Abstract
MicroRNAs (miRNAs) are small non-coding RNAs that regulate gene expression at the transcriptional or posttranscriptional level. Many miRNAs are found to play a significant role in cancer development either as tumor suppressor genes or as oncogenes. Examination of tumor-specific miRNA expression profiles in diverse cancers has revealed widespread deregulation of these molecules, whose loss and overexpression respectively have diagnostic and prognostic significance. Genetic variations, mostly single-nucleotide polymorphisms (SNPs) within miRNA sequences or their target sites, have been found to be associated with many kinds of cancers. In this review, we summarize the current knowledge of miRNAs including their biogenesis and role in cancer development, and finally, how SNPs among miRNAs affect miRNA biogenesis and contribute to cancer.
Keywords: Breast cancer, microRNA, target sequence, SNP, cancer risk
In 1993, Lee et al.[1] discovered a gene, lin-4, that affects development in Caenorhabditis elegans and found that its product is a small non–protein-coding RNA. After this seminal finding, the cloning and characterization of small, 20 to 22 nucleotide–long members of the non–protein-coding RNA family have led to the identification of thousands of microRNAs (miRNAs). The term “miRNA” was first introduced in 2001 in Science[2]. miRNAs are now emerging as one of the most interesting small regulatory, non-coding RNAs in molecular biology[3],[4]. They perform functions as key trans-acting factors by switching off or fine tuning expression of target genes[5],[6]. They are believed to function largely, although not exclusively, through base-pairing to the complementary sequences in the 3′ -untranslated region (3′UTR) of target genes to suppress gene expression at the mRNA or protein levels[7],[8]. There are also some exceptions. For example, microRNA-346 (miR-346) targets the 5′-untranslated region (5′UTR) of receptor-interacting protein 140 mRNA and up-regulates its protein expression[9]. miRNA regulation can be highly pleiotropic, with one miRNA able to target several hundred different transcripts, and one mRNA able to be cooperatively targeted by several miRNAs. In fact, bioinformatic prediction indicates that 30% of animal genes may be directly regulated by miRNAs[10]. Recent miRNA transfection experiments show strong evidence that miRNAs influence not only their targets but also non-target genes through a two-layer regulatory network in which transcription factors function as important mediators of miRNA-initiated regulatory effects[11]. miRNAs critically regulate almost all biological processes involving gene expression[12],[13]. Therefore, alterations in miRNA expression have been implicated in various human diseases, including cancer[14]–[16]. To date, the miRNA registry (miRBase) contains more than one thousand human miRNAs. Thus far, most registered miRNAs are widely expressed and highly conserved. Deep sequencing of small RNA libraries and cell type–specific analyses are currently valuable approaches to uncovering miRNAs with less abundant expression[17]. Thus, the catalog of miRNAs is expected to grow substantially in the future, and the exploration of miRNAs in cancer development will shed more light on the causes of cancer.
MiRNA Biogenesis
miRNA genes are scattered among each of the chromosomes in humans, except for the Y chromosome. miRNAs can be encoded in independent transcription units, in polycistronic clusters, or within the introns of protein-coding genes[18]. They are primarily derived from capped, polyadenylated RNA polymerase II transcripts, termed “primary” miRNAs (pri-miRNA)[19]. A typical metazoan pri-miRNA consists of a double-stranded RNA (dsRNA) stem of about 33 bp, with a terminal loop on one end and single-stranded RNA (ssRNA) flanking segments on the other end. The ssRNA-dsRNA junction (SD junction) is very important for pri-miRNA processing. Mature miRNAs are generated by a two-step processing mechanism, the “cropping” step and the “dicing” step[20].
In the cropping step, pri-miRNAs are first processed to stem-loop structured precursor miRNAs (pre-miRNA), typically 55 to 80 nt in length, in the nucleus. Aside from a small group of pre-miRNAs that are generated through mRNA splicing/debranching machinery termed the “miRtron” pathway[21], most pre-miRNAs are processed from pri-miRNAs by the nuclear ribonuclease (RNase) III Drosha[22], which partners with the RNA-binding protein DiGeorge syndrome critical region gene 8 (DGCR8)[23]. Drosha is a large nuclear protein of 160 kDa in humans with two tandem RNase III domains and a dsRNA-binding domain. Drosha forms a large complex, weighing approximately 650 kDa in humans, known as the “microprocessor, ” in which it interacts with its co-factor, DGCR8 [24]. Neither recombinant DGCR8 nor Drosha alone is active in pri-miRNA processing, whereas the combination of these two proteins possesses pri-miRNA processing activity, indicating that both proteins are critical for processing.
Pri-miRNA processing consists of two sequential steps: substrate recognition and catalytic reaction. DGCR8 recognizes the substrate by anchoring tightly at the SD junction and interacting with the approximately 33-bp stem. The sequence preceding the 5′ terminus or trailing the 3′ terminus of the pre-miRNAs forms an approximately 11-bp imperfect stem that is recognized by DGCR8 as part of the required structure for Drosha cutting. Drosha may not be in direct contact with RNA at this stage. After the substrate recognition, Drosha may interact transiently with the stem, and catalytic reaction happens on the stem at about 11 bp from the SD junction, releasing the pre-miRNA[25],[26].
A small terminal loop of pri-miRNA may impose structural constraints on the stem and affect processing, so pri-miRNA with a small loop is less active in processing. On the contrary, a large terminal loop may act as a flexible ssRNA and may contribute to tight binding by the microprocessor[27],[28]. Because some large terminal loops can be seen as unstructured ssRNA segments, pri-miRNAs may be considered to have “ssRNA-dsRNA-ssRNA” structure. Because of the symmetry properties of this structure, the microprocessor appears to recognize the terminal loop as ssRNA and binds to the stem-loop in an opposite orientation. In this case, cleavage can occur at an alternative site, about 11 bp from the terminal loop. The processing at this alternative site would be “abortive” because the cleavage product does not contain miRNA sequences in full.
Pre-miRNAs are exported to the cytoplasm by exportin-5/RAN-GTP for the dicing step[29]. In this step, the pre-miRNAs are further processed in the cytoplasm by another RNase III enzyme, Dicer, that cuts the pre-miRNA at 22 bp from the terminus set by Drosha to generate a double-stranded miRNA duplex[30]. The production of miRNA/miRNA* duplexes is an essential step in miRNA biogenesis and precisely defines the termini of the mature miRNAs for preferential loading of the guide strand[31]. Recruitment of co-factors such as transactivation response element RNA-binding protein (TRBP) and protein kinase R–activating protein (PACT) increases the efficiency of pre-miRNA processing by Dicer[32]. Human Dicer is composed of an amino-terminal RNA helicase domain, followed by a PAZ (Piwi/Argonaute/Zwille) domain, two RNase III domains, and a carboxy-terminal RNA-binding domain[33]. Despite the similarities in their basic modes of action, RNase III proteins are different in their substrate specificities. Human Dicer tends to act on any dsRNA with a simple preference toward the terminus of the molecule. The PAZ domain of Dicer interacts with the 3′ terminus and determines the processing site in a ruler-like fashion that measures approximately 22 nucleotide segments from the terminus[34],[35].
For most miRNAs, only one strand (the guide strand) of the double-stranded miRNA duplex is loaded into miRNA-induced silencing complex (miRISC)[36]. The choice of the guide strand is dependent in part on the thermodynamic properties of the duplex, with the strand possessing the least thermodynamically stable 5′ terminus usually being chosen as the guide strand and eventually becoming the mature miRNA, whereas the other strand, labeled miRNA*, is usually degraded [37]. However, in cases where both the 5′ and 3′ termini of each miRNA have similar stabilities, such as in miR-142, miR-125, miR-126, and miR-342, both miRNAs have an equal chance of being selected as the guide strand[38]. Also, the fates of the miRNA guide and miRNA* strands have been shown to be tissue-dependent, with both strands being functionally active under specific conditions[39].
The mature miRNAs are used to guide miRISC to the complementary sequences in the 3′UTR of targeted transcripts[40]. The core component of every RISC is a member of the Argonaute (Ago) protein family, containing a central PAZ domain and a carboxy-terminal PIWI domain. Structural studies have shown that the PIWI domain binds to small RNAs at their 5′ terminus, whereas the PAZ domain binds to the 3′ terminus of ssRNAs[41]. Argonaute 2 (AG02), sometimes in cooperation with elF6[42], is the target-cleaving endonuclease of the RISC[43],[44]. The result is site-specific mRNA cleavage when the pairing is nearly complete (mostly in plants) or translational inhibition when imperfect base pairing occurs (mostly in animals)[45]. Several lines of evidence suggests that miRNAs may perform their function in the RNA processing bodies (P-bodies) by sequestering target transcripts to P-bodies for storage, decapping, deadenylation, and degradation[46].
For effective translational suppression, Watson-Crick base pairing between nucleotides 2 to 7 or 2 to 8 (the “seed region”) of the miRNA 5′ terminus and the 3′UTR of the target mRNA is ubiquitously required[47]. The critical role played by the seed region in most of the miRNA/mRNA interactions implies that changes in the seed region, or shift of the processing sites during biogenesis of the miRNA/miRNA* duplex could result in a novel miRNA with alternative target spectra. Therefore, both the 5′ terminus of the mature miRNA that is generated from the 5′ arm of the pre-miRNA (5p) by Drosha and the 5′ terminus of the mature miRNA that is produced by Dicer from the 3′ arm of the pre-miRNA (3p) should be highly conserved. Sequences outside of the seed in the mature miRNA sequence can also impact, although less dramatically, the strength of inhibition and the spectra of targeted transcripts.
Generally, the cropping step is the most important step for miRNA biogenesis. The precise position and orientation of Drosha cleavage are critically important for the generation of mature miRNA because the second cleavage step occurs at a defined distance from the free end generated by Drosha. Therefore, Drosha cleavage determines the identity of both the 5′ and 3′ termini of the mature miRNA. The slightest error in Drosha cleavage could not only alter the seed sequence, but also invert the relative stability of the two strands, resulting in the incorporation of the improper miRNA strand into the RISC complex. Since Drosha is present in a large protein complex and the association of Drosha with co-factors besides DGCR8 present within this complex promotes the fidelity and activity of Drosha cleavage, it will be important to identify additional factors involved in the biogenesis pathway.
Most miRNAs are under the control of developmental and/or tissue-specific signaling[48],[49]. miRNA expression may be regulated at multiple steps of RNA biogenesis[50]. Although RNA pol II-mediated transcription provides a major point of control for miRNA biogenesis, most miRNAs seem to be controlled at the posttranscriptional level. For example, the maturation of pre-miR-38 may be temporally regulated in C. elegans since miR-38 is expressed only in the embryo, whereas the pre-miR-38 is ubiquitously detected. It is also possible that the nuclear export of pre-miR-38 may be controlled by a specific developmental signal or that Dicer processing may be repressed until a certain stage; some of these deregulations have been found to contribute to cancer development[51],[52]. Indeed, a number of miRNAs are down-regulated in cancer though their pri-miRNAs are expressed at a relatively high level, indicating that the cropping step may be controlled dynamically during cell differentiation and tumorigenesis.
MiRNA and Cancer
Over the past several years, many miRNAs have been implicated in various human cancers[53],[54]. Both losses and gains of miRNA function have been shown to contribute to cancer development. Over half of all known human miRNA genes are located at fragile sites and genomic regions involved in cancers[55]. Similarly, mouse miRNA genes are also frequently located near mouse cancer susceptibility loci. High-resolution array-based comparative genomic hybridization has revealed that the number of miRNA copies is quite abnormal in human cancers[56]. miRNA profiling has revealed that most of miRNAs are significantly down-regulated in human cancers.
The first evidence of miRNA involvement in cancer was reported in 2002. During their attempts to clone a tumor suppressor gene at chromosome 13q14, a region that is frequently lost in chronic lymphocytic leukemia (CLL), Calinet al.[57] found that all of the protein-coding genes present in the region of interest were not specifically altered in CLL, suggesting that they were not involved in the disease. However, they found two miRNA genes, miR-15a and miR-16-1, in the deletion. The translocation breakpoint cut the precursors of these two miRNA genes which reside in the same polycistronic RNA. The loss of miR-15a and miR-16-1 was observed in nearly 70% of CLLs, indicating that miR-15a and miR-16-1 can function as tumor suppressor genes in CLL, as their loss is associated with the development of CLL. Calin's group mapped the chromosomal location of all known miRNA genes and discovered that many are located in regions that are frequently involved in chromosomal alterations, such as deletions or amplifications, in many types of human cancers[55]. Neither the activation of oncogenes nor the loss of tumor suppressor genes has been observed in extensive studies of these cancers, suggesting that the role of miRNAs in cancer could extend far beyond CLL. In fact, the expression of miRNAs was deregulated in many human cancers, including leukemia, lymphoma, and glioblastoma, as well as colon, lung, breast, prostate, thyroid, cervical, and ovarian cancers[58]–[64].
The development of high-throughput miRNA quantification technologies, such as miRNA microarray, bead-based flow Cytometry, RNA-primed array-based Klenow enzyme (RAKE) assay, miRNA serial analysis of gene expression (miRAGE), and real-time RT-PCR based TaqMan miRNA assay, has facilitated the study of the global miRNA profile in the whole cancer genome. Important questions on miRNA deregulation in cancer are being addressed with the advent of these new technologies. Deregulated miRNA expression in human cancer may prove to be a powerful tool for diagnosis, classification, and prediction of clinical behavior and prognosis[65]; specific findings on miRNA expression and cancer may be categorized into the following.
First, there is a differential global expression of miRNAs between tumors and their corresponding normal tissues in most, if not all, types of cancer. Interestingly, miRNA expression is more likely to be repressed in tumors than in normal tissues. For example, colon cancer is associated with alteration in miRNA expression. Michael et al.[53] identified different miRNAs in colonic adenocarcinoma and normal mucosa and found that the expression of two mature miRNAs, miR-143 and miR-145, were consistently reduced in colorectal neoplasia. Using miRNA array analyses for age-matched normal cervix and cervical cancer tissues, in combination with Northern blot verification, Wanget al.[58] identified significantly deregulated miRNAs in cervical cancer tissues, with miR-126, miR-143, and miR-145 down-regulated and miR-15b, miR-16, miR-146a, and miR-155 up-regulated.
Second, miRNA expression signatures are informative enough to identify and classify human cancers. Rosenfeldet al.[66] suggested that miRNAs can accurately identify cancer tissue origin. In their study using miRNA microarray data from 253 samples, they were able to classify 22 different types of cancer using a 48 miRNA classifier with over 90% accuracy. This classification is of great clinical importance because miRNAs may be used to identify the tissue in which cancers of unknown primary origin arose. miRNA expression profiles can also be used to distinguish two subtypes of diffuse large B cell lymphoma (DLBCL), germinal center B cell–like (GCB) and activated B cell–like (ABC) DLBCL, as miR-21, miR-155, and miR-221 were more highly expressed in ABC-type than in GCB-type DLBCU[67].
Third, miRNA expression signatures can predict biological and clinical behavior within the same cancer type. In a study regarding breast cancer, lorio et al.[62] found that the miRNA expression patterns were significantly different between normal and neoplastic breast tissues, with miR-125b, miR-145, miR-21, and miR-155 significantly reduced in breast cancer tissues. They also observed that the expression of miRNAs was correlated with specific breast cancer clinicopathologic features, such as tumor stage, proliferation index, estrogen and progesterone receptor expression, and vascular invasion[62].
Last but not least, miRNA expression is associated with the prognosis and progression of human cancer. Takamizawa et al.[68] observed that the expression levels of let-7 were down-regulated in lung cancer, and reduced let-7 expression was significantly associated with shortened postoperative survival, independent of disease stage. Another study revealed that high expression of miR-21 was associated with poor survival and poor therapeutical outcome in colon adenocarcinomas[69]. Thus, from all the above examples we can see the important function of miRNA in cancer development, progression, and prognosis.
The underlying mechanisms of miRNA deregulation in human cancer remain a challenging topic. Recent laboratory findings have provided evidence of multiple mechanisms of miRNA deregulation in human cancers[70]. Most primary miRNAs have been well-documented to be transcripted from RNA Pol II promoter and regulated by transcription factors. Several examples showing miRNA deregulation in cancer by transcriptional deregulation have been reported [71]. Recent studies suggest that miRNA deregulation may also be attributed to the following: epigenetic alterations, such as DNA methylation and histone deacetylation[72]; single-nucleotide polymorphism (SNP) and mutation, which may affect mature miRNAs[73]; DNA copy-number abnormalities[75]. and dysfunction or deregulation of key proteins in the miRNA biogenesis pathway, which may enhance tumorigenesis[75]. In addition to these mechanisms, “dynamic” factors that modulate miRNA activity without altering mature miRNA concentrations may also contribute miRNA deregulation. Examples of dynamic factors include 3′UTRs, which have alternative forms with different complements of miRNA target sites that can be used by mRNAs. Expression of 3′UTR binding proteins that affect accessibility of miRNAs[76] even to those target sites that are present in a given 3′UTR; “decoy” RNAs that sequester miRNAs away from their target or, most dramatically, a switch from target repression to target activation by miRISC itself[77],[78]. Thus, taken together, these diverse mechanisms might each contribute alone or, more likely, together to miRNA deregulation in human cancer[79].
SNPs among MiRNA
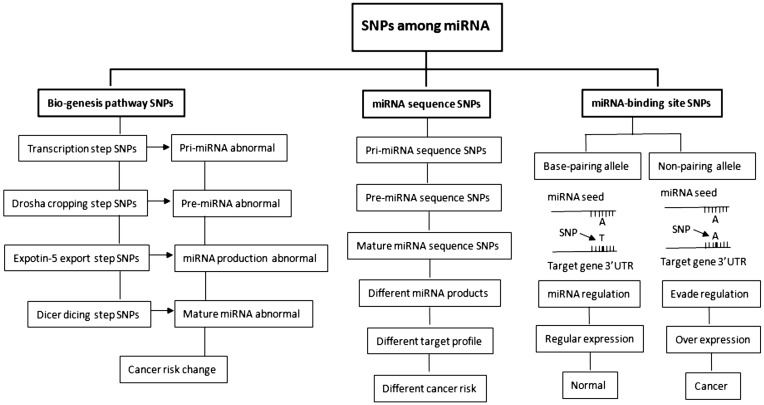
Genetic variations range from large chromosomal anomalies to single nucleotide changes. SNPs are the most frequently identified variants in DNA sequences. miRNA-related SNPs could potentially affect the maturation of miRNAs, the silencing machinery, the structure or the expression level of mature miRNA, and the base pairing at the target site; they may also have functional role in miRNA-mediated gene regulation, thereby affecting cancer risk[80],[81] (Figure 1). That this is indeed the case as has been demonstrated by a germline DNA variant (C/T) in the primary sequence of the miRNA cluster encoding miR-16-1 and miR-15a (7 bp in the 3′ direction after the precursor). This DNA variant, which results in reduced mature expression of miR-16-1 and miR-15 in vitro and in vivo and is associated with deletion of the chromosome region containing this miRNA cluster, was found in 11 of 75 patients with CLL but not observed in 160 subjects without cancer[82],[83]. Such genetic variations are not rare, and some are proven functional. For example, a mutation in the seed region of human miR-96 is responsible for nonsyndromic progressive hearing loss[84]; DICER1 mutations are associated with familial pleuropulmonary blastoma[85]; and one SNP in the mature sequence of miR-30c-2 is likely to affect stem integrity[86]. Furthermore, a G/U polymorphism (rs12975333), located at the eighth nucleotide within the mature sequence of miR-125a, has been functionally characterized to block the processing of pri-miRNA into pre-miRNA and alter the translation suppression on the lin-28 target mRNA[87],[88]. All these are good examples for the importance of miRNA related SNP.
Figure 1. Effects of single-nucleotide polymorphisms (SNPs) among microRNA (miRNA) sequences, miRNA processing genes, and miRNA-binding sites that affect cancer risk.
Polymorphisms in pre-miRNA may influence miRNA maturation and thereby modulate miRNA expression. Several groups have tried to identify SNPs within or flanking the pre-miRNA sequences using experimental or bioinformatic approaches. In one study, 173 human pre-miRNAs in 96 Japanese individuals were sequenced, and 10 SNPs in 10 pre-miRNA hairpins were identified[86]. In another study, a bioinformatics search identified 12 known SNPs located within 227 human pre-miRNA sequences[87]. In a similar study, researchers screened the dbSNP database for common SNPs in 474 human pre-miRNAs. Sixty-five SNPs in 49 pre-miRNAs were found, and 3, hsa-miR-125a, hsa-miR-627, and hsa-miR-662, were located within seed regions rs12975333, rs2620381, and rs9745376, respectively[89], indicating that SNPs within miRNA seed region are very rare.
To assist in the identification of DNA sequence polymorphisms (DSPs) that affect miRNA-mediated regulation, Hiard et al. [90] have searched the public domain databases for SNPs and other polymorphisms in the 3 sequence compartments involved in miRNA control: targets, miRNA precursors, and silencing machinery. The outcome of this search is browsable via the Patrocles website (http://www.patrocles.org/). DSPs are sorted depending on whether they affect the seed region, the mature miRNA outside the seed, or other parts of the pre-miRNA. In humans, for instance, 184 DSPs affecting 136 out of 676 pre-miRNAs were identified. Twelve of these mapped to the miRNA seed and 26 to the mature miRNA outside the seed. However, the 12 human miRNAs with a DSP in their seed sequence were either members of a seed-sharing miRNA family or more recently discovered miRNAs that are likely to be expressed at lower levels. The effect of the DSPs on pre-miRNA structure was evaluated using RNAfold and the predicted secondary structures are viewable in Patrocles[90].
The roles that SNPs among miRNA play in miRNA processing and cancer development have been studied in recent years, with the miR-146a SNP (rs2910164) within the pre-miR-146a sequence one of the most thoroughly studied examples[81]. This SNP reduced both the amount of pre-miR-146a and mature miR-146a, and affected the Drosha/DGCR8 cropping step, and was associated with papillary thyroid carcinoma (PTC), familial/sporadic breast cancer, ovarian cancer, prostate cancer, and hepatocellular carcinoma[91]–[95]. In an association study of 608 PTC patients and 901 controls, Jazdzewski et al.[96] found marked differences in genotype distribution of rs2910164 (P < 0.001). The GC heterozygous state was associated with an increased risk of acquiring PTC (OR = 1.62, P < 0.001) compared with both homozygous states. Further functional analysis showed that GC heterozygotes differed from both the GG and CC homozygotes by producing three mature miRNAs, one from the leading strand (miR-146a) and the other two from the passenger strand (miR-146a*G and miR-146a*C), each with a distinct set of target genes. TaqMan analysis of paired tumor and normal samples revealed a 1.5- to 2.6-fold overexpression of polymorphic miR-146a* in 7 of 8 tumors compared with the unaffected part of the same gland. Microarray data showed that widely different transcriptomes occurred in the tumors and in unaffected parts of the thyroid from patients with GC and GG homozygotes. The affected genes were mainly involved in the regulation of apoptosis, leading to exaggerated DNA-damage response in heterozygotes and potentially explaining the predisposition to cancer[96].
We summarized in this review the SNPs within miRNA sequences, miRNA pathway genes, and miRNA binding sites that have been found to be significantly associated with cancer susceptibility[91],[97]–[112] (Table 1). Systematic analysis of the association between miRNA-related SNPs and cancer risk has been reported predominantly in two studies. Horikawa et al. [107] evaluated SNPs in miRNA-related genes and the risk of bladder cancer and renal cell carcinoma. In their studies, in which they assessed the effects of a total of 41 SNPs in genes of the miRNA biogenesis pathway (24 SNPs), pre-miRNAs (7 SNPs), and pri-miRNAs (10 SNPs) on cancer predisposition, they found that a non-synonymous SNP in GEMIN3 and a common haplotype of GEMIN4 are associated with bladder cancer risk, whereas two SNPs in the GEMIN4 gene are associated with altered renal cell carcinoma risk. Both GEMIN3 and GEMIN4 proteins are core components of a large macromolecular complex that plays an essential role in pre-miRNA splicing and ribonucleoprotein assembly. In addition to the significant SNP/haplotype identified for the GEMIN genes, borderline significant associations with bladder cancer risk were also identified for SNPs in several other genes, including TRBP, miR-423, miR-492, miR-26a-1, and miR-124-1. In particular, the variant allele of rs784567, which is located in the 5′UTR of the TRBP gene, is associated with a 20% risk reduction (P = 0.07) [106],[107]. Through a thorough searching of available bioinformatic databases, Tian et al.[99] found 4 SNPs within the pre-miRNA region (miR-146a, rs2910164; miR-149, rs2292832; miR-196a2, rs11614913; miR-499, rs3746444), with a minor allele frequency more than 0.05, from about 400 known miRNAs. They tested the association of these 4 SNPs with the risk of lung cancer[99] and breast cancer[95], as well as the prognosis of lung cancer[113]. They found that the rs11614913 SNP in miR-196a2 is associated with increased risk and shortened survival time of non–small cell lung cancer as it alters the expression of mature miR-196a and the binding activity of target mRNA. miR-196a2 rs11614913 and miR-499 rs3746444 are associated with significantly increased risk of breast cancer. Although both of these studies involved a systematic search for SNPs among miRNAs known at that time, the findings do not include some common SNPs within pre-miRNA or the miRNA biogenosis pathway, mainly because of the expanding sum of human miRNAs identified after these studies were conducted.
Table 1. Single-nucleotide polymorphisms (SNPs) among microRNA (miRNA) sequences, miRNA processing genes, and miRNA-binding sites that are significantly associated with cancer risk.
| SNP category | Gene | SNP ID | Variants | Cancer | Association |
| miRNA sequence | |||||
| miR-146a | rs2910164 | G/C | Thyroid | 0.50(0.28, 0.89) | |
| Esophageal | 2.39(1.36,4.20) | ||||
| miR-219-1 | rs213210 | T/C | Esophageal | 1.66(1.00,2.74) | |
| miR-124-1 | rs531564 | C/G | Esophageal | 8.79(1.06, 73.17) | |
| miR-26a-1 | rs7372209 | C/T | Esophageal | 1.35(1.04, 1.76) | |
| miR-631 | rs5745925 | CT/- | Esophageal | 1.57(1.03,2.41) | |
| miR-423 | rs6505162 | C/A | Esophageal | 0.57(0.44, 0.73) | |
| miR-196a2 | rs11614913 | C/T | Esophageal | 1.73(1.16, 2.56) | |
| Lung | 1.25(1.01, 1.54) | ||||
| Glioma | 0.74(0.56, 0.98) | ||||
| miR-27a | rs895819 | G/A | Breast | 0.88(0.78, 0.99) | |
| Gastric | 1.48(1.06, 2.05) | ||||
| miR-218 | rs11134527 | A/G | Cervical | 0.72(0.52, 0.99) | |
| miR-34b/c | rs4938723 | T/C | Hepatocellular | 1.37(1.06, 1.78) | |
| Bio-genesis pathway | miR-499 | rs3746444 | A/G | Head and neck | 0.83(0.69, 0.99) |
| GEMIN3 | rs197414 | A/C | Bladder | 2.40(1.04, 5.56) | |
| Esophageal | 1.45(1.02,2.06) | ||||
| GEMIN4 | rs7813 | T/C | Renal cell | 0.68(0.47, 0.98) | |
| Ovarian | 0.71(0.57, 0.88) | ||||
| GEMIN4 | rs2740348 | G/C | Renal cell | 0.67(0.47, 0.96) | |
| GEMIN4 | rs2740349 | A/G | Ovarian | 0.70(0.51, 0.96) | |
| GEMIN4 | rs2740351 | A/G | Ovarian | 0.71(0.57, 0.87) | |
| XP05 | rs11077 | A/C | Esophageal | 1.84(1.16,2.93) | |
| XP05 | rs2257082 | G/A | Ovarian | 0.73(0.54, 0.99) | |
| RAN | rs14035 | C/T | Esophageal | 1.93(1.09,3.40) | |
| AG01 | rs636832 | A/G | Lung | 0.67(0.53, 0.84) | |
| DDX20 | rs197383 | A/G | Ovarian | 0.69(0.48, 0.99) | |
| Binding sites | RNASEN | rs4867329 | G/A | Ovarian | 0.71(0.51,0.99) |
| CD86 | rs17281995 | G/C | Colorectal | 2.93(1.29, 6.67) | |
| LAMB3 | rs2566 | C/T | Cervical | 1.57(1.25, 1.96) | |
| SET8 | rs16917496 | C/T | Breast | 1.66(1.06,2.61) | |
| ATG4A | rs5973822 | A/G | Ovarian | 0.42(0.24, 0.75) | |
| UGT2A3 | rs17147016 | T/A | Ovarian | 1.47(1.08,2.01) | |
| COL18A1 | rs7499 | G/A | Ovarian | 1.47(1.07, 2.02) | |
| CAV1 | rs9920 | A/G | Ovarian | 1.50(1.04,2.17) | |
| IL1R1 | rs3917328 | G/A | Ovarian | 1.65(1.03, 2.64) | |
| KRAS | rs10771184 | T/A | Ovarian | 1.26(1.01, 1.57) | |
| E2F2 | rs2075993 | A/G | Ovarian | 1.24(1.00, 1.54) | |
| IQGAP1 | rs1042538 | A/T | Breast | 0.42(0.61,0.99) |
The data for association are presented as odds ratio (95% confidence interval).
We previously reviewed SNPs within miRNA binding sites and cancer risk[114], followed by case control studies in this field[108]. Recently, Liang et al.[109] reported their findings that miRNA binding site SNPs may impact ovarian cancer predisposition and clinical outcome both individually and jointly with genetic variants in miRNA biosynthesis pathways. To test the hypothesis that disruption of miRNA target binding by SNPs is a widespread mechanism relevant to cancer susceptibility, Nicoloso et al.[115] analyzed SNPs known to be associated with breast cancer risk, in silico and in vitro, for their ability to modify miRNA binding sites and miRNA gene regulation. They proposed that transcribed target SNPs alter miRNA gene regulation and, consequently, protein expression, thereby contributing to the likelihood of cancer susceptibility by a novel mechanism of subtle gene regulation[115]. All of these studies have contributed to our knowledge about miRNA binding site SNPs and cancer susceptibility. However, binding site SNPs represent a large group of genetic variation that can be observed from online searching software such as Patrocles and PolymiRTS. Studies to date have been limited in the investigation of this kind of SNPs. Further systematic studies are warranted for a general view of the role this group of SNPs plays in the development of cancer.
miRNAs can also be post-transcriptionally modified, such as by RNA editing via adenosine deaminases that act on RNA (ADAR)[116] A to I–editing of pre-miR-151 blocks its processing by Dicer/TRBP [117]. ADAR-edited pri-miR-142 is more easily degraded by Tudor-SN [118]. Edited miR-376a-5p within the middle of the seed region alters the set of targets regulated by this miRNA[119]. A survey of RNA editing of miRNAs from 10 human tissues implies RNA editing of miRNA happens quite often and is a mechanism to increase the diversity of miRNAs and their targets[120]. All the above examples show that mutant or post-transcriptional editing of miRNAs can result in alterations of processing and function and are often involved in cancer development. Hence, SNPs that occur in sequences downstream or upstream of the pre-miRNA, sequences in the terminal loop of pre-miRNA, sequences in the miRNA and miRNA* duplexes, and sequences of genes in the miRNA biogenesis pathway are potentially valuable predictors of cancer risk and prognosis[121],[122].
In conclusion, miRNAs play a substantial role in cancer development. miRNA related SNPs are important biomarkers for their function, and the amount of this kind of SNPs is very large, most of them have not been evaluated in association studies. Therefore, we currently have little knowledge of the overall contribution of this kind of SNPs to the development of cancer. In future researches, studies specifically designed to examine the relationship between miRNA related SNPs in the whole genome and cancer risks are warranted.
Acknowledgments
This study was jointly supported by grants from the National Natural Science Foundation of China to K.C. (No. 30872172) and the Tianjin Science and Technology Committee Foundation (No. 08ZCGH202000, 09ZCZDSF04400).
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14 [J] Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Lau NC, Lim LP, Weinstein EG, et al. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans [J] Science. 2001;294(5543):858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 3.Hannon GJ, Rivas FV, Murchison EP, et al. The expanding universe of noncoding RNAs [J] Cold Spring Harb Symp Quant Biol. 2006;71:551–564. doi: 10.1101/sqb.2006.71.064. [DOI] [PubMed] [Google Scholar]
- 4.Ruvkun G, Wightman B, Ha I. The 20 years it took to recognize the importance of tiny RNAs [J] Cell. 2004;116(2 Suppl):S93–96; 2 p following S6. doi: 10.1016/s0092-8674(04)00034-0. [DOI] [PubMed] [Google Scholar]
- 5.Selbach M, Schwanhausser B, Thierfelder N, et al. Widespread changes in protein synthesis induced by microRNAs [J] Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 6.Meister G. miRNAs get an early start on translational silencing [J] Cell. 2007;131(1):25–28. doi: 10.1016/j.cell.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 7.Baek D, Villen J, Shin C, et al. The impact of microRNAs on protein output [J] Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing [J] Cell. 2008;132(1):9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 9.Tsai NP, Lin YL, Wei LN. MicroRNA mir-346 targets the 5′-untranslated region of receptor-interacting protein 140(RIP140) mRNA and up-regulates its protein expression [J] Biochem J. 2009;424(3):411–418. doi: 10.1042/BJ20090915. [DOI] [PubMed] [Google Scholar]
- 10.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets [J] Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 11.Tu K, Yu H, Hua YJ, et al. Combinatorial network of primary and secondary microRNA-driven regulatory mechanisms [J] Nucleic Acids Res. 2009;37(18):5969–5980. doi: 10.1093/nar/gkp638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bicker S, Schratt G. microRNAs: tiny regulators of synapse function in development and disease [J] J Cell Mol Med. 2008;12(5A):1466–1476. doi: 10.1111/j.1582-4934.2008.00400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapsimali M, Kloosterman WP, de Bruijn E, et al. MicroRNAs show a wide diversity of expression profiles in the developing and mature central nervous system [J] Genome Biol. 2007;8(8):R173. doi: 10.1186/gb-2007-8-8-r173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L, Huang J, Yang N, et al. microRNAs exhibit high frequency genomic alterations in human cancer [J] Proc Natl Acad Sci USA. 2006;103(24):9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen PY, Meister G. microRNA-guided posttranscriptional gene regulation [J] Biol Chem. 2005;386(12):1205–1218. doi: 10.1515/BC.2005.139. [DOI] [PubMed] [Google Scholar]
- 16.Drakaki A, Iliopoulos D. MicroRNA Gene Networks in Oncogenesis [J] Curr Genomics. 2009;10(1):35–41. doi: 10.2174/138920209787581299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang J, Hao P, Chen H, et al. Genome-wide identification of schistosoma japonicum microRNAs using a Deep-sequencing approach [J] PLoS One. 2009;4(12):e8206. doi: 10.1371/journal.pone.0008206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodriguez A, Griffiths-Jones S, Ashurst JL, et al. Identification of mammalian microRNA host genes and transcription units [J] Genome Res. 2004;14(10A):1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II [J] EMBO J. 2004;23(20):4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing [J] Nat Rev Mol Cell Biol. 2005;6(5):376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 21.Shomron N, Levy C. MicroRNA-biogenesis and pre-mRNA splicing crosstalk [J] J Biomed Biotechnol. 2009;2009:594678. doi: 10.1155/2009/594678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing [J] Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 23.Tomari Y, Zamore PD. MicroRNA biogenesis: drosha can't cut it without a partner [J] Curr Biol. 2005;15(2):R61–64. doi: 10.1016/j.cub.2004.12.057. [DOI] [PubMed] [Google Scholar]
- 24.Gregory RI, Yan KP, Amuthan G, et al. The Microprocessor complex mediates the genesis of microRNAs [J] Nature. 2004;432(7014):235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 25.Han J, Lee Y, Yeom KH, et al. The Drosha-DGCR8 complex in primary microRNA processing [J] Genes Dev. 2004;18(24):3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han J, Lee Y, Yeom KH, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex [J] Cell. 2006;125(5):887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 27.Lund E, Dahlberg JE. Substrate selectivity of exportin 5 and Dicer in the biogenesis of microRNAs [J] Cold Spring Harb Symp Quant Biol. 2006;71:59–66. doi: 10.1101/sqb.2006.71.050. [DOI] [PubMed] [Google Scholar]
- 28.Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha [J] Embo J. 2005;24(1):138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs [J] Rna. 2004;10(2):185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flores-Jasso CF, Arenas-Huertero C, Reyes JL, et al. First step in pre-miRNAs processing by human Dicer [J] Acta Pharmacol Sin. 2009;30(8):1177–1185. doi: 10.1038/aps.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vermeulen A, Robertson B, Dalby AB, et al. Double – stranded regions are essential design components of potent inhibitors of RISC function [J] Rna. 2007;13(5):723–730. doi: 10.1261/rna.448107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chendrimada TP, Gregory RI, Kumaraswamy E, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing [J] Nature. 2005;436(7051):740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel DJ, Ma JB, Yuan YR, et al. Structural biology of RNA silencing and its functional implications [J] Cold Spring Harb Symp Quant Biol. 2006;71:81–93. doi: 10.1101/sqb.2006.71.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cullen BR. Transcription and processing of human microRNA precursors [J] Mol Cell. 2004;16(6):861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 35.Murphy D, Dancis B, Brown JR. The evolution of core proteins involved in microRNA biogenesis [J] BMC Evol Biol. 2008;8:92. doi: 10.1186/1471-2148-8-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parker JS, Roe SM, Barford D. Molecular mechanism of target RNA transcript recognition by Argonaute-guide complexes [J] Cold Spring Harb Symp Quant Biol. 2006;71:45–50. doi: 10.1101/sqb.2006.71.029. [DOI] [PubMed] [Google Scholar]
- 37.Leung AK, Sharp PA. Function and localization of microRNAs in mammalian cells [J] Cold Spring Harb Symp Quant Biol. 2006;71:29–38. doi: 10.1101/sqb.2006.71.049. [DOI] [PubMed] [Google Scholar]
- 38.Wu H, Ye C, Ramirez D, et al. Alternative processing of primary microRNA transcripts by Drosha generates 5′ end variation of mature microRNA [J] PLoS One. 2009;4(10):e7566. doi: 10.1371/journal.pone.0007566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ro S, Park C, Young D, et al. Tissue-dependent paired expression of miRNAs [J] Nucleic Acids Res. 2007;35(17):5944–5953. doi: 10.1093/nar/gkm641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gregory RI, Chendrimada TP, Cooch N, et al. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing [J] Cell. 2005;123(4):631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 41.Miyoshi K, Tsukumo H, Nagami T, et al. Slicer function of Drosophila Argonautes and its involvement in RISC formation [J] Genes Dev. 2005;19(23):2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chendrimada TP, Finn KJ, Ji X, et al. MicroRNA silencing through RISC recruitment of elF6 [J] Nature. 2007;447(7146):823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- 43.Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA [J] Development. 2005;132(21):4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 44.Joshua-Tor L. The Argonautes [J] Cold Spring Harb Symp Quant Biol. 2006;71:67–72. doi: 10.1101/sqb.2006.71.048. [DOI] [PubMed] [Google Scholar]
- 45.Behm-Ansmant I, Rehwinkel J, Izaurralde E. MicroRNAs silence gene expression by repressing protein expression and/or by promoting mRNA decay [J] Cold Spring Harb Symp Quant Biol. 2006;71:523–530. doi: 10.1101/sqb.2006.71.013. [DOI] [PubMed] [Google Scholar]
- 46.Maroney PA, Yu Y, Nilsen TW. MicroRNAs, mRNAs, and translation [J] Cold Spring Harb Symp Quant Biol. 2006;71:531–535. doi: 10.1101/sqb.2006.71.043. [DOI] [PubMed] [Google Scholar]
- 47.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? [J] Nat Rev Genet. 2008;9(2):102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 48.Davis CJ, Bohnet SG, Meyerson JM, et al. Sleep loss changes microRNA levels in the brain: a possible mechanism for state-dependent translational regulation [J] Neurosci Lett. 2007;422(1):68–73. doi: 10.1016/j.neulet.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olsen L, Klausen M, Helboe L, et al. MicroRNAs show mutually exclusive expression patterns in the brain of adult male rats [J] PLoS One. 2009;4(10):e7225. doi: 10.1371/journal.pone.0007225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee J, Li Z, Brower-Sinning R, et al. Regulatory circuit of human microRNA biogenesis [J] PLoS Comput Biol. 2007;3(4):e67. doi: 10.1371/journal.pcbi.0030067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gregory RI, Shiekhattar R. MicroRNA biogenesis and cancer [J] Cancer Res. 2005;65(9):3509–3512. doi: 10.1158/0008-5472.CAN-05-0298. [DOI] [PubMed] [Google Scholar]
- 52.Kusenda B, Mraz M, Mayer J, et al. MicroRNA biogenesis, functionality and cancer relevance [J] Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2006;150(2):205–215. doi: 10.5507/bp.2006.029. [DOI] [PubMed] [Google Scholar]
- 53.Michael MZ, SM OC, van Hoist Pellekaan NG, et al. Reduced accumulation of specific microRNAs in colorectal neoplasia [J] Mol Cancer Res. 2003;1(12):882–891. [PubMed] [Google Scholar]
- 54.Cheng C, Fu X, Alves P, et al. mRNA expression profiles show differential regulatory effects of microRNAs between estrogen receptor-positive and estrogen receptor-negative breast cancer [J] Genome Biol. 2009;10(9):R90. doi: 10.1186/gb-2009-10-9-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers [J] Proc Natl Acad Sci USA. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuo KT, Guan B, Feng Y, et al. Analysis of DNA copy number alterations in ovarian serous tumors identifies new molecular genetic changes in low-grade and high-grade carcinomas [J] Cancer Res. 2009;69(9):4036–4042. doi: 10.1158/0008-5472.CAN-08-3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia [J] Proc Natl Acad Sci USA. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang X, Tang S, Le SY, et al. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth [J] PLoS One. 2008;3(7):e2557. doi: 10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Merritt WM, Lin YG, Han LY, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer [J] N Engl J Med. 2008;359(25):2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hanlon K, Rudin CE, Harries LW. Investigating the targets of MIR-15a and MIR-16-1 in patients with chronic lymphocytic leukemia(CLL) [J] PLoS One. 2009;4(9):e7169. doi: 10.1371/journal.pone.0007169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eder M, Scherr M. MicroRNA and lung cancer [J] N Engl J Med. 2005;352(23):2446–2448. doi: 10.1056/NEJMcibr051201. [DOI] [PubMed] [Google Scholar]
- 62.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer [J] Cancer Res. 2005;65(16):7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 63.He H, Jazdzewski K, Li W, et al. The role of microRNA genes in papillary thyroid carcinoma [J] Proc Natl Acad Sci USA. 2005;102(52):19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Croce CM. Causes and consequences of microRNA dysregulation in cancer [J] Nat Rev Genet. 2009;10(10):704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Farazi TA, Spitzer JI, Morozov P, et al. miRNAs in human cancer [J] J Pathol. 223(2):102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rosenfeld N, Aharonov R, Meiri E, et al. MicroRNAs accurately identify cancer tissue origin [J] Nat Biotechnol. 2008;26(4):462–469. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 67.Lawrie CH, Soneji S, Marafioti T, et al. MicroRNA expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma [J] Int J Cancer. 2007;121(5):1156–1161. doi: 10.1002/ijc.22800. [DOI] [PubMed] [Google Scholar]
- 68.Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival [J] Cancer Res. 2004;64(11):3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 69.Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma [J] Jama. 2008;299(4):425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang L, Coukos G. MicroRNAs: a new insight into cancer genome [J] Cell Cycle. 2006;5(19):2216–2219. doi: 10.4161/cc.5.19.3319. [DOI] [PubMed] [Google Scholar]
- 71.Saumet A, Vetter G, Bouttier M, et al. Transcriptional repression of microRNA genes by PML-RARA increases expression of key cancer proteins in acute promyelocytic leukemia [J] Blood. 2009;113(2):412–421. doi: 10.1182/blood-2008-05-158139. [DOI] [PubMed] [Google Scholar]
- 72.Valeri N, Vannini I, Fanini F, et al. Epigenetics, miRNAs, and human cancer: a new chapter in human gene regulation [J] Mamm Genome. 2009;20(9–10):573–580. doi: 10.1007/s00335-009-9206-5. [DOI] [PubMed] [Google Scholar]
- 73.Diederichs S, Haber DA. Sequence variations of microRNAs in human cancer: alterations in predicted secondary structure do not affect processing [J] Cancer Res. 2006;66(12):6097–6104. doi: 10.1158/0008-5472.CAN-06-0537. [DOI] [PubMed] [Google Scholar]
- 74.Li C, Kim SW, Rai D, et al. Copy number abnormalities, MYC activity, and the genetic fingerprint of normal B cells mechanistically define the microRNA profile of diffuse large B-cell lymphoma [J] Blood. 2009;113(26):6681–6690. doi: 10.1182/blood-2009-01-202028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kumar MS, Lu J, Mercer KL, et al. Impaired microRNA processing enhances cellular transformation and tumorigenesis [J] Nat Genet. 2007;39(5):673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 76.Nielsen AF, Gloggnitzer J, Martinez J. MicroRNAs cross the line: the battle for mRNA stability enters the coding sequence [J] Mol Cell. 2009;35(2):139–140. doi: 10.1016/j.molcel.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 77.Bhattacharyya SN, Habermacher R, Martine U, et al. Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells [J] Cold Spring Harb Symp Quant Biol. 2006;71:513–521. doi: 10.1101/sqb.2006.71.038. [DOI] [PubMed] [Google Scholar]
- 78.Ding XC, Weiler J, Grosshans H. Regulating the regulators: mechanisms controlling the maturation of microRNAs [J] Trends Biotechnol. 2009;27(1):27–36. doi: 10.1016/j.tibtech.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 79.Deng S, Calin GA, Croce CM, et al. Mechanisms of microRNA deregulation in human cancer [J] Cell Cycle. 2008;7(17):2643–2646. doi: 10.4161/cc.7.17.6597. [DOI] [PubMed] [Google Scholar]
- 80.Mishra PJ, Mishra PJ, Banerjee D, et al. MiRSNPs or MiR-polymorphisms, new players in microRNA mediated regulation of the cell: Introducing microRNA pharmacogenomics [J] Cell Cycle. 2008;7(7):853–858. doi: 10.4161/cc.7.7.5666. [DOI] [PubMed] [Google Scholar]
- 81.Jazdzewski K, de la Chapelle A. Genomic sequence matters: a SNP in microRNA-146a can turn anti-apoptotic [J] Cell Cycle. 2009;8(11):1642–1643. [PMC free article] [PubMed] [Google Scholar]
- 82.Calin GA, Ferracin M, Cimmino A, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia [J] N Engl J Med. 2005;353(17):1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 83.Aqeilan RI, Calin GA, Croce CM. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives [J] Cell Death Differ. 2009;17(2):215–20. doi: 10.1038/cdd.2009.69. [DOI] [PubMed] [Google Scholar]
- 84.Mencia A, Modamio-Hoybjor S, Redshaw N, et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss [J] Nat Genet. 2009;41(5):609–613. doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- 85.Hill DA, Ivanovich J, Priest JR, et al. DICER1 mutations in familial pleuropulmonary blastoma [J] Science. 2009;325(5943):965. doi: 10.1126/science.1174334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Iwai N, Naraba H. Polymorphisms in human pre-miRNAs [J] Biochem Biophys Res Commun. 2005;331(4):1439–1444. doi: 10.1016/j.bbrc.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 87.Duan R, Pak C, Jin P. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA [J] Hum Mol Genet. 2007;16(9):1124–1131. doi: 10.1093/hmg/ddm062. [DOI] [PubMed] [Google Scholar]
- 88.Wu M, Jolicoeur N, Li Z, et al. Genetic variations of microRNAs in human cancer and their effects on the expression of miRNAs [J] Carcinogenesis. 2008;29(9):1710–1716. doi: 10.1093/carcin/bgn073. [DOI] [PubMed] [Google Scholar]
- 89.Borel C, Antonarakis SE. Functional genetic variation of human miRNAs and phenotypic consequences [J] Mamm Genome. 2008;19(7–8):503–509. doi: 10.1007/s00335-008-9137-6. [DOI] [PubMed] [Google Scholar]
- 90.Hiard S, Charlier C, Coppieters W, et al. Patrocles: a database of polymorphic miRNA-mediated gene regulation in vertebrates [J] Nucleic Acids Res. 2010;38(Database issue):D640–651. doi: 10.1093/nar/gkp926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jazdzewski K, Murray EL, Franssila K, et al. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma [J] Proc Natl Acad Sci USA. 2008;105(20):7269–7274. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xu B, Feng NH, Li PC, et al. A functional polymorphism in Pre-miR-146a gene is associated with prostate cancer risk and mature miR-146a expression in vivo [J] Prostate. 2009;70(5):467–472. doi: 10.1002/pros.21080. [DOI] [PubMed] [Google Scholar]
- 93.Shen J, Ambrosone CB, DiCioccio RA, et al. A functional polymorphism in the miR-146a gene and age of familial breast/ovarian cancer diagnosis [J] Carcinogenesis. 2008;29(10):1963–1966. doi: 10.1093/carcin/bgn172. [DOI] [PubMed] [Google Scholar]
- 94.Xu T, Zhu Y, Wei QK, et al. A functional polymorphism in the miR-146a gene is associated with the risk for hepatocellular carcinoma [J] Carcinogenesis. 2008;29(11):2126–2131. doi: 10.1093/carcin/bgn195. [DOI] [PubMed] [Google Scholar]
- 95.Hu Z, Liang J, Wang Z, et al. Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women [J] Hum Mutat. 2009;30(1):79–84. doi: 10.1002/humu.20837. [DOI] [PubMed] [Google Scholar]
- 96.Jazdzewski K, Liyanarachchi S, Swierniak M, et al. Polymorphic mature microRNAs from passenger strand of pre-miR-146a contribute to thyroid cancer [J] Proc Natl Acad Sci USA. 2009;106(5):1502–1505. doi: 10.1073/pnas.0812591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Guo H, Wang K, Xiong G, et al. A functional varient in microRNA-146a is associated with risk of esophageal squamous cell carcinoma in Chinese Han [J] Fam Cancer. 2010;9(4):599–603. doi: 10.1007/s10689-010-9370-5. [DOI] [PubMed] [Google Scholar]
- 98.Ye Y, Wang KK, Gu J, et al. Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk [J] Cancer Prev Res(Phila) 2008;1(6):460–469. doi: 10.1158/1940-6207.CAPR-08-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tian T, Shu Y, Chen J, et al. A functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in Chinese [J] Cancer Epidemiol Biomarkers Prev. 2009;18(4):1183–1187. doi: 10.1158/1055-9965.EPI-08-0814. [DOI] [PubMed] [Google Scholar]
- 100.Dou T, Wu Q, Chen X, et al. A polymorphism of microRNA196a genome region was associated with decreased risk of glioma in Chinese population [J] J Cancer Res Clin Oncol. 2010;136(12):1853–1859. doi: 10.1007/s00432-010-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang R, Schlehe B, Hemminki K, et al. A genetic variant in the pre-miR-27a Oncogene is associated with a reduced familial breast cancer risk [J] Breast Cancer Res Treat. 2010;121(3):693–702. doi: 10.1007/s10549-009-0633-5. [DOI] [PubMed] [Google Scholar]
- 102.Sun Q, Gu H, Zeng Y, et al. Hsa-mir-27a genetic variant contributes to gastric cancer susceptibility through affecting miR-27a and target gene expression [J] Cancer Sci. 2010;101(10):2241–2247. doi: 10.1111/j.1349-7006.2010.01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhou X, Chen X, Hu L, et al. Polymorphisms involved in the miR-218-LAMB3 pathway and susceptibility of cervical cancer, a case-control study in Chinese women [J] Gynecol Oncol. 2010;117(2):287–290. doi: 10.1016/j.ygyno.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 104.Xu Y, Liu L, Liu J, et al. A potentially functional polymorphism in the promoter region of miR-34b/c is associated with an increased risk for primary hepatocellular carcinoma [J] Int J Cancer. 2011;128(2):412–417. doi: 10.1002/ijc.25342. [DOI] [PubMed] [Google Scholar]
- 105.Liu Z, Li G, Wei S, et al. Genetic variants in selected pre-microRNA genes and the risk of squamous cell carcinoma of the head and neck [J] Cancer. 2010;116(20):4753–4760. doi: 10.1002/cncr.25323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang H, Dinney CP, Ye Y, et al. Evaluation of genetic variants in microRNA-related genes and risk of bladder cancer [J] Cancer Res. 2008;68(7):2530–2537. doi: 10.1158/0008-5472.CAN-07-5991. [DOI] [PubMed] [Google Scholar]
- 107.Horikawa Y, Wood CG, Yang H, et al. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma [J] Clin Cancer Res. 2008;14(23):7956–7962. doi: 10.1158/1078-0432.CCR-08-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Song F, Zheng H, Liu B, et al. An miR-502-binding site single-nucleotide polymorphism in the 3′-untranslated region of the SET8 gene is associated with early age of breast cancer onset [J] Clin Cancer Res. 2009;15(19):6292–6300. doi: 10.1158/1078-0432.CCR-09-0826. [DOI] [PubMed] [Google Scholar]
- 109.Liang D, Meyer L, Chang DW, et al. Genetic variants in MicroRNA biosynthesis pathways and binding sites modify ovarian cancer risk, survival, and treatment response [J] Cancer Res. 2010;70(23):9765–9776. doi: 10.1158/0008-5472.CAN-10-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kim JS, Choi YY, Jin G, et al. Association of a common AGO1 variant with lung cancer risk: a two-stage case-control study [J] Mol Carcinog. 2010;49(10):913–921. doi: 10.1002/mc.20672. [DOI] [PubMed] [Google Scholar]
- 111.Landi D, Gemignani F, Naccarati A, et al. Polymorphisms within micro-RNA-binding sites and risk of sporadic colorectal cancer [J] Carcinogenesis. 2008;29(3):579–584. doi: 10.1093/carcin/bgm304. [DOI] [PubMed] [Google Scholar]
- 112.Zheng H, Song F, Zhang L, et al. Genetic variants at the miR-124 binding site on the cytoskeleton-organizing IQGAP1 gene confer differential predisposition to breast cancer [J] Int J Oncol. 2011;38(4):1153–1161. doi: 10.3892/ijo.2011.940. [DOI] [PubMed] [Google Scholar]
- 113.Hu Z, Chen J, Tian T, et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival [J] J Clin Invest. 2008;118(7):2600–2608. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen K, Song F, Calin GA, et al. Polymorphisms in microRNA targets: a gold mine for molecular epidemiology [J] Carcinogenesis. 2008;29(7):1306–1311. doi: 10.1093/carcin/bgn116. [DOI] [PubMed] [Google Scholar]
- 115.Nicoloso MS, Sun H, Spizzo R, et al. Single-nucleotide polymorphisms inside microRNA target sites influence tumor susceptibility [J] Cancer Res. 70(7):2789–2798. doi: 10.1158/0008-5472.CAN-09-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bass BL. How does RNA editing affect dsRNA-mediated gene silencing? [J] Cold Spring Harb Symp Quant Biol. 2006;71:285–292. doi: 10.1101/sqb.2006.71.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kawahara Y, Zinshteyn B, Chendrimada TP, et al. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex [J] EMBO Rep. 2007;8(8):763–769. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yang W, Chendrimada TP, Wang Q, et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases [J] Nat Struct Mol Biol. 2006;13(1):13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kawahara Y, Zinshteyn B, Sethupathy P, et al. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs [J] Science. 2007;315(5815):1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Blow MJ, Grocock RJ, van Dongen S, et al. RNA editing of human microRNAs [J] Genome Biol. 2006;7(4):R27. doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sun G, Yan J, Noltner K, et al. SNPs in human miRNA genes affect biogenesis and function [J] RNA. 2009;15(9):1640–1651. doi: 10.1261/rna.1560209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ryan BM, Robles Al, Harris CC. Genetic variation in microRNA networks: the implications for cancer research [J] Nature reviews. 10(6):389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]