Abstract
Mycosis fungoides (MF), the most common type of cutaneous T-cell lymphoma, has various unspecific clinical and histological characteristics. Its early diagnosis is challenging. The application of T-cell receptor (TCR) gene clonal rearrangement to the diagnosis of MF has been widely studied. In this study, we used polymerase chain reaction (PCR) to investigate the diagnostic significance of detecting TCR-γ and -β gene clonal rearrangement in the early diagnosis of mycosis fungoides. PCR for TCR-γ and TCR-β gene rearrangement was performed on 19 patients with suspected early MF, 6 with typical MF, and 6 with chronic dermatitis. Of the 19 patients with suspected early MF, 13 had TCR-γ gene clonal rearrangement, whereas none had TCR-β gene clonal rearrangement. All patients with typical MF had TCR gene clonal rearrangement, in which 4 showed TCR-γ clonal rearrangement, 1 showed TCR-β gene clonal rearrangements, and 1 showed both. No patients with chronic dermatitis had TCR gene clonal rearrangement. These results indicate that TCR gene clonal rearrangement analysis is a useful tool in diagnosing early MF. TCR-γ gene is recommended to the routine analysis, whereas TCR-β gene has potential in combination toward intractable cases.
Keywords: Lymphoma, T-cell, mycosis fungoides, Immunophenotype, gene rearrangement, T-cell receptor
Mycosis fungoides (MF) is the most common type of primary cutaneous T-cell lymphoma. Most patients have an infiltration of small to medium size mature helper CD4 T cells with cerebriform nuclei; a few patients have an infiltration of cytotoxic CD8 T cells. MF often starts as an unspecific erythema (patch phase) similar to many inflammatory skin diseases. Progression from patch phase to plaque and tumor phase often takes many years. The diagnosis of MF depends on a combination of clinical and pathologic examination. However, early MF (patch phase) pathology is unspecific and may simulate a variety of benign inflammatory skin diseases, such as chronic eczema, neurodermatitis, and lichen planus. Due to a lack of specific clinicopathologic presentation, the early diagnosis of MF is a great challenge.
During the differentiation of stem cells to T cells, non-transcriptional gene fragments are rearranged into a complete gene in a process catalyzed by DNA recombinase. The new gene gains transcriptional activity, expressing antigen-specific receptor as T cells continue differentiating. In contrast, if gene rearrangement fails, cells undergo apoptosis. This process is termed T-cell receptor (TCR) gene rearrangement. Polyclonal rearrangement happens during normal T-cell development. However, all hematopoietic and lymphoid tumor cells share the same TCR gene rearrangement, namely monoclonal rearrangement. Accordingly, by using Southern blot and polymerase chain reaction (PCR), researchers analyze clonality by detecting TCR gene rearrangement of T cells in skin lesions. Previous gene rearrangement studies have relied heavily on Southern blot analysis. However, PCR has several advantages (simple technique, short cycle, no radioactive contamination, compatible with paraffin-embedded tissue samples, and high detection rate) and has been widely used in clinical and experimental detection to replace Southern blot analysis.
As T cells (including α/β and γ/δ T cells) completely keep the rearranged TCR-γ gene with relatively simple structure, designing PCR primers to obtain a high detection rate is easy. Therefore, most investigators choose TCR-γ as a target gene for lymphoma clonal gene rearrangement [1]. As a part of the European BIOMED-2 program, the emergence of specific primers for TCR-β gene has greatly enhanced the detection rate of clonal TCR-β gene rearrangement[2],[3].
In recent years, investigators have gradually applied PCR to analyze TCR gene rearrangement in cutaneous lymphoma[4]–[8], primarily detecting the rearrangement of TCR-β and TCR-γ. Because of the complexity and diversity of TCR gene rearrangement and differences in primer design and numbers, samples and PCR products, results of these studies have discrepancies, showing 50% to 90% of T-cell lymphomas had TCR-γ gene rearrangement. When narrowed down to the MF stages, the detection rates were 52% to 75% for patch phase, 73% to 83% for plaque phase, and 90% to 100% for the tumor[4]–[8].
In this study, we detected TCR-β and TCR-γ gene rearrangement in paraffin-embedded tissue samples from patients with early suspected MF to further explore the application of TCR gene rearrangement to early diagnosis of MF.
Materials and Methods
Patients and samples
The specimens were from 19 patients with suspected early MF, 6 patients with typical MF (2 plaque phase and 4 at tumor phase as positive control), and 6 patients with chronic inflammatory skin disease (3 lichen planus, 2 psoriasis, and 1 neurodermatitis as negative control) examined in the Pathology Division at the Department of Dermatology and the Department of Pathology in the West China Hospital of Sichuan University between 2001 and 2007. Histological diagnosis of MF was confirmed by a senior cutaneous pathologist and a lymphatic hematopoietic pathologist according to Lever's diagnostic criteria[9] and the 2005 WHO-EORTC classification of cutaneous lymphoma[10].
Early suspected patients were 9 men and 10 women, aged 15 to 79 years with a mean age of 53 and median age of 59. All 19 patients had multiple skin lesions in the upper limbs (16 cases), trunk (15 cases), and lower limbs (14 cases). Some patients had skin lesions in the face, neck, and head. The lesions mainly characterized as multiple erythema and papule covered with small scales. A few patients had pigment changes, skin atrophy, and ichthyosis-like skin lesions. Most patients had varying degrees of itching and 1 had a burning sensation/itch.
Histological examination and immunophenotyping
HE staining was conducted on paraffin sections of 19 early MF suspected cases and 6 typical MF cases to observe morphologic characteristics, such as epidermotrophism, Pautrier microabscess, involvement of dermis and subcutaneous tissue, and heteromorphism of infiltrated cells. Ten immunophenotypes (PF1, CD2, CD3e, CD4, CD7, CD8, CD20, CD45RO, CD68, and CD79a) were examined. βF1 and CD4 were examined with Elivision and EnVision immunohistochemical staining; other immunophenotypes were stained with the SP method.
Detection of TCR gene rearrangement
DNA was phenol-chloroform extracted from paraffin tissue with a concentration range of 0.5 to 2.6 µg/µL and an A260/280 of 1.7 to 2.1. Three TCR-γ gene rearrangement primer sets (TVG/TJX[11], V2–5V8–12/JGT1[12], and the BIOMED-2-TCR-γ multiplex PCR amplification system[13]), two TCR-β gene rearrangement primer sets (D1J2[14] and the BIOMED-2-TCR-β multiplex PCR amplification system[13]), and the PCR targets are listed in Tables 1 and 2. PCR was conducted in a 25 µL reaction volume containing 0.2 mmol/L dNTP, 1.5 mmol/L MgCl2, 1 µmol/L primers, 1.25 U Taq enzyme (Takara Bio Inc.), and 0.5 to 1 µg template DNA. Amplifications with primer TVG/TJX were done with touchdown PCR. The amplifying conditions for V2–5/V8–12/JGT1 were as follows: denaturation at 95°C for 3 min, 30 cycles of 94°C for 30 s, 60°C for 45 s, and 72°C for 45 s, followed by an extension at 72°C for 10 min; the amplifying condition for BIOMED-2-TCR-γ/β and D1J2 were as follows: denaturation at 95°C for 3 min, 35 cycles of 95°C for 30 s, 60°C for 40 s, and 72°C for 1 min, followed by an extension at 72°C for 10 min. The PTC-200 thermal cycler from MJ Research Inc. was used for all PCR reactions. β-globin was amplified as an internal control. A DNA sample from Jurkat cells, a T-cell lymphoma line, was used as a positive control. PCR products were electrophoresed on an 8% polyacrylamide gel and analyzed after silver staining. For heteroduplex analysis of gene rearrangement, 10 to 20 µL of PCR product from BIOMED-2 system was denatured at 95°C for 5 min and then annealed at 4°C for 60 min. The resulting PCR products were electrophoresed on a 10% polyacrylamide gel and analyzed after silver staining.
Table 1. Primer sequences of TCR-γ and their product length.
| Name | Sequence 5′→3′ | Product length |
| β-globin | PC03: ACACAACTGTGTTCACTAGC | 110 bp |
| PC04: CAACTTCATCCACGTTCACC | ||
| Tvg: AGGGTTGTGTTGGAATCAGG | 160–190 bp | |
| TJX: CGTCGACAACAAGTGTTGTTCCAC | ||
| V2–5/V8–12/JGT1 | V2: CTTCCTGCAGATGACTCCTACAACTCCAAGGTTG | |
| V3: CTTCCTGCAGATGACGTCTCCACCGCAAGGGATG | 170–230 bp | |
| V4: CTTCCTGCAGATGACTCCTACACCTCCAGCGTTG | ||
| V5: TTCCTGCAGATGACGTCTCCAACTCAAAGGATG | ||
| V8: CTTCCTGCAGATGACTCCTACAACTCCAGGGTTG | ||
| V9: GG(A/T/G/C)ACTGCAGGAAAGGAATCTGGCATTCCG | ||
| V10: CTCTGCAGAATCCGCAGCTCGACGCAGCA | ||
| V11: CACTGCAGGCTCAAGATTGCTCAGGTGGG | ||
| V12: ACTCTGCAGCCTCTTGGGCACTGCTCTAAA | ||
| JGT1: AAGTGTTGTTCCACTGCCAAA | ||
| BIOMED-2-TCR-γ | Vrlf: GGAAGGCCCCACAGCRTCTT | Tube A: |
| Vr9: CGGCACTGTCAGAAAGGAATC | Vγ1f+Vγ10+Jγ1.1/2.1+ Jγ1.3/2.3 | |
| Vr10: AGCATGGGTAAGACAAGCAA | 145–255 bp | |
| Vr11: CTTCCACTTCCACTTTGAAA | ||
| Jr1.1–1.2: TTACCAGGCGAAGTTACTATGAGC | Tube B: | |
| Jr1.3–2.3: GTGTTGTTCCACTGCCAAAGAG | Vγ9+Vγ11+Jγ1.1/2.1+Jγ1.3/2.3 | |
| 80–140 or 160–220 bp |
Table 2. Primer sequences of TCR-β and their product length.
| Name | Sequence 5′→3′ | Product length |
| D1J2 | D1: CAAAGCTGTAACATTGTGGGGAC | 50–110 bp |
| J2: AGCACCGTGAGCCTGGTGCC | ||
| BIOMED-2-TCR-β | Vβ2: AACTATGTTTTGGTATCGTCA | Tube A: |
| Vβ4: CACGATGTTCTGGTACCGTCAGCA | Vβ + Jβ1.1–1.6, 2.2, 2.6, 2.7 | |
| Vβ5/1: CAGTGTGTCCTGGTACCAACAG | 240–285 bp | |
| Vβ6a/11: AACCCTTTATTGGTACCGACA | ||
| Vβ6b/25: ATCCCTTTTTTGGTACCAACAG | ||
| Vβ6c: AACCCTTTATTGGTATCAACAG | Tube B: | |
| Vβ7: CGCTATGTATTGGTACAAGCA | Vβ + Jβ2.1, 2.3, 2.4, 2.5 | |
| Vβ8a: CTCCCGTTTTCTGGTACAGACAGAC | 240–285 bp | |
| Vβ9: CGCTATGTATTGGTATAAACAG | ||
| Vβ10: TTATGTTTACTGGTATCGTAAGAAGC | ||
| Vβ11: CAAAATGTACTGGTATCAACAA | ||
| Vβ12a/3/13a/15:ATACATGTACTGGTATCGACAAGAC | ||
| Vβ13b: GGCCATGTACTGGTATAGACAAG | ||
| Vβ13c/12b/14:GTATATGTCCTGGTATCGACAAGA | ||
| Vβ16: TAACCTTTATTGGTATCGACGTGT | ||
| Vβ17: GGCCATGTACTGGTACCGACA | ||
| Vβ18: TCATGTTTACTGGTATCGGCAG | ||
| Vβ19: TTATGTTTATTGGTATCAACAGAATCA | ||
| Vβ20: CAACCTATACTGGTACCGACA | ||
| Vβ21: TACCCTTTACTGGTACCGGCAG | ||
| Vβ22: ATACTTCTATTGGTACAGACAAATCT | ||
| Vβ23/8b: CACGGTCTACTGGTACCAGCA | ||
| Vβ24: CGTCATGTACTGGTACCAGCA | ||
| Jβ1.1: CTTACCTACAACTGTGAATCTGGTG | ||
| Jβ1.2: CTTACCTACAACGGTTAACCTGGTC | ||
| Jβ1.3: CTTACCTACAACAGTGAGCCAACTT | ||
| Jβ1.4: CATACCCAAGACAGAGAGCTGGGTTC | ||
| Jβ1.5: CTTACCTAGGATGGAGAGTCGAGTC | ||
| Jβ1.6: CATACCTGTCACAGTGAGCCTG | ||
| Jβ2.1: CCTTCTTACCTAGCACGGTGA | ||
| Jβ2.2: CTTACCCAGTACGGTCAGCCT | ||
| Jβ2.3: CCCGCTTACCGAGCACTGTCA | ||
| Jβ2.4: CCAGCTTACCCAGCACTGAGA | ||
| Jβ2.5: CGCGCACACCGAGCAC | ||
| Jβ2.6: CTCGCCCAGCACGGTCAGCCT | ||
| Jβ2.7: CTTACCTGTAACCGTGAGCCTG |
DNA sequencing
The PCR product of a case of TCR gene rearrangement was randomly selected for DNA sequencing. The PCR product was purified with DNA Gel Extraction Kit (Axygen Biosciences Inc), cloned into a T-vector (PMD 20-T Vector, Takara Bio Inc.) and sequenced with an ABI 377 DNA sequencer. The sequencing result was analyzed by the BLASTing gene database in NCBI for gene rearrangement.
Statistical methods
Statistical analysis was done with SPSS13.0 software. The Fisher exact test was applied and the difference was statistically significant when P < 0.05.
Results
Morphology and Immunophenotype
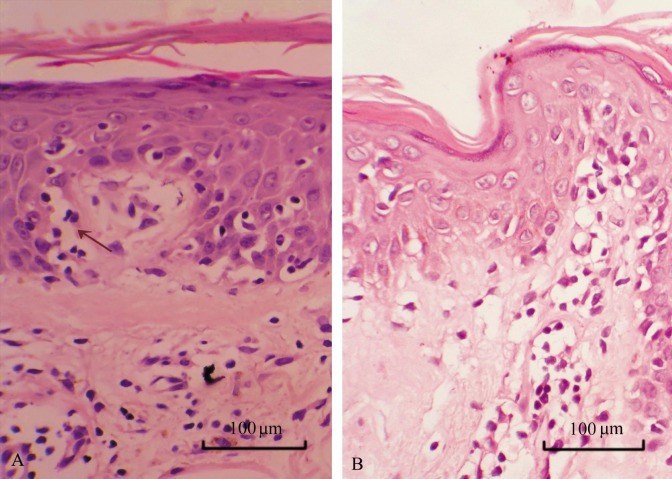
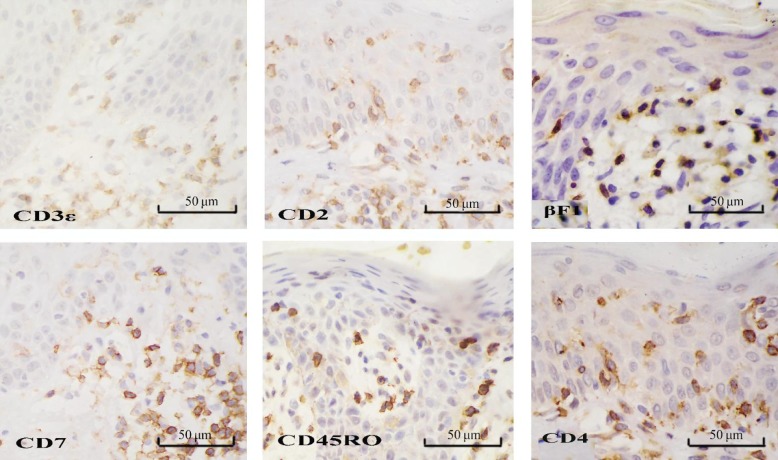
Nineteen patients with suspected early MF predominantly had lesions within the epidermis and shallow middle derm is. Eleven patients had epidermal hyperkeratosis and/or parakeratosis, acanthosis, and epidermal protuberance extension, including 5 had edema in the spinous layer and 3 had focal basal liquefaction degeneration. Fifteen had epidermotrophism of lymphoid cells, including 4 had linear arrangement of lymphoid cells in the basal layer (Figure 1A). Epidermal infiltrated cells had distorted or cerebriform nuclei and perinuclear halo which were deeply stained. All patients had lymphoid cell infiltration in superficial middle dermis: 9 had perivascular infiltration; 2 had striated infiltration in superficial dermal; 8 had scattered infiltration in the dermis. Dermal infiltrating cells were of small to medium size and individually were relatively large. In 2 cases, dermal infiltrating cells were smaller, had less cytoplasm and were stained deeper than those in the epidermis; some cells had distorted and heterogeneous nuclei (Figure 1B). Three cases had papillary dermal fibrosis. All suspected cases expressed two or more T cell markers (CD2, CD3e, CD7, and CD45RO) (Figure 2); 17 (89.5%) expressed α/β T-cell marker βF1 (Figure 2); 14 (73.7%) expressed CD4 (Figure 2); 13 (68.4%) expressed CD8 in some cells (10% to 80%) but not B-cell markers (CD20 and CD79a) or monocyte/macrophage marker CD68.
Figure 1. Histopathologic changes of a skin lesion of suspected early mycosis fungoides (MF). The skin lesion from a patient with suspected early MF was stained with HE and observed under a microscope. A, medium-sized epidermal lymphocytes with prominent cytoplasmic halos are aligned within the basal layer (arrow). Lymphocytes with pleomorphic nuclei infiltrated in the dermis are smaller than those in the epidermis. B, presence of haloed cells with round or hyperconvoluted cerebriform nuclei infiltrate in the epidermis; particularly, single cells are present at the rete ridges with epidermotropism.
Figure 2. Immunophenotype of lymphocytes in a skin lesion of suspected early MF. The skin lesion from a patient with suspected early MF was stained with respective antibodies by the SP, Elivision, or EnVision method. The lymphocytes infiltrated in the epidermis express T-cell-associated antigens CD2, CD3eϵ, CD7, and CD45RO (SP method). Lymphocytes express βF1 (Elivision) and CD4 (EnVision) derived from αβ cells with a helper T cell profile.
PCR amplification and DNA sequence
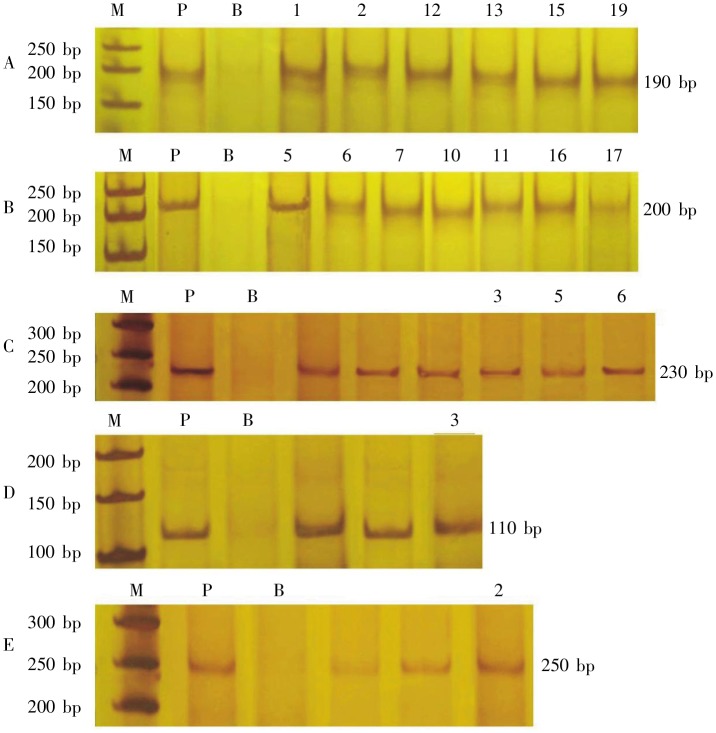
PCR results are shown in Table 3. Of the 19 patients with suspected early MF, 13 (68.4%) had TCR-γ gene clonal rearrangement (Figures 3, 4A and 4B), none had TCR-β gene rearrangement. All patients with typical MF had TCR gene rearrangement: 4 had TCR-γ gene rearrangement (Figure 4C), 2 had TCR-β gene rearrangement (Figures 4D and 4E), and 1 had both. No TCR gene rearrangement was detected in the 6 patients with chronic inflammatory skin diseases. The PCR product from a case of TCR-γ gene rearrangement amplified with primers TVG/Tjx was sequenced and the sequence was analyzed in NCBI BLAST gene database. The results showed a 99.4% homology (170/171) between the PCR product and the chromosome 7 genomic contig, alternate assembly RA_TCAGchr7v2 gene (GI: 079592) in the NCBI Genbank. Comparison of TCR-γ gene rearrangement in suspected cases and typical MF cases and χ2 test showed that the positive rates of the two groups had no significant difference (P > 0.05).
Table 3. Detecting results of TCR gene clonal rearrangement in 19 suspected cases of early mycosis fungoides (MF).
| Case number | TCR-γ | TCR-β |
Total | ||||||
| Tvg/Tjx | V2–5.8–12/JGT1 | BIOMED/γ | Total (TCR-γ) | D1J2 | BIOMED/β | Total (TCR-β) | |||
| 1 | R | R | N | R | N | N | N | R | |
| 2 | R | R | N | R | N | N | N | R | |
| 3 | N | N | N | N | N | N | N | N | |
| 4 | N | N | N | N | N | N | N | N | |
| 5 | N | R | N | R | N | N | N | R | |
| 6 | N | R | N | R | N | N | N | R | |
| 7 | N | R | N | R | N | N | N | R | |
| 8 | N | N | N | N | N | N | N | N | |
| 9 | N | N | N | N | N | N | N | N | |
| 10 | N | R | N | R | N | N | N | R | |
| 11 | N | R | N | R | N | N | N | R | |
| 12 | R | R | N | R | N | N | N | R | |
| 13 | R | R | N | R | N | N | N | R | |
| 14 | N | N | N | N | N | N | N | N | |
| 15 | R | R | N | R | N | N | N | R | |
| 16 | N | R | N | R | N | N | N | R | |
| 17 | N | R | N | R | N | N | N | R | |
| 18 | N | N | N | N | N | N | N | N | |
| 19 | R | R | N | R | N | N | N | R | |
| Summary | 6 | 13 | 0 | 13 | 0 | 0 | 0 | 13 | |
| Positive rate | 31.6% | 68.4% | 0 | 68.4% | 0 | 0 | 0 | 68.4% | |
R, rearrangement; N, no rearrangement.
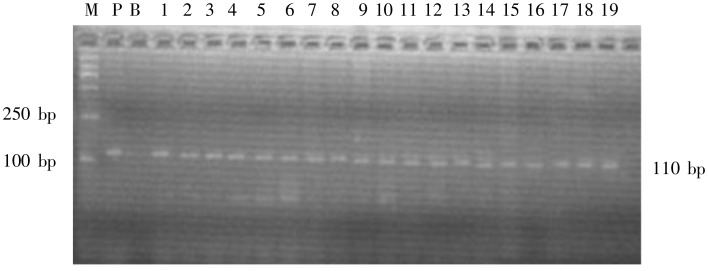
Figure 3. Agarose electrophoresis of β-globin PCR products of 19 patients with suspected early MF. β-globin was amplified using primer PC03/PC04. Une M, molecular weight marker DL2000; lane P, positive control; lane B, blank control; lanes 1-19, PCR products of the 19 patients, β-globin gene, with a single band of 110 bp, was detected in all cases of suspected early MF. DNA extraction with phenol-chloroform was qualified by the results.
Figure 4. Poly-acrylamide gel electrophoresis (PAGE) of PCR products of TCR gene clonal rearrangement in the 19 patients with suspected early MF and the 6 patients with typical MF. Lane M, molecular weight marker DL50; lane P, positive control; lane B, blank control; lanes 1–19, PCR products of the 19 patients. A, TVG/TJX, with a single band of 190 bp, was detected in the suspected early MF cases 1,2, 12, 13, 15 and 19, confirming TCR-γ clonal rearrangement. B, V2–5/V8–12/JGT1, with a single band of 200 bp, was detected in the suspected early MF cases 5, 6, 7, 10, 11, 16 and 17, confirming TCR-γ clonal rearrangement. C, BIOMED-2-TCR-γ, with a single band of 230 bp, was detected in cases 3, 5 and 6 of 6 typical cases of MF, confirming TCR--γ clonal rearrangement. D, D1J2, with a single band of 110 bp, was detected in typical MF case 3, confirming TCR-β clonal rearrangement. E, BIOMED-2-TCR-β, with a single band of 250 bp, was detected in typical MF case 2, confirming TCR-β clonal rearrangement.
Discussion
Early MF has a variety of clinical manifestations and unspecific pathologic characteristics. Typical characteristics include focal perivascular dermal infiltration of small to medium-sized lymphoid cells with cerebriform nuclei, or arrangement along the junction of the epidermis and dermis and edema in the spinous layer. Pautrier microabscesses and epidermotrophism, which have important diagnostic value, are not obvious or are missing in early MF[15]. MF may simulate chronic eczema, neurodermatitis, or other inflammatory skin diseases and requires long-term follow-up and repeated biopsy. As an important diagnostic method for MF, immunohistochemistry can determine the origin of tumor cells but can not determine the clonality of proliferating cells. Moreover, as early MF is often accompanied by reactive lymphocytes, many cases do not show a characteristic immunophenotype. For these cases, immunohistochemistry has a limited diagnostic value. Thus, molecular diagnosis is needed.
Analysis of clonal gene rearrangement plays an important role in the diagnosis of lymphoproliferative disease. Combining this approach with pathologic and immunohistochemical examinations may aid in the diagnosis of difficult cases. Currently, detecting TCR-γ gene rearrangement by PCR has become an important adjunct to the diagnosis of cutaneous T-cell lymphoma[16]. In this study, TCR gene rearrangement was detected in all typical MF cases but not detected in all chronic inflammatory skin disease cases, indicating that detecting TCR rearrangement by PCR is an effective adjunct to MF diagnosis.
In addition, when PCR was used to analyze gene rearrangement in tissues of atypical cutaneous T lymphocyte proliferative diseases, the sensitivity was 83.5% and specificity was 97.7%[8]. The positive rate of TCR-γ gene rearrangement in early MF suspected cases was reported to be 40% to 71%[6],[7], which is useful for early diagnosis. Stevens et al. [17] conducted logistic regression analysis on some clinical and laboratory data and established a simple clinical scoring system to facilitate MF diagnosis. They found that the histological diagnosis of MF and TCR gene rearrangement are closely related (odds ratio = 14.4) and proposed a scoring program: TCR gene rearrangement positive scores 2.5, classical morphology scores 2.0, and classic clinical manifestation scores 1.5; a total score equal to or greater than 3.5 indicates a high probability of MF (> 85%). In our study, the clinical manifestations, histopathologic and immunologic phenotypes suggest all suspected cases were of possible MF. TCR-γ gene rearrangement was detected in 13 patients (68.4%). Thus, we believe that these 13 cases can be diagnosed as early MF (patch phase). Four of the 13 patients were followed up and diagnosed with MF according to disease development and treatment response. The diagnosis of 6 patients was still unclear and further observation and follow-up was needed.
This study showed that the detection rate of clonal TCR-γ gene rearrangement was higher than that of TCR-β. No clonal TCR-β gene rearrangement was detected in the suspected cases. Currently, universal PCR primers can be designed at the V and J regions of α, β, δ and γ chains. However, all T cells (including α/β and γ/δ T cells) keep the rearranged TCR-γ gene. TCR-γ gene structure is relatively simple and has no variable D region, which facilitates primer design and PCR amplification. For this reason, researchers usually choose TCR-γ when analyzing TCR clonality. Therefore, TCR-γ gene is still a good target for the detection of clonal gene rearrangement in early MF.
In addition, no TCR-β gene rearrangement was detected in the suspected cases. But false negative could not be ruled out because the PCR may have been affected by pre-treatment of the archived paraffin-embedded tissue samples. Although amplification of control β-globin (110 bp) was good, some target genes of greater or lesser size than β-globin (such as 250 to 300 bp or 80 to 100 bp fragments) may have been destroyed. In this study, we used primers to amplify 80 to 285 bp long fragments, so damaged DNA fragments could not be amplified in our PCR reaction, possibly causing false negative results.
False results are also inevitable in the interpretation of PCR results. Through optimizing PCR primer design and selecting a higher resolution technique, one can reduce false results. In this study, we used the higher resolution techniques of polyacrylamide gel electrophoresis and silver staining to display rearrangement bands and used heteroduplex analysis to verify gene rearrangement, both of which could reduce false positive results. In addition, the sensitivity of PCR has a certain range and false negatives can arise when the ratio of DNA from monoclonal proliferating lymphocytes to that from reactive lymphocytes is less than 1%. If the amount of template DNA is too small, DNA from a single or several lymphocytes is highly amplified, similar bands like a monoclonal rearrangement may arise, namely pseudoclonality. Therefore, for paraffin-embedded tissue samples, the initial amount of template DNA should be no less than 50 ng, of which DNA from reactive lymphocytes must be more than 5 ng[18]. Under the premise of not affecting PCR, an appropriate increase of the amount of template DNA can reduce pseudoclonality. In this study, template DNA was within a 200 to 500 ng range and the proportion of tumor cells was higher than 1%, thereby reducing false positive results.
Based on this study, we believe that, when MF is suspected and diagnosis cannot be made on clinical manifestations, regular histopathologic examination, and immunohistochemistry, TCR gene rearrangement is a valuable diagnostic adjunct. The detection of clonal TCR-γ gene rearrangement is recommended and the analysis of TCR-β rearrangement can be performedalong with TCR-γ rearrangement for a few difficult cases.
However, it must be noted that gene rearrangement of antigen receptor is not involved in the malignant transformation of lymphocytes. These two processes occur independently. In general, gene rearrangement occurs before tumor transformation. On one side, clonally proliferating tumor cells harbor the same antigen gene rearrangement (monoclonality), whereas each T or B cell from benign lesions carries a unique type of gene rearrangement (polyclonality). Therefore, in theory, the genetic rearrangement analysis of antigen receptor gene is helpful to distinguish proliferating lymphoma and reactive lesions. On the other side, due to the independence of each other, the occurrence of clonal gene rearrangement does not indicate a malignant lesions, because some benign or reactive inflammatory lesions may also have clonal rearrangement. For example, clonal gene rearrangements could be detected in benign monoclonal γ-globulin disease, acute smallpox-like moss-like rash, other skin inflammatory diseases, and immune deficiency disease with severe Epstein-Barr virus infection [19]. Conversely, the lack of monoclonal gene rearrangement is not equivalent to benign lesions. In many conditions, such as the low coverage of primers, the absence of a tested gene rearrangement in the tumor genome, DNA degradation in the sample, and the number of tumor cells below the detection sensitivity threshold, clonal gene rearrangement may not be detected. Therefore, the early diagnosis of MF must be based on pathologic changes, with immunohistochemistry and gene rearrangement analysis as important adjunct diagnostic tools. For some suspected cases with no clonal gene rearrangement, long-term follow-up and multiple pathologic examinations should be done to confirm the diagnosis[20].
Acknowledgments
This work was supported by grant from Sichuan Provincial Sci-Tech Research Foundation (No. 07SG004009).
References
- 1.Cozzio A, French LE. T-cell clonality assays: how do they compare? [J] J Invest Dermatol. 2008;128(4):771–773. doi: 10.1038/jid.2008.49. [DOI] [PubMed] [Google Scholar]
- 2.Sandberg Y, van Gastel-Mol EJ, Verhaaf B, et al. BIOMED-2 multiplex multiplex immunoglobulin/T-cell receptor polymerase chain reaction protocols can reliably replace Southern blot analysis in routine clonality diagnostics [J] J Mol Diagn. 2005;7(4):495–503. doi: 10.1016/S1525-1578(10)60580-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan QG, Huang GS, Wang Z, et al. Application of BIOMED-2 standardized clonality analysis system for immunoglobulin/T cell receptor gene rearrangement [J] Chin J Diag Pathol. 2008;4(15):307–309. [in Chinese] [Google Scholar]
- 4.Wood GS. T-cell receptor and immunoglobulin gene rearrangements in diagnosing skin disease [J] Arch Dermatol. 2001;137(11):1503–1506. doi: 10.1001/archderm.137.11.1503. [DOI] [PubMed] [Google Scholar]
- 5.Hodges E, Krishna MT, Pickard C, et al. Diagnostic role of tests for T cell receptor (TCR) genes [J] J Clin Pathol. 2003;56(1):1–11. doi: 10.1136/jcp.56.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kandolf L, Cikota B, Stojadinović O, et al. TCR gamma gene rearrangement analysis in skin samples and peripheral blood of mycosis fungoides patients [J] Acta Dermatoven Alp Panonica Adriat. 2007;16(4):149–155. [PubMed] [Google Scholar]
- 7.Hsiao PF, Hsiao CH, Lin YC, et al. Histopathologic-molecular correlation in early mycosis fungoides using T-cell receptor gamma gene rearrangement by polymerase chain reaction with laser capture microdissection [J] J Formos Med Assoc. 2007;106(4):265–272. doi: 10.1016/S0929-6646(09)60251-5. [DOI] [PubMed] [Google Scholar]
- 8.Ponti R, Fierro MT, Quaglino P, et al. TCR gamma-chain gene rearrangement by PCR-based GeneScan: diagnostic accuracy improvement and clonal heterogeneity analysis in multiple cutaneous T-cell lymphoma samples [J] J Invest Dermatol. 2008;128(4):1030–1038. doi: 10.1038/sj.jid.5701109. [DOI] [PubMed] [Google Scholar]
- 9.Lever WF, Schaumburg-Lever G. 6th edition. Philadelphia: JB Lippincott; 1983. Histopathology of the Skin [M] pp. 697–700. [Google Scholar]
- 10.Willemze R, Jaffe ES, Burg G, et al. WHO-EORTC classification for cutaneous lymphomas [J] Blood. 2005;105(10):3768–3785. doi: 10.1182/blood-2004-09-3502. [DOI] [PubMed] [Google Scholar]
- 11.Benhattar J, Delacretaz F, Martin P, et al. Improved polymerase chain reaction detection of clonal T-cell lymphoid neoplasms [J] Diagn Mol Pathol. 1995;4(2):108–112. doi: 10.1097/00019606-199506000-00006. [DOI] [PubMed] [Google Scholar]
- 12.Guo HQ, Zhang MZ, Wang GY, et al. The diagnostic value for lymphoproliferative diseases with gene rearrangements [J] Chin J Misdiag. 2004;4(9):1377–1379. [in Chinese] [Google Scholar]
- 13.Van Dongen JJ, Langerak AW, Brüggemann M, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombination in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936 [J] Leukemia. 2003;17(12):2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 14.Zhong BN, Zhang XH, Li M, et al. Study of the pathology, Immunophenotype, etiology and genetic marker of NK/T-cell lymphoma [J] Chin J Hematol. 2003;24(10):505–509. [in Chinese] [PubMed] [Google Scholar]
- 15.LeBoit PE, Burg G, Weedon D, et al. Pathology and Genetics of Skin Tumours [M] Lyon: IARC Press; 2006. World Health Organization Classification of Tumours; p. 170. [Google Scholar]
- 16.Hwang ST, Janik JE, Jaffe ES, et al. Mycosis fungoides and Sézary syndrome [J] Lancet. 2008;371(9616):945–957. doi: 10.1016/S0140-6736(08)60420-1. [DOI] [PubMed] [Google Scholar]
- 17.Stevens SR, Ke MS, Birol A, et al. A simple clinical scoring system to improve the sensitivity and standardization of the diagnosis of mycosis fungoides type cutaneous T-cell lymphoma: logistic regression of clinical and laboratory data [J] Br J Dermatol. 2003;149(3):513–522. doi: 10.1046/j.1365-2133.2003.05458.x. [DOI] [PubMed] [Google Scholar]
- 18.Medeiros LJ, Carr J. Overview of the role of molecular methods in the diagnosis of malignant [J] Arch Pathol Lab Med. 1999;123(12):1189–1207. doi: 10.5858/1999-123-1189-OOTROM. [DOI] [PubMed] [Google Scholar]
- 19.Wood GS, Crooks CF, Uluer AZ. Lymphomatoid papulosis and associated cutaneous lymphoproliferative disorders exhibit a common clonal origin [J] J Invest Dermatol. 1995;105(1):51–55. doi: 10.1111/1523-1747.ep12312548. [DOI] [PubMed] [Google Scholar]
- 20.Eros N, Károlyi Z, Marschalkó M, et al. Clinical, histopathological, immunophenotypic and molecular analysis of 60 patients with cutaneous T-cell infiltrates with follow up of indeterminate cases to identify T-cell lymphoma [J] Pathol Oncol Res. 2008;14(1):63–67. doi: 10.1007/s12253-008-9014-3. [DOI] [PubMed] [Google Scholar]