Abstract
Proteins fold into their functional 3-dimensional structures from a linear amino acid sequence. In vitro this process is spontaneous; while in vivo it is orchestrated by a specialized set of proteins, called chaperones. Protein folding is an ongoing cellular process, as cellular proteins constantly undergo synthesis and degradation. Here emerging links between this process and cancer are reviewed. This perspective both yields insights into the current struggle to develop novel cancer chemotherapeutics and has implications for future chemotherapy discovery.
Keywords: Protein folding, proteostasis, chaperones, heat-shock proteins, proteasome, chemotherapy
Introduction
Citing statistics from the American Cancer Society, Harold Varmus noted that we have made depressingly little progress in combating cancer[1]. While over the past 50 years dramatic strides have been made against cardiovascular and infectious diseases, age-adjusted mortality in patients with cancer has declined only slightly, with the decrease mostly related to the drop in lung cancer-caused deaths due to aggressive efforts to discourage cigarette smoking.
The discovery of the first Oncogene in 1976[2] triggered a wave of discoveries into oncogenes (genes which increase the risk of developing cancer when mutated) and tumor suppressor genes (TSGs, genes which typically appear in pathways controlling cell growth and regulation). In succeeding decades, these insights were exploited by drug discovery researchers, aiming to displace standard chemotherapeutic drugs (empirically discovered “cytotoxics”) with agents targeting oncogenes (molecular targeted drugs). This approach led to one notable success, Novartis' Gleevec for chronic myelogenous leukemia (CML), a disease linked to the Philadelphia chromosome, an aberration of chromosomes 9 and 22. Overall, however, the new wave of chemotherapeutic research has been described by researchers at the Dana-Farber Cancer Institute as “neocytotoxics”[3], a reference to their tendency to show unacceptable toxicity at therapeutic doses like the older drugs.
This relative lack of success in discovering novel chemotherapeutic drugs prompts the questions of how well we really understand the basic mechanisms underlying cancer and whether there are other avenues of exploration worthy of greater attention. In this review, I focus on the intertwined mechanisms of protein folding and proteostasis and on the balance between production and destruction of proteins in cells. What draws our attention here are two distinct advances: first, the recent clinical successes of Velcade (bortemozib), a drug that inhibits the protein destruction pathway, which was approved in 2003 by the Food and Drug Administration (FDA) of the US for use in refractory multiple myeloma[4]; and second, the multiplicity of HSP90 inhibitors in phase II clinical trials (HSP90 is one of the chaperone proteins intimately involved in protein folding mechanisms)[5].
Based on these developments and a better understanding of protein homeostasis, I aim here to provide information that may lead to a novel perspective on cancer, with a focus on the dynamic state of the proteome as it relates to cancer. This perspective yields intriguing insights into why so little progress has been made in the development of cancer chemotherapies and indicates alternative directions. In particular, this perspective provides a focus on the work of Whitesell and Lindquist who have proposed, in essence, a novel theory of cancer[6], asserting that cancer cells acquire a hyper-mutating phenotype and further claiming that control of cancer will best be achieved by modulating cells' ability to adapt and evolve in response to selection pressures.
One can best understand and appreciate this perspective after an overview of basic biochemical and cellular processes. I start with a review of in vitro protein folding, describing the spontaneous process by which a linear chain of amino acids acquires the precise three-dimensional (3D) structure required for the protein's function. Next, I describe how this folding occurs in the crowded and chaotic internal environment of the cell: this is not a simple spontaneous process; rather, this process is mediated by a class of proteins, chaperones or heat-shock proteins (HSPs), as a “quality control” mechanism to reduce the likelihood of misfolding and loss of function[7]–[9]. The third section describes why protein folding is only one part of the broader mechanism of proteostasis (protein homeostasis)[10] (Figure 1), by which proteins in the cell are constantly degraded and created anew, and that degradation is a specific ATP-dependent process occurring in the ubiquitin-proteosome system. The fourth section describes the current evolving understanding linking these cellular processes and cancer. The first clue to this link between proteostasis and cancer emerged in 1981, when HSPs were first linked to cancer by Oppermann et al.[11], who found that HSP90 coimmunoprecipitated with the src Oncogene protein. Another observation linking these proteostasis processes to cancer is that, while most proteins have cellular half-lives of 1–2 h, many Oncogene proteins and TSG proteins have half-lives of a few minutes. Of a particular note, P53, which has been nicknamed the “guardian of the genome” for its role in DNA repair and is a protein that is mutated in over 50% of all cancers, has a half-life of only 20 min. P53 is constantly being synthesized, folded, and degraded, and its half-life is extended in response to various cellular stresses[12]–[15]. The final section concludes by describing how this understanding that linking protein folding, proteostasis, and cancer may point to future directions in the discovery and development of cancer chemotherapy.
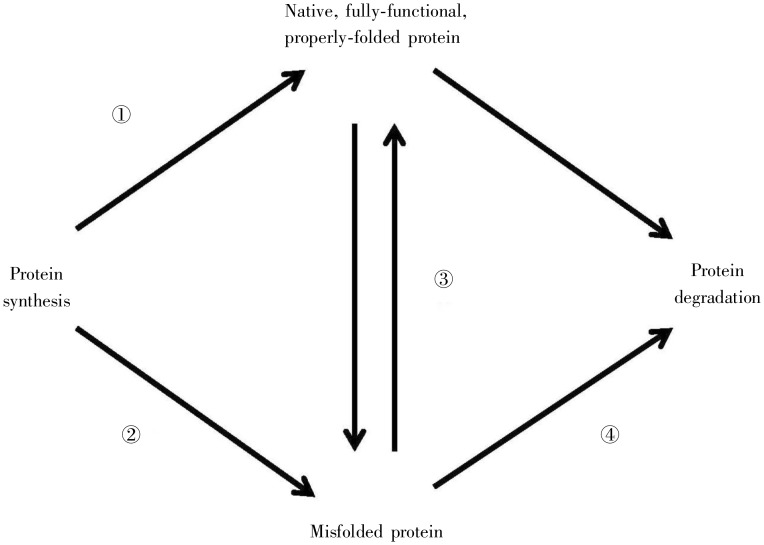
Figure 1. Proteostasis (protein homeostasis). Proteins in a cell are constantly undergoing renewal, as proteins are degraded and synthesized anew. As proteins are synthesized, they must fold properly to acquire their native, functional form (pathway 1). When this protein folding process misfunctions, misfolded proteins arise (pathway 2). Misfolded proteins may either eventually be converted into properly folded proteins (pathway 3) or be degraded (pathway 4). Native, functional proteins can either spontaneously misfold or undergo degradation (unlabeled pathways).
Protein Folding in vitro
The “hallmarks of cancer” are all phenotypic[16]. Yet the standard perspective states “cancer is a disease of the genes”[17], which places the focus purely on the genotype. In the perspective presented here, it does not suffice to focus purely on the genotype—we must also understand the means by which the phenotype results from the genotype, both in the cases where the genotype is normal and in those cases where the genotype has undergone changes that characteristic of cancer[18].
Protein folding and proteostasis are the central mechanisms by which the phenotype emerges from the genotype. In this section, we delve into how protein folding functions in vitro (pathway #1, Figure 1), that is, when the genome is pristine and is in a state that does not yet require the cellular apparatus designed to assist folding under stress conditions. With this as background knowledge, we can proceed in following sections to elaborate how protein folding functions in vivo, under stress conditions such as elevated temperature or mutations, and explore how this may relate to cancer.
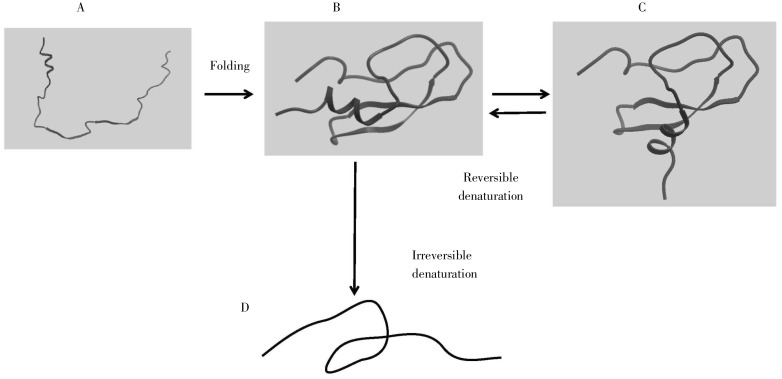
Christian Anfinsen shared the 1972 Nobel Prize in Chemistry for his work on protein folding. His Nobel lecture, “Studies on the principles that govern the folding of protein chains”[19], succinctly describes conclusions that set the paradigm for in vitro protein folding to this day. Figure 2 conveys this general idea of protein folding. A random conformation of a polypeptide chain in an aqueous environment will acquire a specific, unique 3D conformation. With a modest increase in temperature, a protein will lose its original 3D structure but will recover it when the temperature returns to normal. With modest increases in temperature (1°C–2°C), this mild denaturation is reversible. At extremely high temperatures, however, this conformation will turn into a random set of conformations, and it may not revert to the original 3D structure because of denaturation (as in cooking an egg).
Figure 2. Protein folding. Initially, the polypeptide sequence is unstructured (A), and it folds into its native, fully-functional form (B). Through application of mild conditions, it may denature into a form (C) that can revert to the original. Under harsh conditions, it may adopt a new non-functional form (D), which cannot revert to the original.
Anfinsen studied the protein ribonuclease, with its multiple disulfide bridges, and focused on the reversibility of its heat-induced denaturation. Based on these results, he concluded that the primary sequence of a protein completely determines its 3D conformation, and that the process of protein folding was based strictly on thermodynamics. He phrased this the “thermodynamic hypothesis”: “This hypothesis states that the three-dimensional structure of a native protein in its normal physiological milieu (solvent, pH, ionic strength, presence of other components such as metal ions or prosthetic groups, temperature, etc.) is the one in which the Gibbs free energy of the whole system is lowest; that is, that the native conformation is determined by the totality of interatomic interactions and hence by the amino acid sequence, in a given environment.”[19]
From this thermodynamic hypothesis, Anfinsen also concluded that the 3D structure would not be significantly altered by mutations of residues on the surface of a protein, or mutations of an inner residue when changed to a residue of comparable size, hydrophobicity, etc. Conversely, mutations on inner residues which change the size or hydrophobicity (e.g. an Ala → Glu or an Ala → Ser mutation) had potential to alter the folding from the wild-type 3D structure[19].
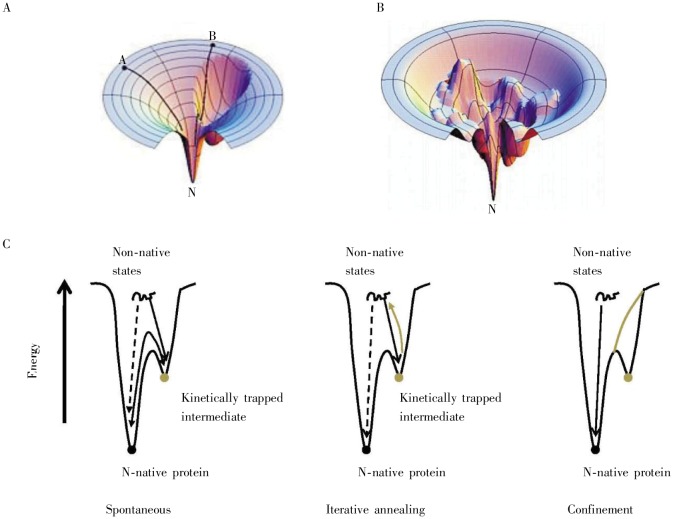
Even with Anfinsen's thermodynamic hypothesis, the kinetics of protein folding, the rate at which this thermodynamic optimum is achieved, remains a puzzle. The number of conformations that should be sampled from a polypeptide chain to find the correct 3D conformation is astronomical, greater than the number of atoms in the universe. The “Levinthal's paradox” is the name given to the puzzling question of how a protein can fold in a few minutes, given the vast number of conformations randomly sampled[20]. Whereas Anfinsen assumed there must be some types of “nucleation” events (small subsequences folding into specific 3D structures which “seed” the formation of the full protein structure), subsequent theoretical[21] and experimental[22] studies on the kinetics of protein folding led to the notion of protein folding funnels (Figure 3A). The notion that a folding pathway is funnel-shaped means that a huge number of conformations are initially explored by many paths in parallel (wide top of the funnel), with a steady winnowing out of the least-promising sets of conformations (middle of the funnel) and final selection of the thermodynamic optimum proceeding via a small number of folding pathways (bottom of the funnel). This notion of folding funnels explains Levinthal's paradox.
Figure 3. Protein-folding funnels. These cartoons depict the energy landscape which a folding polypeptide chain must follow in vitro to achieve its final native form. A, depicts a smooth folding funnel, where folding is all “downhill” with no energy barriers stopping intermediate states from folding spontaneously into the final, native form (“kinetic traps”). Small proteins, which fold very rapidly, generally have this type of folding funnel. B, illustrates a rugged folding funnel with many kinetic traps, i.e. multiple energy barriers exist to stop intermediate states from folding spontaneously into the final form. These rugged landscapes are common for larger proteins. C, shows a one-dimensional slice depicting that kinetically-trapped folding intermediates must surmount the energy barrier through a rugged folding funnel to achieve the final, native form. (Figures A and B were reproduced from reference [21] with permission; figure C was adapted from reference [26].).
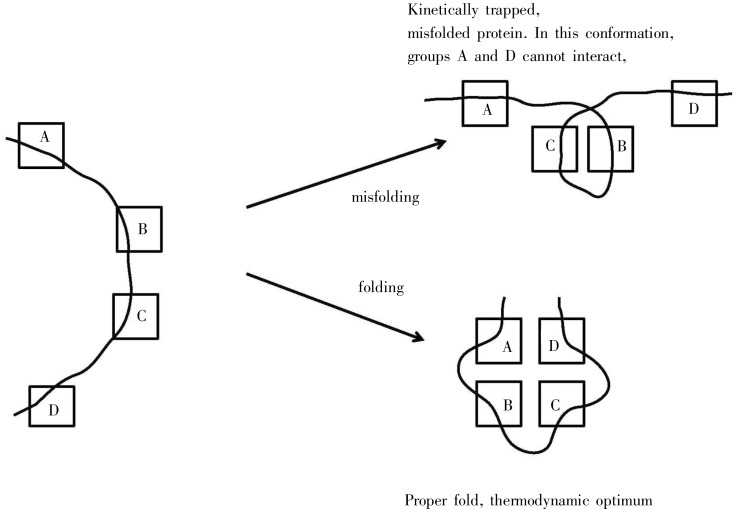
Figure 3A illustrates a smooth folding funnel that pertains to small proteins, ones which fold extremely fast in a few microseconds, near the theoretical folding “speed limit”[23]. The smooth funnel indicates that there are few if any “kinetic traps”, namely conformations that are stable but will not lead to the final global thermodynamic optimum. Figure 3B depicts a “rugged landscape”, a protein folding funnel with many kinetic traps. As shown in Figure 3C, if dependent solely on random motion of molecules, it would take a long time for the rare occurrence that will permit a conformation to jump out of this trap. Figure 4 shows a schematic of what such a kinetic trap might look like; one pair of hydrophobic interactions is satisfied in a way that doesn't permit the other pair to form.
Figure 4. A schematic example of misfolding vs. folding. Hydrophobic elements are shown as squares; the protein backbone is shown as a curve. In an aqueous environment, hydrophobic groups will tend to associate. The left side shows an unfolded protein. The upper right shows a misfolded protein, kinetically trapped. The lower right shows a properly folded protein in its thermodynamic optimum. Note that significant conformational re-arrangement is needed to convert the misfolded conformation into the properly folded one.
Another insight that emerged from recent theoretical studies of protein folding is that the dominant driving force consists of hydrophobic interactions, the tendency for hydrophobic sidechains (Val, Leu, Ile, Ala, Phe, Tyr, Trp) to escape an aqueous environment and seclude themselves with similarly hydrophobic sidechains in a hydrophobic interior—“hydrophobic collapse”. Pauling made the spectacular prediction of the alpha-helix, as a regular motif for protein structures a decade before the first experimental structure was elucidated. This was based on the assumption that hydrogen-bonds would form along backbone peptide units spaced three residues apart. By contrast, it is now thought that helices (and other elements of secondary structures) begin to form as a result of constraints imposed by hydrophobic collapse, with hydrogen-bonds rigidifying these helices but their overall effect being secondary to the much stronger hydrophobic forces[24].
An interesting test of Anfinsen's hypothesis was done in 1992, when a series of D-amino acids (mirror image of standard L-amino acids) were chemically linked according to the sequence of human immunodeficiency virus (HIV) protease. As expected, this led to a protein that was in every way the mirror image of the native HIV protease: it cleaved mirror image substrates with the same specificity, it bound mirror image inhibitors with the same affinity, achiral Physicochemical measurements were the same, and chiral ones were of the opposite sign[25].
This section on protein folding in vitro concludes with the observation that the types of conformations shown in Figure 4 are prone to aggregation, as depicted in Figure 5. When hydrophobic elements are exposed on the surface of a protein, they may interact with hydrophobic elements of other proteins to escape the aqueous environment. When multiple such surface-exposed hydrophobic elements are present, the potential exists for protein aggregation, forming insoluble aggregates that are often toxic to cells[26].
Figure 5. A schematic example of how protein aggregates may form from misfolded conformations when they leave hydrophobic groups exposed on the surface.
Protein Folding in vivo
Protein folding in vivo is quite different from that in vitro.[27] For medium- to large-sized proteins it occurs much faster than one might expect, typically in the range of milliseconds to seconds[7]. It takes place in the crowded internal environment of the cell, where many intermolecular interactions could potentially disrupt the normal protein folding pathway. Protein folding in vivo is facilitated by chaperones, also known as HSPs or stress proteins.
HSPs were first isolated in 1975 by Lindquist et al.[28]. The first speculation about the function of HSPs evidently was published in 1986 by Pelham[29] in a breathtaking leap of insight. He had found that the heat-shocked nucleoli recovered more rapidly after overexpression of HSP70 in an ATP-dependent manner. Pelham's model “proposes that during heat shock, proteins become partially denatured, exposing hydrophobic regions which then interact to form insoluble aggregates. By binding tightly to hydrophobic surfaces, HSP70 limits such interactions and promotes disaggregation”. The term “chaperone” was coined shortly thereafter by Ellis[30]. By the early 1990's, the general importance of this mechanism for protein folding in vivo was clear[27]. The proportion of proteins that spontaneously fold in the cell is thought to be approximately 10%; the rest rely on chaperoned folding[7].
Chaperones are highly conserved, being present in prokaryotes and eukaryotes, and, as I will note below, are important for the folding of the equally conserved oncogenes and tumor suppressor genes. Chaperones are ubiquitous and up-regulated in response to a variety of cell stresses, such as heat, mutations, heavy metals, etc. Chaperones constitute 1%–2% of the total weight of proteins in a cell, and they range in size from the 15–30 kDa “small HSPs”[31], to the over 100-kDa HSP100/CIP family[32].
Chaperoned protein folding occurs in many ways. At the most abstract level, one can think of them as converting a rugged protein folding funnel into a smooth one, eliminating kinetic traps and speeding up achievement of the thermodynamic optimum[7]. This leads to a profound proposition concerning the role of HSPs in cellular evolution, namely that HSPs act as a buffer, hiding the natural phenotypic variations present in cells, which are unmasked in response to stress[8]. In this view, HSPs expedite the cell' s ability to evolve in response to stress. For example, HSP70 overexpression has been shown to correlate with certain types of drug resistance, as a response to the stress of drug therapy[33].
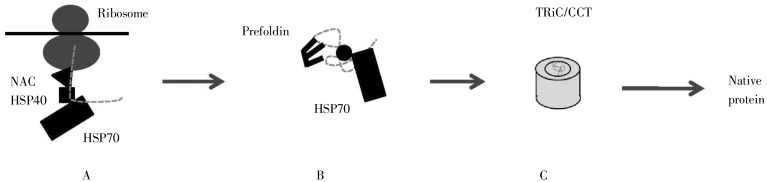
HSPs act as a buffer in four stages[7] (Figure 6): (1) as the nascent polypeptide chain emerges from the ribosome, chaperones protect it from premature folding; (2) the polypeptide chain acquires the approximate overall 3D conformation; (3) the polypeptide chain iteratively folds and re-folds in a confined “cage” until the final 3D conformation is achieved. In addition, it may make sense to describe a fourth category; and (4) chaperones unfold a misfolded and/or aggregated protein. Distinct chaperones are involved in each stage in an ATP-dependent manner. Misfolded proteins that cannot be properly folded are marked for degradation, the topic of the next section.
Figure 6. Three stages of protein folding. A, shows the nascent chain emerging from the ribosome, with the involvement of the nascent chain–associated complex (NAC), heat-shock protein 40 (HSP40), and HSP70. B, shows the intermediate stages of folding, involving HSP70 and prefoldin. C, shows the final stage, folding within the barrel-shaped TRiC/CCT. Not all proteins go through all three stages; for example, short polypeptide chains require only step one. (Adapted from reference [26]).
(1) In bacteria, a chaperone called the Trigger Factor (TF) is found proximal to the ribosome and protects the nascent chain from premature folding. In eukaryotes, the homologous factor is the ribosome-associated complex (RAC), sometimes in association with nascent chain-associated complex (NAC). The protection is a necessary step, as ribosomal synthesis occurs at a rate of about 10 residues/sec, whereas the rate of folding in vitro is 10 residues/microsec.
(2) In eukaryotes, as the full protein is released from the ribosome, HSP40 and HSP70 assist in ensuring the overall 3D conformation is approximately achieved.
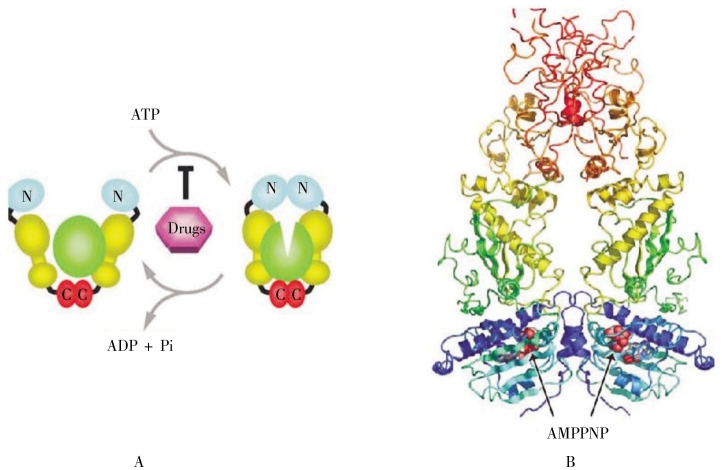
(3) The final stage of folding occurs iteratively, in which the partially-folded protein interacts with a chaperone and is then released. One way this is accomplished is by the pre-folded protein entering the cavity of barrel-shaped complexes termed GroES/GroEL in bacteria and TriC/CCT in eukaryotes (Figure 7). This creates a protected environment in which the protein can properly fold in a way that may be understood conceptually as an “Anfinsen cage”, where the compact environment limits the range of possible extended conformations that may be sampled. The involved chaperones are called chaperonins. Another path in this final stage of folding is via interactions with HSP90, a dimeric protein with each monomer containing three regions: an N-terminal ATP-binding domain, a linker, and a C-terminal dimerization domain (Figure 8). The precise nature of HSP90 interactions with a pre-folded protein has not been resolved.
Figure 7. Structure of GroES/GroEL, which forms a secluded cage-like environment to speed protein folding and protect it from potential aggregation with neighboring proteins. (Image produced from PDB entry 2c7c; data originally from reference [59].).

Figure 8. Structure and action of HSP90. A, depicts the chaperone action of HSP90 for a client protein. The dimeric HSP90 consists of a C-terminal domain (red), a linker (yellow), and an ATP-binding domain at the N-terminus. When ATP is present, it binds to the N-terminal domain, inducing a conformational change in the client protein (green). HSP90 inhibitors block this ATP-binding event, halting the chaperone function. B, depicts a ribbon diagram of a low-resolution X-ray structure of HSP90, with similar color-coding, in an orientation reversed from 8A. AMPPNP is an ATP-mimic inhibitor of HSP90. (Figure 8 was reproduced from reference [60] with permission.).
(4) Members of the HSP100/CIP family of chaperones appear to interact with misfolded and/or aggregated protein, in effect undoing the damage. As part of a broad network of chaperones, they appear to be able to disassemble stable complexes; to unfold highly stable native protein domains; and to help to resolubilize and refold non-native proteins trapped in a high molecular weight aggregate[32]. The function of HSP104 has been described as a “molecular crowbar”[34]: it functions by prying apart aggregates. HSP104, which has multiple binding sites, can bind multiple components within the same aggregate. The HSP104 domains that contact the aggregate undergo an ATP-driven conformational change that separates or further unfolds the misfolded aggregates and then releases them. Newly exposed hydrophobic elements, shielded by chaperones such as HSP70 and HSP40, are transiently protected from reaggregation. HSP104 acts iteratively, eventually resulting in the release of HSP70-bound folding intermediates that have a renewed opportunity to proceed to the native state.
From the perspective of identifying novel drug targets for cancer, this multiplicity of HSPs throughout protein folding provides many opportunities for modulating these stages of folding with therapeutic agents. Agents targeting HSP90 have been tested in phase III clinical trials[5], whereas HSP70 is still in basic research phase[35].
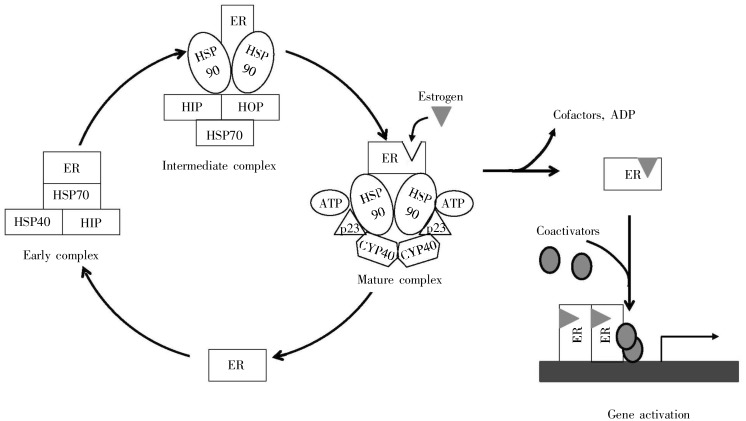
There are seemingly endless variations on the overall protein-folding processes as outlined above. Transporting the protein across membranes may require one pre-folded conformation; final folding then occurs after the protein is in the target organelle (e.g. the endoplasmic reticulum)[36]. Exemplified by estrogen receptor, final folding occurs only when that protein is in complex with HSP90 and other factors, and its ligand, progesterone (Figure 9). HSP90 forms complexes with many distinct factors, probably explaining its ability to assist the folding of a huge variety of proteins, such as p53[6].
Figure 9. Chaperone-facilitated folding of the estrogen receptor (ER). Estrogen only binds with high affinity to a mature complex of the receptor with HSP90 and other cofactors. Binding of estrogen stimulates dissolution of the complex and binding to DNA. (Adapted from reference [6]).
Protein Degradation
The previous sections discussed the processes by which proteins are created, and acquire their functional 3D structure. We now turn to the complementary process of protein degradation. Cellular proteolysis was once viewed as a nonspecific process—the “garbage collection” of the cell. Once it became clear that proteins constantly turn over, with degradation occurring rapidly, specifically, and in an ATP-dependent manner, attention focused on the processes of protein degradation. This ultimately led to the isolation of a cell-free system for ATP-dependent proteolysis in 1978[37], now called the ubiquitin-proteasome system (UPS), which is responsible for most proteolysis in the cell.
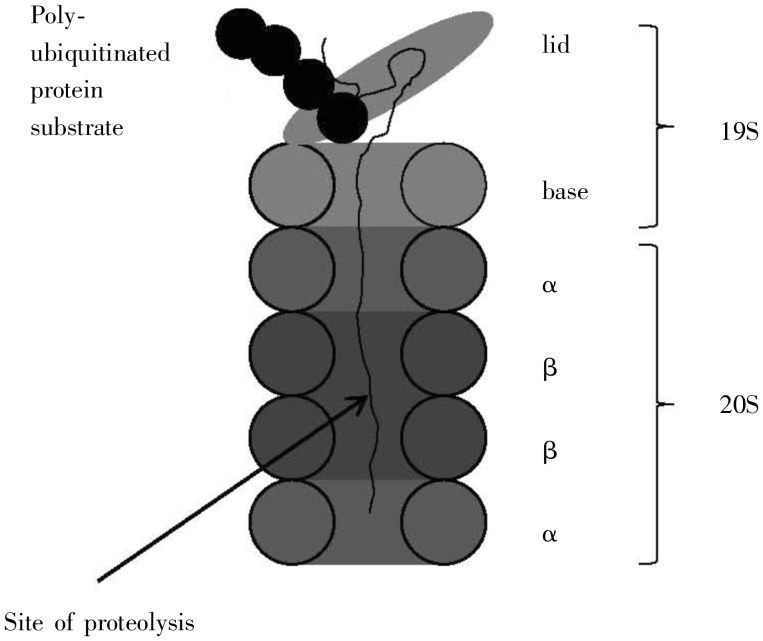
Attention was drawn to the UPS as a cancer drug target by the 2003 approval of the proteosome inhibitor Velcade/bortemozib for multiple myeloma. One year later, the discovery of the UPS was rewarded by the Nobel Prize to Ciechanover, Hershko, and Rose; their lectures represent a concise introduction to this system[38]–[40]. The proteolytic machinery of the UPS is comprised of the proteosome, a 2.5-MDa multicomponent system consisting of a barrel-shaped 20S catalytic core particle with a 19S regulatory particle capping both sides of the core particle (Figure 10). A protein tagged with a poly-ubiquitin tail is recognized by the proteosome and unfolded near the cap, with proteolysis occurring deeply within the core particle[41].
Figure 10. High-level structure of the proteosome. It consists of two domains, a 19S and a 20S. The former consists of a lid and a base, which sits atop the 20S, a barrel–shaped complex composed of four subunits, two α subunits and two β. Proteolytic cleavage occurs deeply within the β subunits. (Adapted from reference [38]).
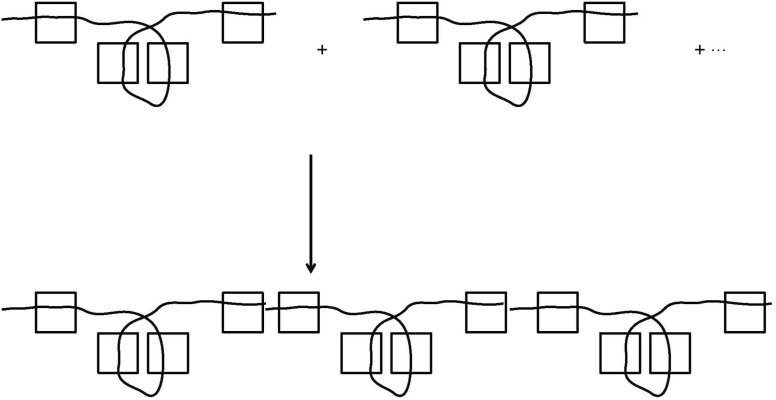
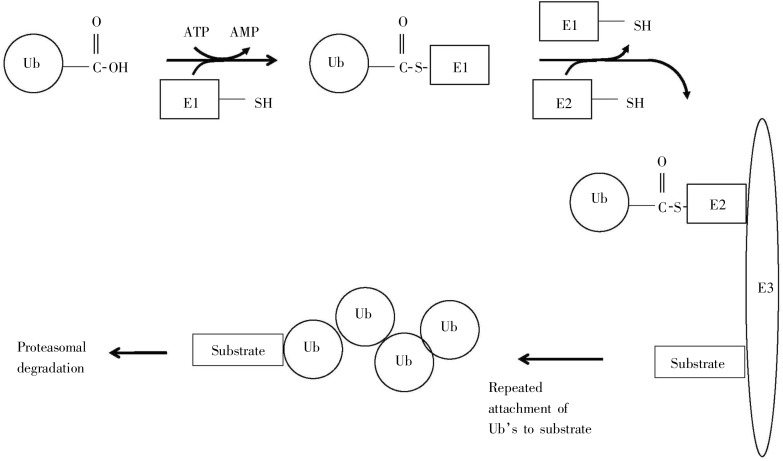
Tagging for protein degradation is accomplished in a series of steps, ultimately resulting in a poly-ubiquitin sequence tag covalently attached to a lysine of the substrate protein[42]. This pathway, summarized in Figure 11, involves a series of enzymes: E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugating enzyme), and E3 (ubiquitin-protein ligase). Step 1 creates a covalent linkage between E1 and Ubiquitin. Step 2 uses the E1-ubiquitin complex, E2, E3, and S (the substrate protein to be tagged for degradation), to form the complex Ubi.E2.E3.S. Step 3 repeatedly appends additional Ubiquitin molecules to the complex Ubi.E2.E3.S. Step 4 cleaves this poly-ubiquitinated complex to form the final tagged substrate, recognized by the proteosome.
Figure 11. Steps leading to the poly-ubiquitin tag. This pathway involves a series of enzymes: E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugating enzyme), and E3 (ubiquitin-protein ligase). Step 1 creates a covalent linkage between E1 and Ubiquitin. Step 2 uses the E1-ubiquitin complex, E2, E3, and S (the substrate protein to be tagged for degradation) to form the complex Ubi.E2.E3.S. Step 3 repeatedly appends additional Ubiquitin molecules to the complex Ubi.E2.E3.S. Step 4 leads to ultimate degradation by the proteosome. (Adapted from reference [42]).
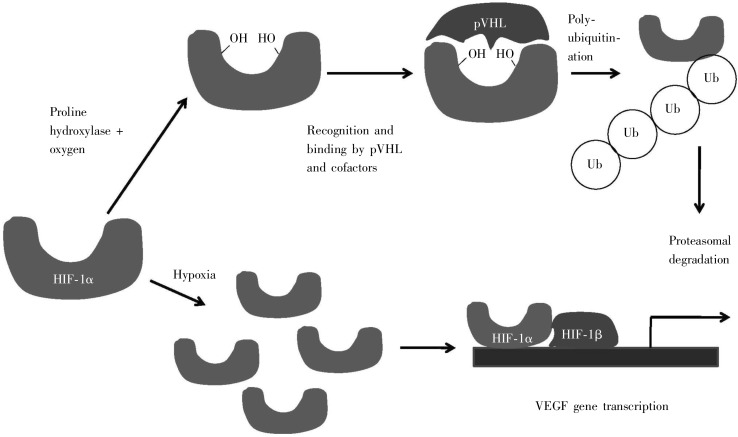
The UPS is also becoming recognized as an important component that is involved in biological processes which are related to a variety of disorders with cancer-related phenotypes. One example is von-Hippel-Lindau (VHL) syndrome, which is a rare autosomal-dominant condition with hemangioblastomas in the kidneys, retina, cerebellum, and spinal cord, resulting from a mutation in the VHL tumor suppressor gene. The mechanism by which VHL gene functions involves the UPS, whereby the protein product of this gene interacts with the hypoxia-inducible factor-1 (HIF-1) in response to the cell stress of insufficient oxygen (hypoxia)[43]. Under normal conditions, in the presence of oxygen, HIF-1 is synthesized, rapidly converted into a form which is recognized by pVHL (the protein product of the VHL gene), and subjected to ubiquitination and proteosomal degradation, with an overall half-life of 10 min. Under hypoxic conditions, HIF-1 remains in a form not recognized by pVHL, and hence is not degraded. Thus HIF-1 is free to promote transcription of genes such as vascular endothelial growth factor (VEGF), a feedback signaling factor that promotes vascularization to facilitate the return to normal conditions (Figure 12). Another point of interest about pVHL degradation is that chaperones can be involved: e.g. chaperone HSP90 is not involved in VHL folding but is essential for its degradation[44]. This is an intriguing example of where chaperones are involved in the degradation process rather than the protein folding process; and, here, degradation of a protein involved in a disease is associated with cancer formation, and links proteostasis with cancer and with chaperones in particular.
Figure 12. Regulation of hypoxia-inducible factor (HIF) by von-Hippel-Lindau (VHL) gene under normal and hypoxic conditions. Under normal conditions, an oxygen-dependent proline hydroxylase attaches a pair of hydroxyl groups to HIF-1α, which allows it to form a complex with pVHL and other co-factors, which leads to HIF-1α being tagged for proteosomal degradation. Under hypoxic conditions, this hydroxylation does not occur, and HIF-1α recruits its partner HIF-1β to up-regulate transcription of the vascular endothelial growth factor (VEGF) gene, to promote vascularization (growth of new blood vessels). (Adapted from reference [61]).
The UPS also plays a key role in regulating cellular levels of P53, with a half-life of only 20 min in normal cells[14]. P53 is the protein which functions as the “guardian of the genome”, and is mutated in over half of all cancers. In response to DNA damage stress such as ultraviolet radiation, p53 levels rise and cells are driven to apoptosis (programmed cell death). P53 acts as a transcription factor that induces the expression of many genes, one of which is mdm2, that codes for a protein Mdm2 that shepherds translocation of P53 from the nucleus to the UPS for degradation. This feedback loop has a transcriptional delay that ensures a brief burst of elevated P53 levels in response to stress.
A final point on the UPS is that misfolded proteins are subjected to a higher rate of degradation than are normal proteins, but these recognition processes are incompletely understood. How the cell distinguishes properly-folded proteins from misfolded ones, given its complexity, is an intellectually fascinating puzzle. It is difficult to even imagine all the types of molecular recognition processes that must be involved to maintain a careful balance.
Connections to Cancer
We recently have been reminded “all cancers arise as a result of changes that have occurred in the DNA sequence of the genomes of cancer cells” [45]. This, however, remains a hypothesis, not a fact. An alternative, and perhaps an important addition, to this hypothesis would be to consider and focus on the dynamic cellular processes centered on the proteome, and the process of proteostasis outlined above. The links between chaperones and cancer were first identified almost 30 years ago, and the evidence continues to grow, indicating that chaperones and protein homeostasis likely play an important role in cancer formation, and thus, present multiple opportunities for therapeutic intervention.
In 1981, shortly after the discovery of oncogenes, researchers observed that HSPs co-immunoprecipitated with the src Oncogene[11]. The authors concluded “it would also seem wise to search for other overlaps between the heat shock response and virus-induced neoplastic transformation”, a suggestion that was not immediately embraced. A dramatic leap forward occurred in 1994 when Whitesell et al.[46] identified the mechanism of geldanamycin-induced reversal of the neoplastic transformation as inhibition of HSP90. Many HSP90 inhibitors are now undergoing evaluation in clinical trials[47]–[49]. The 2003 approval of the proteosome inhibitor Velcade/bortemozib for multiple myeloma further supports the UPS as a cancer drug target[4].
Many other clues support this link to cancer. The list of client proteins of HSP90—those proteins require HSP90 for proper folding—is replete with well-known oncogenes and tumor suppressor genes: src, AKT, Bcr-Abl, etc[50]. HSP90 levels often are higher in cancer cells than in normal cells, and HSP expression in breast or gastric cancer is associated with poor prognosis and resistance to chemotherapy or radiotherapy[51]. HSP90 inhibitors have been shown to be synergistic with other chemotherapeutic agents.[52] The connections between the hemangioblastomas of VHL syndrome and HSP90's involvement in pVHL degradation were mentioned above. Other HSPs are also suspected to be involved in cancer formation: e.g. down-regulation of HSP70 has been shown to increase apoptosis in cancer cells, while leave normal cells unaffected[53]—which raises an intriguing idea of considering HSP70 as a potential cancer-specific target.
Other drug targets are emerging from increased understanding of proteostasis. Mdm2, mentioned above as a protein involved in P53 degradation, is one example: inhibitors (nutlins) of the P53/Mdm2 interaction were reported in 2004 to shrink tumors in xenograft models [54]. NEDD8 is another protein involved in the UPS; inhibitors of the enzyme which activates NEDD8 were recently found to be effective in suppressing the growth of human tumor xenografts in mice[55].
The homeostatic regulation of P53 illuminates how the dual processes of protein synthesis/folding and protein degradation may be directly connected to neoplastic transformation. The 20-min half-life of P53 in the cell is unusually short and its levels, regulated by proteostasis, rise very quickly in response to stressors even though its mRNA levels remain constant[14]. Mutations in p53 gene are found in 30%–50% of all cancers, and the mutations predominate in a particular class, i.e. point mutations in its DNA-binding domain[13]. Chaperoned folding of P53 is especially complex, involving a variety of co-chaperones[6]. It may be that chaperoned folding protects mutant p53 from degradation as well. Cancer cells, even those homozygous for mutations in p53, often have high levels of P53, which may facilitate a variety of mutation-induced mechanisms to run amok[14]. In this regard, application of HSP90 inhibitors has been demonstrated to lower P53 levels and induce apoptosis.
In 1994, Whitesell et al.[46] identified the mechanism of geldanamycin-induced reversal of the neoplastic transformation as inhibition of HSP90. In 1998, Rutherford and Lindquist[8] suggested that HSPs serve as a capacitor to buffer phenotypic variation. Taking these two findings together, Whitesell and Lindquist[6] have proposed a novel theory of cancer, centered on the role of chaperones. HSP90 can conceal inherent genetic variation within populations of cells, allowing polymorphic variants of crucial signaling pathways to accumulate cryptically while the overall normal phenotype is maintained. This “buffering” capacity of HSP90 funnels complex developmental processes into discrete outcomes despite underlying genotypic variations. Under stress, some of the unstable HSP90 clients may become more unstable, increasing the demand for HSP90 to facilitate refolding of its usual clients and that of these new stress-destabilized proteins. New phenotypes can emerge when this buffering capacity of HSP90 is exceeded, exposing previously hidden genetic variations to natural selection. In cancer, HSP90 might function as a buffer of the extensive genetic heterogeneity common to cancer. Furthermore, Whitesell and Lindquist[6] explain “HSP90 has a more complex role in facilitating neoplastic transformation than simply inhibiting apoptosis. The dynamic, low-affinity interactions of HSP90 with its client proteins—such as hormone receptors, transcription factors and kinases—maintain them in a latent but readily activated state. Oncogenic mutation of such clients, however, leads to higher requirements for HSP90 function, presumably because of an exaggerated conformational instability of the mutant.”
Implications for Chemotherapy
“Molecular targeting”—using genetic information that links specific proteins to cancer—has led to one spectacular success: Novartis' Gleevec is effective against CML, a disease linked to the Philadelphia chromosome with a frequency exceeding 95%[56]. Approved in 2001 to much justifiable acclaim, Gleevec promised to be a harbinger of a new wave of molecularly-targeted therapeutics—the “magic bullet” for CML, designed to inhibit the tyrosine kinase Bcr-Abl, the Oncogene activated via the Philadelphia chromosome. Even oncology researchers at Novartis now conclude that Gleevec may be more an outlier than a standard-bearer for the next wave of therapy[57]. Gleevec is the great exception; most new chemotherapies could be considered to be “neocytotoxics”[3], an unflattering reference to their tendency to show unacceptable toxicity at therapeutic doses (i.e. they have a marginal therapeutic index). Chemotherapy for cancer has the highest attrition rate of any therapeutic area: fewer than 10% of drug candidates entering phase I trials are ultimately approved for commercial use[58]. Fallout in the clinic is primarily due to drug toxicities, i.e. their inability to achieve an advantageous therapeutic index.
If we are to make significant progress against cancer, we need to move beyond developing more neocytotoxics. The standard paradigm often leads to drug candidates that inhibit rapidly dividing cells, which hits both cancer cells and the rapidly dividing cells of the epithelia, leading to the harsh side-effects of chemotherapy. This approach naturally leads to a low therapeutic index and the duration of efficacy is frequently only 6–12 months, as cancer cells appear to evolve in response to the selection pressure of the drug.
Whitesell and Lindquist[6] clearly recognized the implications of the theory of cancer: “Such an evolutionary view of the malignant progression problem suggests that definitive control of a cancer will probably be achieved most effectively by altering the key determinants that shape its ability to adapt and evolve. Consequently, HSP90 might provide a broader, more effective target for anticancer therapies than single, oncogenically activated but dispensable signaling pathways that are the focus of most current drug-discovery efforts.” This is a very provocative suggestion—saying that the key to controlling cancer is to modulate a cell's ability to evolve. This is a testable hypothesis, and one that, if true, would significantly alter our approach towards the discovery and development of chemotherapeutic agents. That cancer cells are hyper-evolving may also explain the difficulty in finding chemotherapies whose effectiveness extends beyond 6–12 months: cancer cells may be evolving away from the selection pressure of a drug.
Conclusions
Appreciating the role of proteostasis in the cell, including the role of chaperones in facilitating proper protein folding and the role of the UPS in protein degradation, yields a novel perspective on cancer, which focuses on the dynamic state of the proteome and the evolving phenotypes of cancer cells. Research on this proteostasis network has yielded one approved drug, with many others undergoing clinical evaluation. We may well be in the early days of fully exploiting protein folding and proteostasis to devise better cancer chemotherapeutic agents.
There are many outstanding questions in this area, the subjects of much current researches:
How do chaperones recognize improperly folded proteins?
What is the precise mechanism by which HSP90 inhibition by geldanamycin diminishes P53 levels?
Would the other HSPs also be attractive drug targets? (HSP70, from early studies, certainly seems of potential interest.)
Of all possible intervention points in the proteostasis network, which are best for chemotherapy? Which have sites most amenable to inhibition by an orally-available “small molecule”? Which will provide the best therapy with the least toxicity?
What is the origin of the therapeutic index of Velcade? What lessons are to be learned by its success as a chemotherapeutic agent?
What are the full implications of the notion of HSPs' buffering evolutionary change? How can this hypothesis be properly evaluated as a theory of the origin of cancer?
What is the precise mechanism of the neoplastic transformation, and what roles do chaperones specifically play?
Understanding protein misfolding and proteostasis should lead to their exploitation in the development of novel and improved cancer chemotherapeutics.
Acknowledgments
The author is grateful to a number of people who reviewed early versions of this manuscript and provided suggestions for improvement, especially L. T. Webster. The author also acknowledges the 2009 Asilomar retreat of the Dill group at UCSF, which indirectly sparked the writing of this review.
References
- 1.Varmus H. The new era in cancer research [J] Science. 2006;312(5777):1162–1165. doi: 10.1126/science.1126758. [DOI] [PubMed] [Google Scholar]
- 2.Stehelin D, Varmus HE, Bishop JM, et al. DNA related to the transforming gene (s) of avian sarcoma viruses is present in normal avian DNA [J] Nature. 1976;260(5547):170–173. doi: 10.1038/260170a0. [DOI] [PubMed] [Google Scholar]
- 3.Chabner BA, Roberts TG., Jr Chemotherapy and the war on cancer [J] Nat Rev Cancer. 2005;5(1):65–72. doi: 10.1038/nrc1529. [DOI] [PubMed] [Google Scholar]
- 4.Adams J, Kauffman M. Development of the proteasome inhibitor Velcade (Bortezomib) [J] Cancer Invest. 2004;22(2):304–311. doi: 10.1081/cnv-120030218. [DOI] [PubMed] [Google Scholar]
- 5.Messaoudi S, Peyrat JF, Brion JD, et al. Recent advances in Hsp90 inhibitors as antitumor agents [J] Anticancer Agents Med Chem. 2008;8(7):761–782. doi: 10.2174/187152008785914824. [DOI] [PubMed] [Google Scholar]
- 6.Whitesell L, Lindquist S. HSP90 and the chaperoning of cancer [J] Nat Rev Cancer. 2005;5(10):761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 7.Hartl FU, Hayer-Hartl M. Converging concepts of protein folding in vitro and in vivo [J] Nat Struct Biol. 2009;16(6):574–581. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- 8.Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution [J] Nature. 1998;396(6709):336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 9.Wegele H, Muller L, Buchner J. Hsp70 and Hsp90—a relay team for protein folding [J] Re. Physiol Biochem Pharmacol. 2004;151:1–44. doi: 10.1007/s10254-003-0021-1. [DOI] [PubMed] [Google Scholar]
- 10.Balch WE, Morimoto RI, Dillin A, et al. Adapting proteostasis for disease intervention [J] Science. 2008;319(5865):916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 11.Oppermann H, Levinson W, Bishop JM. A cellular protein that associates with the transforming protein of Rous sarcoma virus is also a heat-shock protein [J] Proc Natl Acad Sci USA. 1981;78(2):1067–1071. doi: 10.1073/pnas.78.2.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scott MD, Frydman J. Aberrant protein folding as the molecular basis of cancer [M] In: Bross P, Gregersen N, editors. Methods in Molecular Biology, vol 232: Protein misfolding and disease: principles and protocols. Totowa, NJ: Humana Press; 2003. pp. 67–76. [DOI] [PubMed] [Google Scholar]
- 13.Bullock AN, Fersht AR. Rescuing the function of mutant p53 [J] Nat Rev Cancer. 2001;1(1):68–76. doi: 10.1038/35094077. [DOI] [PubMed] [Google Scholar]
- 14.Oren M. Regulation of the p53 tumor suppressor protein [J] J Biol Chem. 1999;274(51):36031–36034. doi: 10.1074/jbc.274.51.36031. [DOI] [PubMed] [Google Scholar]
- 15.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network [J] Nature. 2000;408(6810):307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 16.Hanahan D, Weinberg R. The hallmarks of cancer [J] Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 17.Bishop JM. The molecular genetics of cancer [J] Science. 1987;235(4786):305–311. doi: 10.1126/science.3541204. [DOI] [PubMed] [Google Scholar]
- 18.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers [J] Nature. 1998;396(6712):643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 19.Anfinsen C. Studies on the principles that govern the folding of protein chains. 1972 Nobel Prize lecture. http://nobelprize.org/nobel_prizes/chemistry/laureates/1972/anfinsen-lecture.pdf. [DOI] [PubMed]
- 20.Levinthal C. Are there pathways for protein folding? [J] Extrait du J de Chimie Physique. 1968;65(1):44–45. [Google Scholar]
- 21.Dill KA, Chan HS. From Levinthal to pathways to funnels [J] Nat Struct Biol. 1997;4(1):10–19. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 22.Creighton TE. Toward a better understanding of protein folding pathways [J] Proc Nat Acad Sci USA. 1988;85(14):5082–5086. doi: 10.1073/pnas.85.14.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang IJ, Lee JC, Winkler JR, et al. The protein-folding speed limit: intrachain diffusion times set by electron-transfer rates in denatured Ru(NH3)5(His-33)-Zn-cytochrome c [J] Proc Nat Acad Sci USA. 2003;100(7):3838–3840. doi: 10.1073/pnas.0637283100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dill KA, Ozkan SB, Shell MS, et al. The protein folding problem [J] Annu Rev Biophys. 2008;37:289–316. doi: 10.1146/annurev.biophys.37.092707.153558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milton RC, Milton SC, Kent SB. Total chemical synthesis of a D-enzyme: the enantiomers of HIV-1 protease show reciprocal chiral substrate specificity [J] Science. 1992;256(5062):1445–1448. doi: 10.1126/science.1604320. [DOI] [PubMed] [Google Scholar]
- 26.Hartl FU, Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein [J] Science. 2002;295(5561):1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 27.Gething MJ, Sambrook J. Protein folding in the cell [J] Nature. 1992;355(6355):33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- 28.Lindquist (McKenzie) S, Henikoff S, Meselson M. Localization of RNA from heat-induced polysomes at puff sites in Drosophila melanogaster [J] Proc Natl Acad Sci USA. 1975;72(3):1117–1121. doi: 10.1073/pnas.72.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pelham HRB. Speculations on the functions of the major heat shock and glucose-regulated proteins [J] Cell. 1986;46(7):959–961. doi: 10.1016/0092-8674(86)90693-8. [DOI] [PubMed] [Google Scholar]
- 30.Ellis J. Proteins as molecular chaperones [J] Nature. 1987;328(6129):378–379. doi: 10.1038/328378a0. [DOI] [PubMed] [Google Scholar]
- 31.Jakob U, Gaestel M, Engel K, et al. Small heat shock proteins are molecular chaperones [J] J Biol Chem. 1993;268(3):1517–1521. [PubMed] [Google Scholar]
- 32.Glover JR, Tkach JM. Crowbars and ratchets: hsp100 chaperones as tools in reversing protein aggregation [J] Biochem Cell Biol. 2001;79(5):557–568. [PubMed] [Google Scholar]
- 33.Sliutz G, Karlseder J, Tempfer C, et al. Drug resistance against gemcitabine and topotecan mediated by constitutive hsp70 overexpression in vitro: implication of quercetin as sensitiser in chemotherapy [J] Br J Cancer. 1996;74(2):172–177. doi: 10.1038/bjc.1996.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins [J] Cell. 1998;94(1):73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 35.Lu S, Tan Z, Wortman M, et al. Regulation of heat shock protein 70-1 expression by androgen receptor and its signaling in human prostate cancer cells [J] Int J Oncol. 2010;36(2):459–467. [PMC free article] [PubMed] [Google Scholar]
- 36.Pelham HRB. The dynamic organisation of the secretory pathway [J] Cell Str Function. 1996;21(5):413–419. doi: 10.1247/csf.21.413. [DOI] [PubMed] [Google Scholar]
- 37.Ciechanover A, Hod Y, Hershko A. A heat-stable polypeptide component of an ATP-dependent proteolytic system from reticulocytes [J] Biochem Biophys Res Comm. 1978;81(4):1100–1105. doi: 10.1016/0006-291x(78)91249-4. [DOI] [PubMed] [Google Scholar]
- 38.Ciechanover A. Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. 2004 Nobel Lecture, http://nobelprize.org/nobeLprizes/chemistry/laureates/2004/ciechanover-lecture.html. [DOI] [PubMed]
- 39.Hersko A. The Ubiquitin system for protein degradation and some of its roles in the control of the cell division cycle. 2004 Nobel Lecture, http://nobelprize.org/nobeLprizes/chemistry/laureates/2004/hershko-lecture.html. [DOI] [PubMed]
- 40.Rose I. Ubiquitin at Fox Chase. 2004 Nobel Lecture, http://nobelprize.org/nobeLprizes/chemistry/laureates/2004/rose-lecture.html. [DOI] [PubMed]
- 41.Tanaka K. The proteasome: overview of structure and functions [J] Proc Jpn Acad. 2009;85(1):12–36. doi: 10.2183/pjab.85.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Y, Kitagaki J, Wang H, et al. Targeting the ubiquitin-proteasome system for cancer therapy [J] Cancer Sci. 2009;100(1):24–28. doi: 10.1111/j.1349-7006.2008.01013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Min JH, Yang H, Ivan M, et al. Structure of an HIF-1alpha -pVHL complex: hydroxyproline recognition in signaling [J] Science. 2002;296(5574):1886–1889. doi: 10.1126/science.1073440. [DOI] [PubMed] [Google Scholar]
- 44.McClellan AJ, Scott MD, Frydman J. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways [J] Cell. 2005;121(5):739–748. doi: 10.1016/j.cell.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 45.Stratton MR, Campbell PJ, Futreal PA. The cancer genome [J] Nature. 2009;458(7239):719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whitesell L, Mimnaugh EG, deCosta B, et al. Inhibition of heat shock protein HSP90-pp6Ov-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation [J] Proc Natl Acad Sci USA. 1994;91(18):8324–8328. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pacey S, Banerji U, Judson I, et al. Hsp90 inhibitors in the clinic [J] Hand Exp Pharmacol. 2006;2006(172):331–358. doi: 10.1007/3-540-29717-0_14. [DOI] [PubMed] [Google Scholar]
- 48.Workman P, Burrows F, Neckers L, et al. Drugging the cancer chaperone HSP90: combinatorial therapeutic exploitation of Oncogene addiction and tumor stress [J] Ann NY Acad Sci. 2007;1113:202–216. doi: 10.1196/annals.1391.012. [DOI] [PubMed] [Google Scholar]
- 49.Whitesell L, Bagatell R, Falsey R. The stress response: implications for the clinical development of hsp90 inhibitors [J] Curr Cancer Drug Targets. 2003;3(5):349–358. doi: 10.2174/1568009033481787. [DOI] [PubMed] [Google Scholar]
- 50.Picard D. 2009. http://www.picard.ch/downloads/Hsp90interactors.pdf.
- 51.Jolly C, Morimoto RI. Role of the heat shock response and molecular chaperones in oncogenesis cell death [J] J Natl Cancer Inst. 2000;92(19):1564. doi: 10.1093/jnci/92.19.1564. [DOI] [PubMed] [Google Scholar]
- 52.Munster PN, Basso A, Solit D, et al. Modulation of Hsp90 function by ansamycins sensitizes breast cancer cells to chemotherapy-induced apoptosis in an RB- and schedule-dependent manner [J] Clin Cancer Res. 2001;7(8):2155–2158. [PubMed] [Google Scholar]
- 53.Nylandsted J, Rohde M, Brand K, et al. Selective depletion of heat shock protein 70 (Hsp70) activates a tumor-specific death program that is independent of caspases and bypasses Bcl-2 [J] Proc Nat Acad Sci USA. 2000;97(14):7871–7876. doi: 10.1073/pnas.97.14.7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2 [J] Science. 2004;303(5659):844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 55.Soucy TA, Smith PG, Milhollen MA, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer [J] Nature. 2009;458(7239):732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 56.O'Dwyer ME, Druker BJ. STI571: an inhibitor of the BCR-ABL tyrosine kinase for the treatment of chronic myelogenous leukaemia [J] Lancet Oncol. 2000;1:207–211. doi: 10.1016/s1470-2045(00)00149-2. [DOI] [PubMed] [Google Scholar]
- 57.Kamb A, Wee S, Lengauer C. Why is cancer drug discovery so difficult? [J] Nat Rev Cancer. 2007;6(2):115–120. doi: 10.1038/nrd2155. [DOI] [PubMed] [Google Scholar]
- 58.Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? [J] Nat Rev Drug Discov. 2004;3(8):711–715. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- 59.Xu Z, Horwich AL, Sigler PB. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex [J] Nature. 1997;388(6644):741–750. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]
- 60.Pearl LH, Prodromou C. Structure and Mechanism of the Hsp90 molecular chaperone machinery [J] Ann Rev Bioch. 2006;75:271–294. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- 61.Weinberg RA. New York: Garland Science; 2007. The biology of cancer [M] p. 244. [Google Scholar]