epithelial cells was initiated as described before[22]. Full-length CENP-H was cloned into pMSCV vector and retroviruses were generated as described [22]. The CENP-H gene was introduced into mammary epithelial cell lines by infection with retroviruses containing pMSCV-CENP-H for 48 h. Cells were selected and maintained in 0.5 µg/mL puromycin for 4 days. Western blot was performed to confirm the expression of CENP-H.
RNA extraction and real-time reverse transcription-polymerase chain reaction
Total RNA from cultured cell lines or frozen tissue biopsies were extracted using Trizol reagent (Invitrogen) according to the manufacturer's instructions. For quantitative real-time RT-PCR analysis, 0.2 µg RNA from each sample was reversely transcribed with TaqMan reverse transcription reagents and random hexamer primers (Applied Biosystems, Inc., Foster City, Calif). The primers and probes for the real-time RT-PCR were designed with Primer Express version 2.0 (Applied Biosystems, Inc.) as described previously[19]. Expression data were normalized to the geometric mean of housekeeping gene GAPDH to control the variability in expression levels and were analyzed using the 2 ΔΔCT method.
Western blotting
Cultured cells were subjected to standard Western blot analysis as described previously[18]. A polyclonal rabbit antibody against CENP-H (1:500, Bethyl Laboratories, Montgomery, TX) was used to detect CENP-H protein expression in breast cancer cell lines as well as normal and immortalized mammary epithelial cell lines. Protein levels of anti–α-tubulin mouse monoclonal antibody (1:1000, Santa Cruz, Santa Cruz, CA) were examined by reprobing the same blot and served as loading control.
Immunohistochemistry
CENP-H expression was determined by immunohistochemistry on paraffin-embedded tissue samples as described earlier[18]. Briefly, 4-µm sections were cut from paraffin-embedded tissue blocks. Paraffin sections on the silane-coated slides were deparaffinized with xylene, rehydrated through a graded ethanol series, and underwent antigen retrieval by microwave boiling in EDTA antigenic retrieval buffer. Subsequently, sections were blocked with 3% hydrogen peroxide for 5 min to quench endogenous peroxidase activity. The sections were incubated with 1% BSA to block non-specific binding and were then incubated overnight at 4°C with rabbit polyclonal anti-CENP-H (1:500, Bethyl Laboratories, Montgomery, TX) or mouse monoclonal anti-Ki-67 (1:200, Santa Cruz Biotechnology, Santa Cruz, CA) in a humidified chamber. For the negative control, normal goat serum was used to replace anti–CENP-H antibody. Counterstaining was performed using 10% Mayer's hematoxylin. The degree of CENP-H staining in the sections was observed and evaluated as described previously[18]. The proportion of CENP-H–expressing cells varied from 0 to 100%. The cells at each intensity of staining were recorded on a scale of 0 (no staining), 1 (weak staining, light yellow), 2 (moderate staining, yellowish brown), and 3 (strong staining, brown). An optimal cutoff value was identified on the basis of a measure of heterogeneity with the log-rank test statistic with respect to overall survival. Tumors with an intensity score of ≥2 and at least 50% of malignant cells with positive CENP-H staining were classified with high expression (++ to +++), and others were classified with low expression (– to +) of CENP-H antigen.
Statistical analysis
The Mann-Whitney U test was used to analyze the CENP-H expression level and clinicopathologic parameters. The Kaplan-Meier method was use to plot survival curves, and the observed differences in survival were compared by the log-rank test. The Cox proportional hazards regression model in multivariate analysis was used to analyze the significance of various clinicopathologic parameters. A P value less than 0.05 was considered statistically significant.
Results
CENP-H expression is elevated in breast cancer
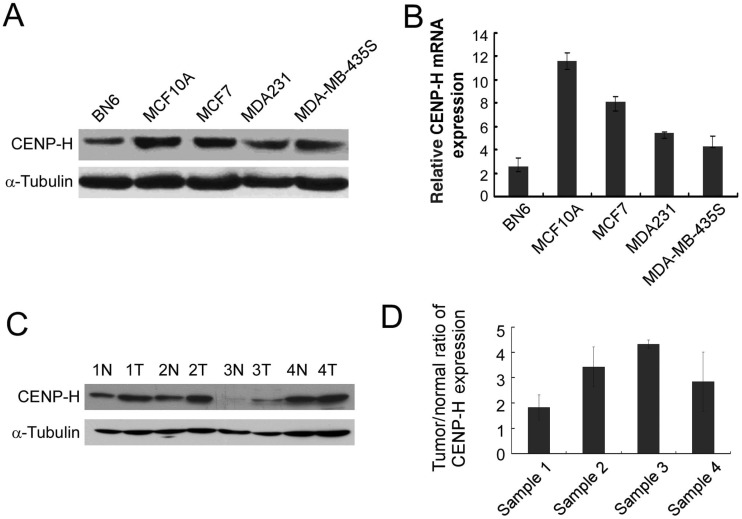
We examined CENP-H mRNA and protein expression in normal mammary epithelial cell line BN6, immortalized mammary epithelial cell line MCF10A, and breast cancer cell lines MCF7, MDA231, and MDA-MB-435S. MCF10A, MCF7, MDA231, and MDA-MB-435S cells showed higher CENP-H expression than BN6 cells (Figure 1A). To determine if CENP-H was also up-regulated at the mRNA level, real-time RT-PCR was performed. Concomitant with up-regulation of the CENP-H protein, there was 14-fold up-regulation of CENP-H mRNA in MCF10A, MCF7, MDA231, and MDA-MB-435S cells compared with that in BN6 cells (Figure 1B).
Figure 1. CENP-H expression is elevated in breast cancer.
A, CENP-H expression in normal mammary epithelial cell line BN6, immortalized mammary epithelial cell line MCF10A, and breast cancer cell lines MCF7, MDA231, and MDA-MB-435S was analyzed by Western blot. B, relative expression level of CENP-H in BN6, MCF10A, MCF7, MDA231, and MDA-MB-435S cell lines quantified by real-time RT-PCR. Columns, means from 3 parallel experiments; bars, standard deviation (SD). C, CENP-H expression in paired primary breast tumors (T) and normal breast tissues (N) from the same patients detected by Western blot. D, average tumor/normal (T/N) ratios of CENP-H expression quantified by real-time RT-PCR. Bars, SD.
To verify whether CENP-H up-regulation is clinically correlated with breast cancer development and progression, comparative analysis of CENP-H expression was conducted on 4 cases of paired primary breast cancer tissues and their adjacent non-cancerous tissues. Western blot revealed that CENP-H expression was up-regulated in all 4 human primary breast cancer tissues compared with that in the matched adjacent non-cancerous tissues (Figure 1C). Real-time RT-PCR analysis showed that the tumor/normal (T/N) ratio of CENP-H mRNA expression was as high as 35-fold in one of the 4 paired primary breast cancer tissues (Figure 1D).
CENP-H expression in archived breast cancer tissues
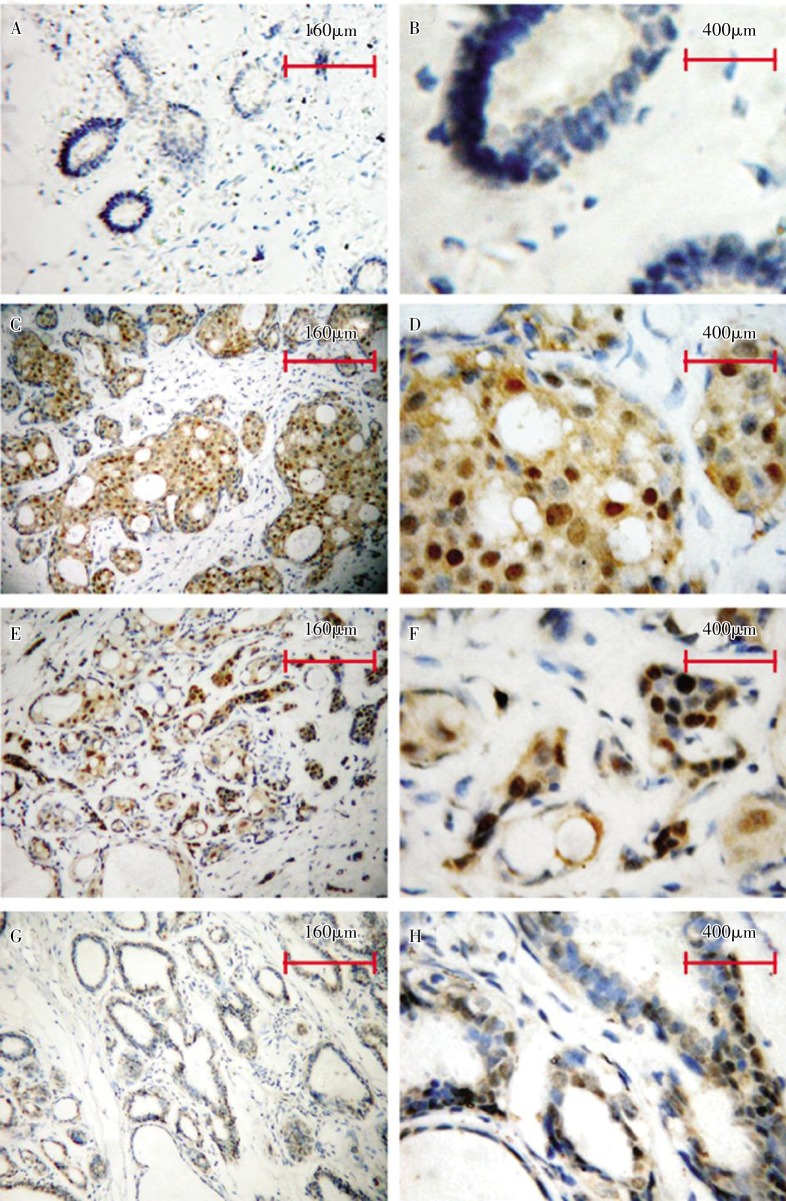
Further, to investigate CENP-H expression in breast cancer tissues and to evaluate its clinicopathologic significance, 307 cases of paraffin-embedded, archived breast cancer tissues and 30 cases of adjacent normal tissues were examined by immunohistochemistry using an antibody against human CENP-H. Only 40.0% (12/30) of adjacent normal tissues showed weak positive signals (Figure 2A and 2B). On the other hand, 81.1% (249/307) of breast cancer tissues showed weak or strong CENP-H staining, which was mainly observed in tumor epithelial cells, not in surrounding stromal cells (Figure 2C-2F). In our previous study[23], CENP-H was found to be dominantly expressed in the nuclei. Notably, CENP-H was also detected in some areas of the hyperplasia of the mammary glands (Figure 2G and 2H).
Figure 2. Representative images from immunohistochemical assays of breast cancer cases.
CENP-H shows weak or undetectable staining in normal epithelial cells (A,B), strong nuclear or nuclear/cytoplasmic staining in breast cancer with low invasive ability (C,D) and in that with high invasive ability(E, F). CENP-H shows moderate nuclear staining in the hyperplasia areas of the mammary glands(G, H).
The relationship of CENP-H up-regulation with the clinicopathologic parameters of breast cancer
The Mann-Whitney U test was used to evaluate the clinicopathologic significance of CENP-H expression in breast cancer. As shown in Table 2, CENP-H expression was related with clinical stage (P = 0.001), T classification (P = 0.032), and N classification (P = 0.018). Spearman correlation analysis revealed that CENP-H expression was positively correlated with clinical stage (r = 0.192, P = 0.001), T classification (r = 0.123, P = 0.031), and N classification (r = 0.135, P = 0.018). No significant correlation was found between CENP-H expression and age or distant metastasis.
Table 2. Correlation of centromere protein H (CENP-H) expression to clinicopathologic characteristics of patients with breast cancer.
| Characteristic | CENP-H expression [cases (%)] |
P | Spearman's r value | ||
| Low | High | ||||
| Age (years) | ≤45 | 69 (22.5) | 55 (17.9) | 0.508 | –0.006 |
| >45 | 104 (33.9) | 79 (25.8) | |||
| Clinical stage | I | 21 (39.4) | 14 (4.6) | 0.001 | 0.192 |
| II | 96 (31.3) | 50 (16.3) | |||
| III | 44 (14.3) | 46 (15.0) | |||
| IV | 12 (3.9) | 24 (7.8) | |||
| T classification | T1 | 41 (13.4) | 25 (8.1) | 0.032 | 0.123 |
| T2 | 92 (30.0) | 65 (21.2) | |||
| T3 | 33 (10.7) | 26 (8.5) | |||
| T4 | 7 (2.3) | 18 (5.9) | |||
| N classification | N0 | 80 (26.1) | 48 (15.6) | 0.018 | 0.135 |
| N1 | 66 (21.5) | 51 (16.6) | |||
| N2 | 22 (7.2) | 27 (8.8) | |||
| N3 | 5 (1.6) | 8 (2.2) | |||
| Distant metastasis | No | 168 (54.7) | 125 (40.7) | 0.112 | 0.091 |
| Yes | 5 (1.6) | 9 (2.9) | |||
| Ki-67 expression | Low | 121 (39.4) | 75 (24.4) | <0.001 | 0.241 |
| High | 40 (13.1) | 71 (23.1) | |||
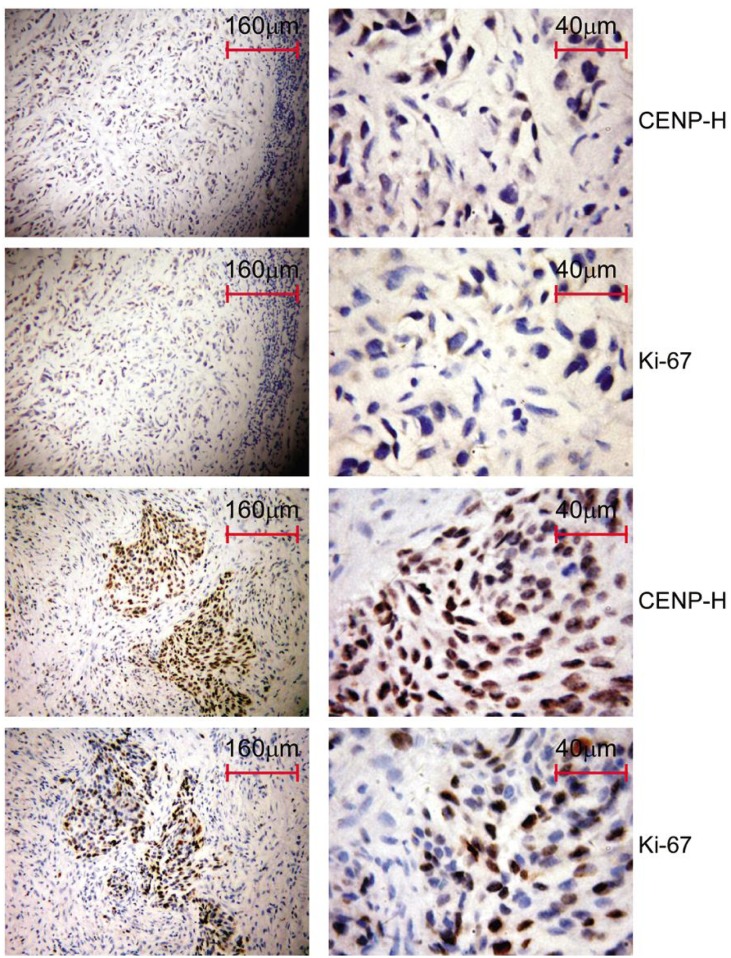
We further investigated the relationship between CENP-H expression and breast cancer proliferation using immunohistochemistry to analyze Ki-67 expression. Areas with low CENP-H expression had asteroid detectable staining for Ki-67 (Figure 3A and 3B), whereas those with high CENP-H expression also had strong Ki-67 staining signals (Figure 3C and 3D). The Mann-Whitney U test revealed that Ki-67 expression was positively correlated with CENP-H expression in breast cancer (r = 0.241, P < 0.001) (Table 2).
Figure 3. CENP-H expression in breast cancer tissues is positively correlated with Ki-67 expression.
Immunohistochemical staining of CENP-H and Ki-67 in a series of sections from paraffin blocks of breast cancer reveals weak CENP-H expression, weak Ki-67 expression, strong CENP-H expression, and strong Ki-67 expression.
Survival analysis
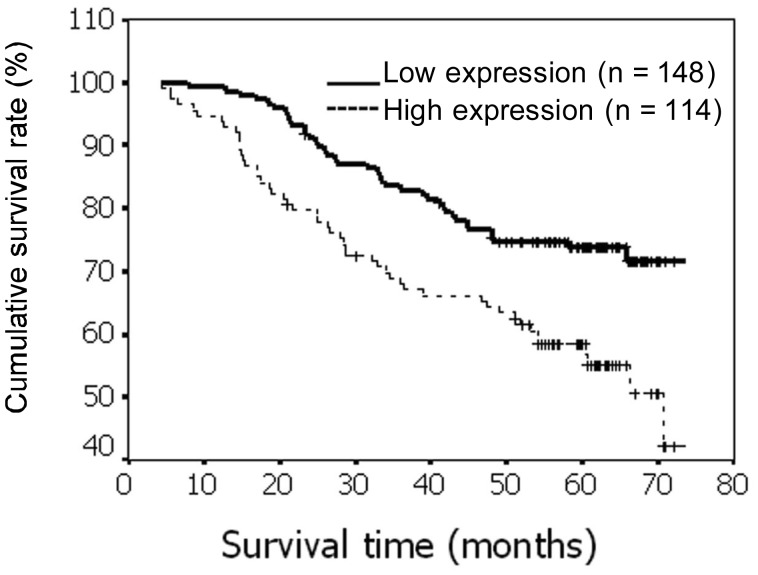
Kaplan-Meier univariate survival analysis was performed on the 262 breast cancer patients for whom follow-up data were available. A log-rank test showed that the survival time was significantly longer in the low CENP-H expression group than in the high expression group (P = 0.001) (Figure 4). The cumulative 5-year survival rate was 72.3% (95% CI, 65.1%–79.5%) in the low CENP-H protein expression group (n = 148), but it was only 51.5% (95% CI, 42.2%–60.8%) in the high expression group (n = 114). In addition to CENP-H expression, the impact of classical clinicopathologic characteristics (including clinical stage, T classification, N classification, and distant metastasis) on survival was analyzed, and these characteristics were all significantly correlated with survival in the Kaplan-Meier analysis and the log-rank test (P < 0.001). Therefore, multivariate survival analysis, including CENP-H expression level, clinical stage, T classification, N classification, and distant metastasis, was performed. This analysis indicated that T classification, N classification, distant metastasis, and CENP-H expression level were independent prognostic indicators of patient survival (Table 3).
Figure 4. Kaplan-Meier survival curves for breast cancer patients with low CENP-H expression (bold line) versus high CENP-H expression (dotted line).
The difference was compared by the log-rank test. The survival rate was significantly higher in the high expression group than in the low expression group (P = 0.0012).
Table 3. Cox univariate and multivariate analysis of different prognostic parameters in patients with breast cancer.
| Variate | No. of patients | Univariate analysis |
Multivariate analysis |
|||
| P | Regression coefficient (SE) | P | Relative risk | 95% confidence interval | ||
| T classification | <0.001 | 0.598 (0.125) | 0.001 | 1.193 | 1.193–1.969 | |
| T1–T2 | 190 | |||||
| T3–T4 | 72 | |||||
| N classification | <0.001 | 0.652 (0.110) | <0.001 | 1.354 | 1.354–2.143 | |
| N0–N1 | 208 | |||||
| N2–N3 | 54 | |||||
| Distant metastasis | <0.001 | 1.773 (0.327) | <0.001 | 4.224 | 2.167–8.235 | |
| No | 253 | |||||
| Yes | 9 | |||||
| CENP–H expression | 0.002 | 0.668 (0.212) | 0.023 | 1.635 | 1.070–2.498 | |
| Low | 148 | |||||
| High | 114 | |||||
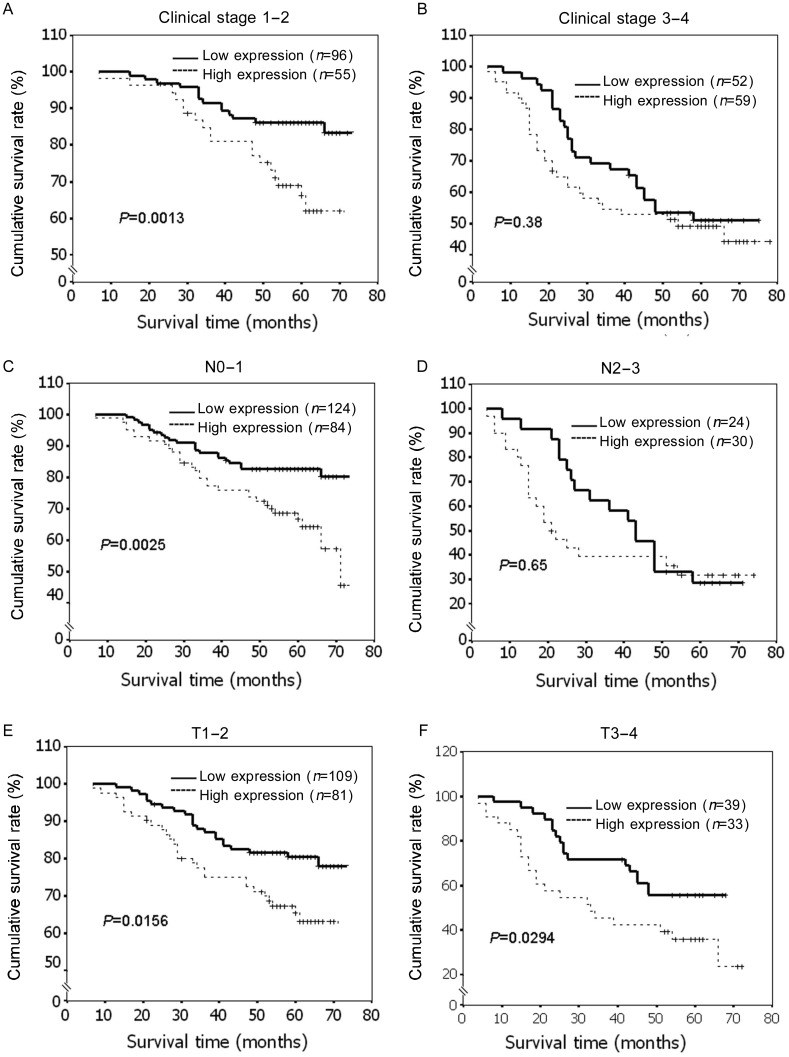
Because CENP-H was a potential prognostic marker for patients with early stage cancers in our previous study[18]–[20], we also analyzed the significance of CENP-H expression in the subgroups stratified on the basis of tumor clinical stage. We found that patients with tumors exhibiting high CENP-H expression had significantly lower overall survival rates in the subgroups of clinical stage I–II (P = 0.001) (Figure 5A), whereas there was no significant difference in survival in the subgroup of clinical stage III–IV (P = 0.38) (Figure 5B). Similarly, patients with high CENP-H expression had poorer overall survival rates in the N0–1 subgroup (P = 0.001) (Figure 5C), but not in the N2–3 subgroup (P = 0.65) (Figure 5D). However, the expression levels of CENP-H are strongly correlated with patients' survival in both T1–2 and T3–4 subgroups (Figure 5E and 5F). These results suggest that CENP-H may be a valuable prognostic marker for early-stage breast cancer patients.
Figure 5. Kaplan-Meier analysis showing the overall survival of patients categorized according to different stages/classifications and status of CENP-H expression.
The curves of patients with high and low expression of CENP-H were compared in the following patient subgroups: stage I–II and stage III–IV (A, B), T classification, N0–N1 and N2–N3 (C, D) and N classification T0–T2 and T3–4 (E, F). In subgroups of stage 1–2, N0–1, T1–2 and T3–4, the survival rates were significantly higher in patients with low CENP-H expression than in those with high CENP-H expression (P < 0.05).
Discussion
In this study, we investigated the clinicopathologic significance and prognostic value of CENP-H in breast cancer. By using RT-PCR and Western blot, we found that CENP-H was overexpressed in immortalized mammary epithelial cell lines and breast cancer cell lines, but not in normal mammary epithelial cells. Additionally, the expression of CENP-H mRNA was higher in breast cancer biopsies than in adjacent normal tissues. These findings suggested that CENP-H was up-regulated in breast cancer cells and breast cancer tissues at both the transcriptional and translational levels. These results are similar to the findings of a previous study suggesting that CENP-H is up-regulated in colorectal cancer[15].
To explore the clinicopathologic significance and prognostic value of CENP-H in breast cancer, we examined CENP-H expression in 307 breast cancers and 30 adjacent normal tissues using immunohistochemistry and compared these findings with clinicopathologic parameters. Over 80% of breast cancers showed weak or strong CENP-H expression, whereas only 40% of adjacent normal tissues showed weak CENP-H staining. Statistical analysis showed that CENP-H expression level was positively correlated with clinical stage, T classification, and N classification, suggesting that CENP-H may play an important role in the development and progression of breast cancer and may be a new marker to identify patients with aggressive breast cancers.
This study also showed that CENP-H was overexpressed in the hyperplasia areas of the mammary epithelium during the early stages of breast cancer and hence was an indicator of the early stage of cancer development. Additionally, high CENP-H expression primarily indicated poor survival in early-stage breast cancer patients. Compared with the expression in normal epithelial cells, CENP-H expression level is much higher in immortalized cell lines, which represent the early stage of Carcinogenesis; hence, we propose that CENP-H may play an important role in the early stage of breast cancer development. CENP-H is essential for cell growth and mitotic progression[14].Since our previous data indicated that CENP-H may be a marker of cancer development [19],[20], our goal was to analyze the relationship between CENP-H and Ki-67, one of the most frequently used markers of proliferating cells. We found a significant correlation between the gene expression of CENP-H and that of Ki-67, indicating that CENP-H is associated with cell proliferation in vivo. This finding provides more evidence that CENP-H may play an important role in the early stage of tumorigenesis. We have previously published similar results, showing that CENP-H may be an early marker of nasopharyngeal carcinoma[18].These results suggest a crucial role for CENP-H in the early development of epithelium-originated cancers and provide evidence that CENP-H can be used as a potential molecular marker for early cancer diagnosis and treatment. However, the other functions and mechanisms underlying CENP-H action remain to be elucidated.
Early stage breast cancer has a highly variable prognostic status and benefits to individual patients from available treatments are critically unpredictable. Although traditional prognostic factors such as lymph node status, tumor grade, histological grade, and distant metastasis are helpful, attention must be paid to the molecular prognostic markers to help identify low-risk from high-risk patients. Recently, CENP-F, another kinetochore component protein, was reported to be associated with poor prognosis and chromosomal instability in patients with primary breast cancer[24]. This suggests that kinetochore proteins can be used to predict the prognosis of patients suffering from breast cancer. In this study, we showed the prognostic value of CENP-H expression in breast cancer patients. Higher CENP-H protein expression was correlated with reduced overall survival in both univariate and multivariate analyses, highlighting the value of CENP-H as a marker of poor prognosis for breast cancer. CENP-H has a particularly great value in predicting the prognosis of early-stage breast cancer patients. These findings are also in agreement with our previous study, where we analyzed CENP-H expression and overall survival in patients with nasopharyngeal carcinoma[18]. Our findings not only indicate the prognostic significance of CENP-H in these cancers but also suggest a potential role for CENP-H in the early stage of cancer development.
References
- 1.Bieche I, Tozlu S, Girault I, et al. Identification of a three–gene expression signature of poor-prognosis breast carcinoma [J] Mol Cancer. 2004;3(1):37. doi: 10.1186/1476-4598-3-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y, Klijn JG, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer [J] Lancet. 2005;365(9460):671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 3.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications [J] Proc Natl Acad Sci USA. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers [J] Nature. 1997;386(6625):623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 5.Yoon DS, Wersto RP, Zhou W, et al. Variable levels of chromosomal instability and mitotic spindle checkpoint defects in breast cancer [J] Am J Pathol. 2002;161(2):391–397. doi: 10.1016/S0002-9440(10)64194-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von Rosen A. Aneuploidy as a prognostic factor in breast cancer [J] Med Oncol Tumor Pharmacother. 1989;6(2):117–120. doi: 10.1007/BF02985233. [DOI] [PubMed] [Google Scholar]
- 7.Nakopoulou L, Tsirmpa I, Giannopoulou I, et al. Aneuploidy of chromosome 20 in invasive breast cancer correlates with poor outcome [J] Cancer Genet Cytogenet. 2002;134(2):127–132. doi: 10.1016/s0165-4608(01)00614-8. [DOI] [PubMed] [Google Scholar]
- 8.Hainsworth PJ, Raphael KL, Stillwell RG, et al. Cytogenetic features of twenty-six primary breast cancers [J] Cancer Genet Cytogenet. 1991;53(2):205–218. doi: 10.1016/0165-4608(91)90097-e. [DOI] [PubMed] [Google Scholar]
- 9.Pandis N, Heim S, Bardi G, et al. Chromosome analysis of 20 breast carcinomas: cytogenetic multiclonality and karyotypic-pathologic correlations [J] Genes Chromosomes Cancer. 1993;6(1):51–57. doi: 10.1002/gcc.2870060110. [DOI] [PubMed] [Google Scholar]
- 10.Pandis N, Idvall I, Bardi G, et al. Correlation between karyotypic pattern and clincopathologic features in 125 breast cancer cases [J] Int J Cancer. 1996;66(2):191–196. doi: 10.1002/(SICI)1097-0215(19960410)66:2<191::AID-IJC9>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 11.Sugata N, Munekata E, Todokoro K. Characterization of a novel kinetochore protein, CENP-H[J] J Biol Chem. 1999;274(39):27343–27346. doi: 10.1074/jbc.274.39.27343. [DOI] [PubMed] [Google Scholar]
- 12.Maiato H, DeLuca J, Salmon ED, et al. The dynamic kinetochore-microtubule interface [J] J Cell Sci. 2004;117(Pt 23):5461–5477. doi: 10.1242/jcs.01536. [DOI] [PubMed] [Google Scholar]
- 13.Rajagopalan H, Lengauer C. Aneuploidy and cancer [J] Nature. 2004;432(7015):338–341. doi: 10.1038/nature03099. [DOI] [PubMed] [Google Scholar]
- 14.Sugata N, Li S, Earnshaw WC, et al. Human CENP-H multimers colocalize with CENP-A and CENP-C at active centromere—kinetochore complexes [J] Hum Mol Genet. 2000;9(19):2919–2926. doi: 10.1093/hmg/9.19.2919. [DOI] [PubMed] [Google Scholar]
- 15.Tomonaga T, Matsushita K, Ishibashi M, et al. Centromere protein H is up-regulated in primary human colorectal cancer and its overexpression induces aneuploidy [J] Cancer Res. 2005;65(11):4683–4689. doi: 10.1158/0008-5472.CAN-04-3613. [DOI] [PubMed] [Google Scholar]
- 16.Shigeishi H, Higashikawa K, Ono S, et al. Increased expression of CENP-H gene in human oral squamous cell carcinomas harboring high-proliferative activity [J] Oncol Rep. 2006;16(5):1071–1075. [PubMed] [Google Scholar]
- 17.Guo XZ, Zhang G, Wang JY, et al. Prognostic relevance of centromere protein H expression in esophageal carcinoma [J] BMC Cancer. 2008;8:233. doi: 10.1186/1471-2407-8-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao WT, Song LB, Zhang HZ, et al. Centromere protein H is a novel prognostic marker for nasopharyngeal carcinoma progression and overall patient survival [J] Clin Cancer Res. 2007;13(2 Pt 1):508–514. doi: 10.1158/1078-0432.CCR-06-1512. [DOI] [PubMed] [Google Scholar]
- 19.Liao WT, Wang X, Xu LH, et al. Centromere protein H is a novel prognostic marker for human nonsmall cell lung cancer progression and overall patient survival [J] Cancer. 2009;115(7):1507–1517. doi: 10.1002/cncr.24128. [DOI] [PubMed] [Google Scholar]
- 20.Liao WT, Yu CP, Wu DH, et al. Upregulation of CENP-H in tongue cancer correlates with poor prognosis and progression [J] J Exp Clin Cancer Res. 2009;28:74. doi: 10.1186/1756-9966-28-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures [J] Methods. 2003;30(3):256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 22.Song LB, Zeng MS, Liao WT, et al. Bmi-1 is a novel molecular marker of nasopharyngeal carcinoma progression and immortalizes primary human nasopharyngeal epithelial cells [J] Cancer Res. 2006;66(12):6225–6232. doi: 10.1158/0008-5472.CAN-06-0094. [DOI] [PubMed] [Google Scholar]
- 23.Zhou H, Kuang J, Zhong L, et al. Tumour amplified kinase stk15/btak induces centrosome amplification, aneuploidy and transformation [J] Nat Genet. 1998;20(2):189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 24.O'Brien SL, Fagan A, Fox EJ, et al. CENP-F expression is associated with poor prognosis and chromosomal instability in patients with primary breast cancer [J] Int J Cancer. 2007;120(7):1434–1443. doi: 10.1002/ijc.22413. [DOI] [PMC free article] [PubMed] [Google Scholar]