Abstract
Platinum-based chemotherapy remains the main treatment of advanced lung cancer. However, platinum resistance has become a major treatment obstacle. Novel therapies, particularly tyrosine kinase inhibitors of the epidermal growth factor receptor (EGFR-TKI) and agents that target vascular endothelial growth factor (VEGF), have improved the treatment. Both chemotherapy and targeted therapy have their molecular mechanisms. This study aimed to determine the mutation, amplification, or expression status and interrelationships of the epidermal growth factor receptor (EGFR), K-Ras proto-oncogene, excision repair cross-complementation group 1 (ERCC1), and VEGF genes as well as their correlations to prognosis of large cell lung carcinoma (LCLC) after EGFR-targeted therapy, chemotherapy, and anti-VEGF therapy. EGFR and K-Ras mutations in 60 specimens of LCLC were detected by direct DNA sequencing. EGFR, ERCC1, and VEGF protein expression was detected by immunohistochemistry (IHC). EGFR gene copy number was detected by fluorescence in situ hybridization (FISH). One (1.7%) patient had an EGFR L858M point mutation in exon 21, 3 (5.0%) had K-Ras mutations, and 10 (19.6%) had EGFR amplification (FISH positive). Positive rates of EGFR, ERCC1, and VEGF proteins were 38.3%, 56.7%, and 70.0%, respectively. EGFR amplification was positively correlated to EGFR protein expression (r = 0.390, P = 0.005). The positive rate of VEGF protein was significantly higher in patients with lymph node metastasis than in those without (84.6% vs. 58.8%, P = 0.046). No significant correlations were observed among the EGFR, K-Ras, ERCC1, and VEGF genes. EGFR gene amplification and the low rate of EGFR mutation suggest that patients with LCLC are likely to obtain little benefit from anti-EGFR therapies.
Keywords: Large cell lung carcinoma, EGFR, KRAS, ERCC1, VEGF
Lung cancer is the number one cause of deaths among all cancers[1]. Platinum-based chemotherapy, one of main treatments for this disease, often fails due to drug resistance of tumors. The excision repair cross-complementing 1 (ERCC1) protein plays a key role in the nucleotide excision repair system, which is primarily responsible for cancer cell resistance to platinum-based drugs. The expression level of the ERCC1 protein is an important marker in predicting platinum drug resistance in tumors; ERCC1-positive patients are refractory to platinum-based chemotherapy[2]–[4].
When the efficacy of chem otherapy reaches to a plateaus[5], lung cancer-targeting drugs, such as tyrosine kinase inhibitors of the epidermal growth factor receptor (EGFR-TKIs)[6]–[9] and anti-vascular endothelial growth factor (VEGF) drugs[10],[11], have achieved good outcomes. Increasing evidence suggests that potential biological markers, including EGFR gene mutation, amplification, or protein expression, are associated with EGFR-TKI drug efficacy[6]–[9], whereas K-Ras mutations are closely related with drug resistance [12],[13]. VEGF is the most important regulatory factor for tumor angiogenesis, and its protein expression is associated with poor prognosis[14],[15] and better efficacy of anti-angiogenic drugs[16] on non–small cell lung cancer (NSCLC). Therefore, targeted therapy and chemotherapy for cancer are based on different molecular mechanisms.
In the future, treatment options for NSCLC patients will be more dependent on precise molecular characterization. The National Comprehensive Cancer Network (NCCN) guidelines state that NSCLC patients with mutated EGFR and wild-type K-Ras should be treated with EGFR-TKIs in clinic. However, the majority of patients treated with EGFR-TKIs were non-smoking female patients with adenocarcinoma. Moreover, molecular studies of EGFR-TKI treatment have been mainly focused on squamous cell carcinoma and adenocarcinoma[7]–[9]. Large cell lung carcinoma (LCLC), a pathologic type of NSCLC, has been rarely studied. How targeted therapy and chemotherapy are correlated with their associated genes, and how patients with optimal outcomes for targeted therapy are related to those with optimal outcomes for chemotherapy remain unknown. In addition, the correlations of these drug-associated genes to the prognosis after targeted therapy and chemotherapy are still unclear. In this study, we correlated the gene statuses of EGFR, K-Ras, ERCC1, and VEGF in 60 biopsies from LCLC patients with their prognoses, explored the molecular basis of targeted therapy, and discussed relationship between targeted therapy and chemotherapy.
Materials and Methods
Patients
Sixty paraffin-embedded tissue samples were collected from patients with LCLC treated between February 1993 and July 2009 at the Tianjin Medical University Cancer Institute and Hospital. The pathology of each patient was re-reviewed. No patient received targeted drug therapy. Patients consisted of 44 men and 16 women who were 30 to 76 years old with a median age of 58 years. Forty-five (75.0%) patients had a smoking history, which was defined as smoking one or more cigarettes per day continuously for 3 or more months.
DNA extraction and mutation analysis
DNA was isolated from the paraffin-embedded tissues using an E.Z.N.A.™ Tissue DNA kit (OMEGA, US) according to the manufacturer's instructions. Nested polymerase chain reaction (PCR) was used to amplify EGFR exons 18–21 and K-Ras exon 2[17]. The PCR was run at 94°C for 60 s, 35 cycles of 58°C for 30 s and 72°C for 30 s, followed by an extension step at 72°C for 10 min. After purification, DNA was directly sequenced using the ABI Prism 3730 DNA sequencer (Applied Biosystems, Foster City, CA).
FISH detection of EGFR gene amplification
The LSI EGFR SpectrumOrange/chromosome 7 (CEP7) SpectrumGreen probe (Vysis; Abbott Laboratories, Downers Grove, IL) was used according to the manufacturer's instructions. Paraffin-embedded LCLC sections (4 µm) were baked, deparaffined by washing in xylene, and dehydrated in 100% ethanol, and then incubated in pretreatment liquid (paraffin pretreatment kit) for 20 to 25 min at 80°C. Next, sections were digested with proteinase K (0.25 mg/mL in 2 × SSC; pH7.0) for 15 to 25 min at 37°C, after which EGFR/CEP7 probes were placed at the tumor regions for hybridization in the instrument. The hybridization was performed as denaturation at 73°C for 10 min, incubation at 37°C 16 h, followed by DAPI staining (0.15 mg/mL, Vector Laboratories, Burlingame, CA). Evaluation was made according to reported literature[7]. Specifically, samples with high copy number (more than 40% of cells containing 4 or more gene copies) and gene amplification (cells containing EGFR gene clusters and with a ratio of EGFR gene and chromosomes ≥ 2, or more than 10% of cells containing 15 or more gene copies) were regarded as EGFR FISH-positive.
Immunohistochemical detection of protein expression
The 4-µm sections were baked, dewaxed, and rehydrated with gradient ethanol followed by overnight incubation at 4°C with primary mouse anti-human EGFR monoclonal antibody (31G7, Zymed Laboratories, South San Francisco, CA), mouse anti-human VEGF monoclonal antibody (1:100 dilution, VG-1, abcam, Hong Kong), and mouse anti-human ERCC1 monoclonal antibody (1:50 dilution, 8F1, abcam, Hong Kong), respectively, in a final volume of 100 µL antibody dilution solution. Next, the samples were incubated with 100 µL anti-mouse or anti-rabbit secondary antibody (ChemMate™ EnVision™ Detection Kit, Dako, USA) for 30 min at 37°C. After staining, the samples were sealed. EGFR [8], ERCC1 [18], and VEGF [14] expression were evaluated according to reported literature. A sample which had more than 10% of cells with membrane EGFR staining was regarded as EGFR-positive.
Statistical methods
Pearson's Chi-square or Fisher's method in SPSS 13.0 software was used to examine relationship between biological markers and clinicopathologic features. The Kaplan-Meier method was used for survival analysis, and the log-rank method was used to test difference. The Cox proportional regression model was used for multivariate analysis, and the Spearman rank correlation coefficient analysis was used to analyze marker correlation. P < 0.05 was considered statistically significant.
Results
Clinicopathologic features and biomarkers of LCLC and their correlations with prognoses
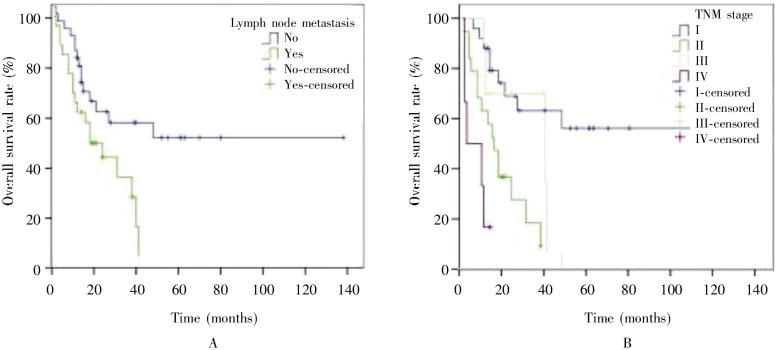
A better prognosis trend was found in the patients with smoking history (P = 0.067), tumor diameter < 3 cm (P = 0.083), or low VEGF expression (P = 0.098), though these findings were not statistically significant. Univariate analysis showed that lymph node metastasis (P = 0.049) and advanced stage (P < 0.001) were associated with poor prognosis (Table 1, Figure 1). Multivariate analysis showed that TNM stage (HR = 3.347, 95% CI = 1.791–6.254, P < 0.001) was the only independent prognosis factor. The biomarker features had no correlation with prognosis (Table 1).
Table 1. The basic clinicopathologic features and survival of patients with large cell lung cancer (LCLC).
| Variable | Cases (%) | Median OS (months) | Survival rate (%) |
P value | ||
| 1-year | 3-year | 5-year | ||||
| Sex | 0.590 | |||||
| Male | 44 (73.3) | 27 | 68.2 | 32.0 | 27.0 | |
| Female | 16 (26.7) | 23 | 70.8 | 38.2 | 30.2 | |
| Age | 0.744 | |||||
| ≥ 60 years | 25(41.7) | 31 | 76.0 | 41.7 | 26.0 | |
| < 60 years | 35 (58.3) | 21 | 65.7 | 42.0 | 34.0 | |
| Smoking history | 0.067 | |||||
| Smokers | 45 (75.0) | 40 | 73.3 | 42.2 | 36.2 | |
| Non-smokers | 15 (25.0) | 18 | 60.0 | 11.4 | 0 | |
| Tumor size | 0.083 | |||||
| < 3 cm | 3 (5.0) | 54 | 100.0 | 100.0 | – | |
| ≥ 3 cm | 57 (95.0) | 24 | 68.4 | 37.9 | 26.5 | |
| Lymph node metastasis | 0.014 | |||||
| No | 34 (56.7) | 48 | 79.4 | 53.0 | 47.5 | |
| Yes | 26 (43.3) | 18 | 57.7 | 23.8 | 0 | |
| TNM stage | 0.001 | |||||
| I | 25(41.7) | – | 87.6 | 63.0 | 56.0 | |
| II | 10 (16.7) | 40 | 70.0 | 35.0 | 0 | |
| III | 19(31.7) | 16 | 57.9 | 9.2 | 0 | |
| IV | 6 (10.0) | 3 | 16.7 | 0 | 0 | |
| EGFR mutation | 0.267 | |||||
| Positive | 1 (1.7) | 65 | 100.0 | 100.0 | 100.0 | |
| Negative | 59 (98.3) | 24 | 69.5 | 40.0 | 28.3 | |
| EGFR expression | 0.943 | |||||
| Positive | 23 (38.3) | 24 | 78.3 | 35.0 | 29.2 | |
| Negative | 37(61.7) | 31 | 64.9 | 39.9 | 31.9 | |
| EGFR amplification * | 0.947 | |||||
| Positive | 10 (19.6) | 18 | 70.0 | 35.5 | 31.2 | |
| Negative | 41 (80.4) | 38 | 70.7 | 46.0 | 26.8 | |
| K-Ras mutation | 0.656 | |||||
| Positive | 3 ( 5.0) | 40 | 66.7 | 66.7 | 33.3 | |
| Negative | 57 (95.0) | 24 | 68.4 | 39.5 | 30.7 | |
| ERCC1 expression | 0.825 | |||||
| Positive | 34 (56.7) | 24 | 67.6 | 38.0 | 33.2 | |
| Negative | 26 (43.3) | 27 | 69.0 | 39.9 | 26.6 | |
| VEGF expression | 0.098 | |||||
| Positive | 42 (70.0) | 21 | 69.0 | 34.0 | 17.0 | |
| Negative | 18 (30.0) | 38 | 66.2 | 48.0 | 34.8 | |
OS, overall survival; EGFR, epidermal growth factor receptor; ERCC1, excision repair cross-complementation group 1; VEGF, vascular endothelial growth factor. “–” indicates that the data have not yet been determined. * Fluorescence In situ hybridization (FISH) failed to detect EGFR in 9 cases, so the total cases were 51.
Figure 1. Kaplan-Meier overall survival curves of patients with large cell lung cancer.
A, the overall survival rate of patients with lymph node metastasis is lower than that of patients without lymph node metastasis; B, the overall survival rate of patients with early TNM stage disease is higher than that of patients with advanced stage disease.
Correlations of biomarkers to clinicopathologic features of LCLC
All samples were examined with related markers, and FISH detection failed in 9 cases. One sample (1.7%) contained EGFR gene L858M mutation, and 3 (5.0%) harbored K-Ras mutations (2 cases of G12C mutation and 1 case of G12D mutation). Ten samples (10/51, 19.6%) were EGFR FISH-positive, exhibiting high Polyploidy (6/51, 11.8%) or gene amplification (4/51, 7.8%). Immunohistochemistry showed that positive rates for EGFR, ERCC1, and VEGF proteins were 38.3%, 56.7%, and 70.0%, respectively (Figure 2). VEGF expression was commonly found in samples with lymph node metastasis (84.6% vs. 58.8%, P = 0.046) and cancer stage III–IV (84.0% vs. 60.0%, P = 0.052). EGFR FISH status, EGFR protein expression, K-Ras mutations, and ERCC1 expression were not correlated with patient sex, age, smoking history, tumor size, TNM stage, and lymph node metastasis (Table 2). The data on EGFR mutations is statistically meaningless due to insufficient mutated samples.
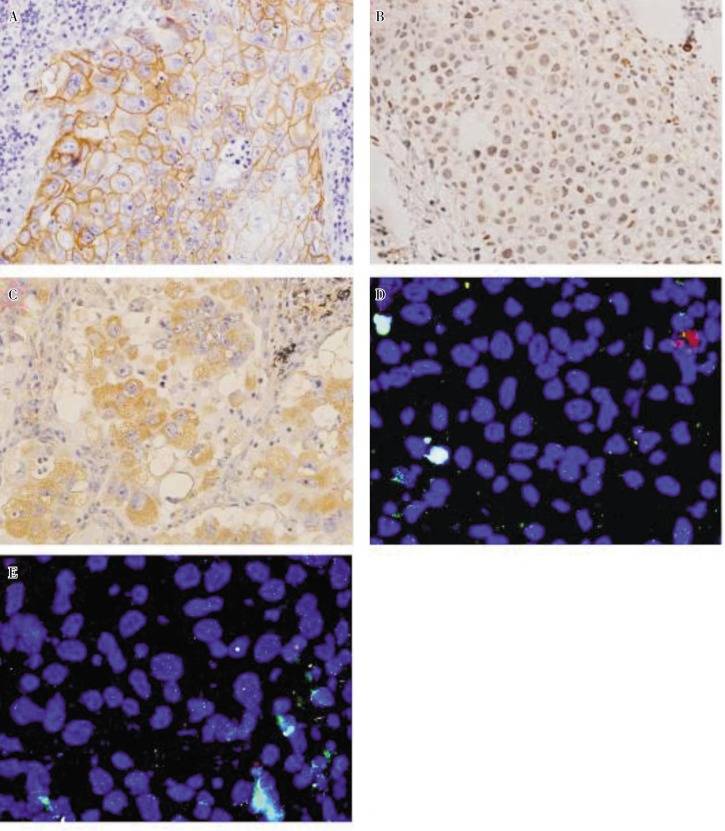
Figure 2. Epidermal growth factor receptor (EGFR), excision repair cross-complementation group 1 (ERCC1), and vascular endothelial growth factor (VEGF) protein expression detected by immunohistochemistry and EGFR amplification detected by fluorescence in situ hybridization in large cell lung cancer.
A, positive immunoreactivity of EGFR (in yellow) is evident in the membrane of tumor cells (×400). B, positive expression of ERCC1 protein was detected in the nuclei of cancer cells (×400). C, immunoreactivity for VEGF was detected predominantly in the cytoplasm of tumor cells (×400). D, EGFR amplification was detected by dual-color FISH assay with probes for EGFR (red) and chromosome 7 (CEP7, green). FISH-negative tumor cells show two red and two green signals (×1000). E, cells with amplification show an excess of red signals (×1000).
Table 2. Association between clinicopathologic variables and biomarkers in patients with large cell lung cancer.
| Variable | EGFR expression [cases (%)] | EGFR FISH [cases (%)] | K-Ras mutation [cases (%)] | ERCC1 expression [cases (%)] | VEGF expression [cases (%)] |
| Sex | P = 0.936 | P = 0.669 | P = 0.171 | P = 0.582 | P = 0.346 |
| Men | 44 (61.4) | 40 (17.5) | 44 (2.3) | 44 (54.5) | 44 (65.9) |
| Women | 16 (62.5) | 11 (27.2) | 16 (12.5) | 16 (62.5) | 16 (81.3) |
| Age | P = 0.445 | P = 0.353 | P = 1.000 | P = 0.660 | P = 0.775 |
| ≥ 60 years | 25 (44.0) | 19 (26.3) | 25 (4.0) | 25 (60.0) | 25 (72.0) |
| < 60 years | 35 (34.3) | 32 (15.6) | 35 (5.7) | 35 (54.3) | 35 (68.5) |
| Smoking history | P = 0.878 | P = 0.769 | P = 1.000 | P = 0.367 | P = 0.192 |
| Yes | 45 (37.8) | 39 (20.5) | 45 (4.4) | 45 (53.3) | 45 (64.4) |
| No | 15 (40.0) | 12 (16.7) | 15 (6.7) | 15 (66.7) | 15 (86.6) |
| Tumor diameter | P = 0.552 | P = 1.000 | P = 1.000 | P = 1.000 | P = 0.212 |
| < 3 cm | 3 (66.7) | 2 (0.0) | 3(0) | 3 (66.7) | 3 (33.3) |
| ≥ 3 cm | 57 (36.8) | 49 (20.4) | 57 (5.3) | 57 (56.1) | 57 (71.9) |
| TNM stage | P = 0.822 | P = 0.316 | P = 0.258 | P = 0.538 | P = 0.052 |
| I–II | 35 (37.1) | 28 (14.3) | 35 (8.6) | 35 (60.0) | 35 (60.0) |
| III–IV | 25 (40.0) | 23 (26.1) | 25(0) | 25 (52.0) | 25 (84.0) |
| Lymph node metastasis | P = 0.604 | P = 0.781 | P = 1.000 | P = 0.700 | P = 0.046 |
| Yes | 26 (34.6) | 23 (17.3) | 26 (3.8) | 26 (53.8) | 26 (84.6) |
| No | 34 (41.1) | 28 (21.4) | 34 (5.8) | 34 (58.8) | 34 (58.8) |
The relationship among markers
EGFR amplification was positively correlated with EGFR protein expression (r = 0.390, P = 0.005). The rate of EGFR amplification was higher in ERCC1-positive samples than in ERCC1-negative samples (23.3% vs. 14.2%). The positive rate of ERCC1 protein expression in samples with EGFR amplification was 70%, suggesting a co-positivity trend of the two markers, but this result was not statistically significant (P = 0.433). In addition, K-Ras mutations were often observed in ERCC1-negative samples (P = 0.411), VEGF-positive samples (P = 0.252), and samples without EGFR amplification (P = 0.486), though, similarly, these data were not statistically significant. The positive rate of VEGF in ERCC1-negative samples was as high as 65.3%, but this result was not statistically significant. A meaningful analysis could not be completed for the low rate of EGFR mutations.
Discussion
LCLC, which has an incidence of about 3%, is associated with poor prognosis[19]. Currently, there is no effective treatment available for this disease. This study showed that LCLC occurred more frequently in male smokers, but sex and smoking history were not correlated with the prognosis. In addition, smokers had longer median overall survival (OS) than did non-smoker patients, presumably because of the small sample size and unevenly collected data.
The impact of EGFR amplification and EGFR protein expression on the prognosis of patients with NSCLC is still unclear. A previous meta-analysis indicated that EGFR-positive patients had worse prognosis than EGFR-negative patients (HR = 1.14, 95% CI = 0.94–1.39)[20], and Jeon et al.[21] reported that non-adenocarcinoma patients with EGFR amplification had poor prognosis. Our study indicated that EGFR amplification and protein expression were not correlated with prognosis of LCLC, consistent with other reports[22],[23]. Mutations of EGFR/K-Ras and protein expression of ERCC1/VEGF had no impact on the prognosis of patients. Only lymph node metastasis (P = 0.014) and TNM stage (P < 0.001) were found to affect prognosis, and multivariate analysis showed that only TNM stage (P < 0.001) affected prognosis.
Suzuki et al.[23] reported that EGFR expression was related to lymph node metastasis (P = 0.028) and pathologic stage (P = 0.0046). Cappuzzo et al. [7] reported that high EGFR gene copy numbers were often observed in females (P = 0.04) and non-smoking patients (P = 0.001). In consistence with most reports [6],[7],[22], however, we did not find that EGFR copy number or protein expression level were correlated with sex, smoking history, TNM stage, and lymph node metastasis, suggesting that the pathologic relationship of EGFR gene copy number and expression level is not as prominent as EGFR mutations mostly found in Asia, women, non-smoking or adenocarcinoma patients[8],[18],[24]. We found that K-Ras mutations and ERCC1 protein expression were not statistically correlated with clinicopathologic features of LCLC. However, we found that VEGF protein expression occurred more commonly in patients with lymph node metastasis (P = 0.052) and poor prognosis (P = 0.098), suggesting that it may be a negative prognostic factor of LCLC.
We further found that the rates of EGFR mutation, amplification, and protein expression were 1.7%, 19.6%, and 38.3%, respectively, and K-Ras mutation rate was 5.0%. Previous studies showed that the rates of EGFR mutation, amplification, and protein expression in patients with lung adenocarcinoma ranged from 15% to 55%, from 32.8% to 47.7%, and from 50.0% to 59.1% [6]–[8],[18],[25],[26], whereas K-Ras mutation rate ranged from 15.2% to 27.7% [12],[13]. In contrast, mutation and amplification rates of EGFR in LCLC patients were far lower than those in patients with lung adenocarcinoma. This may be ascribed to the high percentage of male patients (73.3%) and smokers (75.0%) in our LCLC cohorts because EGFR mutations primarily occur in female non-smoking patients with adenocarcinoma[8],[18],[24].
After statistical analysis of 1335 patients with NSCLC, Mitsudomi et al.[27] found that 70% of patients with EGFR mutations were responsive to EGFR-TKIs, whereas only 10% of patients with wild-type EGFR were responsive to EGFR-TKIs, 35% of 663 patients with high EGFR gene copy number were responsive to EGFR-TKIs, whereas only 9% of patients with low copy number were responsive to the same treatment. High responsiveness of patients with EGFR mutations and amplification to EGFR-TKIs has been confirmed in other related studies[6],[7],[25]. Our study found that only a few patients with LCLC contained EGFR mutations or high gene copy number, suggesting that only a few patients can benefit from EGFR-TKI treatment.
The positive rate of EGFR protein in NSCLC was reported in the range of 40% to 80%[28]. However, we observed EGFR protein in only 38.3% of LCLC patients. This may be due to low EGFR mutation rate or gene copy number, or to protein degradation during long storage time. Few studies suggest that EGFR protein expression is correlated with high patient responsiveness to EGFR- TKIs[6], most results failed to support this conclusion[29],[30]. Although this supposition is still being debated, low EGFR protein expression rate give hint that most LCLC patients can not benefit from the EGFR-TKI treatment.
High level of ERCC1 protein expression is a marker of resistance to platinum chemotherapy in NSCLC patients[3],[4]. Furthermore, VEGF-positive patients generally have higher responsiveness to anti-VEGF drugs than VEGF-negative patients [16]. Correlation of EGFR gene mutation, expression, or amplification status and ERCC1 protein expression have seldom been studied. Andrieux et al.[31] reported that EGFR induced ERCC1 expression via MAPK pathway in human hepatoma cells, and Kim et al.[32] showed that EGFR protein expression was positively correlated with ERCC1 expression in gastric cancer. Nevertheless, we did not find any correlation among EGFR expression, EGFR amplification, and ERCC1 protein expression. EGFR amplification was more frequently observed in patients with ERCC1 expression, suggesting that higher EGFR copy number may induce protein expression of ERCC1 in LCLC patients. In contrast, Lee et al.[18] reported that EGFR mutations were more common in ERCC1-negative NSCLC patients with squamous cell carcinomas or adenocarcinomas. These inconsistent results suggest a complicated relationship between EGFR and ERCC1 protein expression in lung cancer. Different pathologic types and EGFR statuses may result in distinct patterns of ERCC1 protein expression. Further study of larger scale is required to uncover their intrinsic correlations. Samples with EGFR amplification and without ERCC1 expression accounted for only 5.8% (3/51) of all studied cases, suggesting that only a few patients can benefit from both EGFR-TKI and platinum therapy. The fact that EGFR amplification was more commonly observed in ERCC1-positive samples implies that there is no overlap between LCLC patients with optimal response to EGFR-TKIs and those with optimal response to platinum-based chemotherapy. Nevertheless, the significance of this hypothesis was limited statistically by the prevalence of few samples with EGFR mutations. Studying patient samples on a larger scale may provide further insight into the two groups of patients and their optimal treatments.
The positive rate of VEGF in ERCC1-negative patients was as high as 65.3%, suggesting a major overlap between LCLC patients with optimal outcomes for platinum-based chemotherapy and those with optimal outcomes for anti-VEGF treatment. Combination of these two treatments may provide a greater benefit for the patients than single agents. However, it should be noted that the correlation of EGFR gene and VEGF protein expression is statistically meaningless, suggesting that the impact of combination therapy on clinical outcomes of the patients is still unclear.
In conclusion, low EGFR mutation and amplification rates suggest only a few patients with LCLC can benefit from EGFR-TKI treatment. Although correlation analysis did not find significant relationships among EGFR, K-Ras, ERCC1, and VEGF genes or protein expression status, this study suggests that patients with optimal outcomes for platinum-based chemotherapy overlapped with patients with optimal outcomes for anti-VEGF treatment rather than those for EGFR-TKI treatment. Thus, it may be useful to treat patients with lower ERCC1 expression with the combination of anti-angiogenic therapy and chemotherapy, and treat patients with higher ERCC1 expression with EGFR-TKI-targeted therapy, although this suggestion still needs supportive data from large scale clinical trials. In addition, we did not find biomarkers that affect the prognosis of LCLC, and TNM stage was the only factor that affect prognosis.
Acknowledgments
This work was supported by grant from the Wu Jieping Medical Foundation (No. 320670009016).
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008 [J] CA Cancer J Clin. 2008;58(2):71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Wilson MD, Ruttan CC, Koop BF, et al. ERCC1: a comparative genomic perspective [J] Environ Mol Mutagen. 2001;38(2–3):209–215. doi: 10.1002/em.1073. [DOI] [PubMed] [Google Scholar]
- 3.Azuma K, Komohara Y, Sasada T, et al. Excision repair cross-complementation group 1 predicts progression-free and overall survival in non-small cell lung cancer patients treated with platinum-based chemotherapy [J] Cancer Sci. 2007;98(9):1336–1343. doi: 10.1111/j.1349-7006.2007.00557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou W, Gurubhagavatula S, Liu G, et al. Excision repair cross-complementation group 1 polymorphism predicts overall survival in advanced non–small cell lung cancer patients treated with platinum-based chemotherapy [J] Clin Cancer Res. 2004;10(15):4939–4943. doi: 10.1158/1078-0432.CCR-04-0247. [DOI] [PubMed] [Google Scholar]
- 5.Spira A, Ettinger DS. Multidisciplinary management of lung cancer [J] N Engl J Med. 2004;350(4):379–392. doi: 10.1056/NEJMra035536. [DOI] [PubMed] [Google Scholar]
- 6.Tsao MS, Sakurada A, Cutz JC, et al. Erlotinib in lung cancer-molecular and clinical predictors of outcome [J] N Engl J Med. 2005;353(2):133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 7.Cappuzzo F, Hirsch FR, Rossi E, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non–small-cell lung cancer [J] J Natl Cancer Inst. 2005;97(9):643–655. doi: 10.1093/jnci/dji112. [DOI] [PubMed] [Google Scholar]
- 8.Hirsch FR, Varella-Garcia M, Bunn PA, Jr, et al. Molecular predictors of outcome with gefitinib in a phase III placebo-controlled study in advanced non–small-cell lung cancer [J] J Clin Oncol. 2006;24(31):5034–5042. doi: 10.1200/JCO.2006.06.3958. [DOI] [PubMed] [Google Scholar]
- 9.Takano T, Ohe Y, Sakamoto H, et al. Epidermal growth factor receptor gene mutations and increased copy numbers predict gefitinib sensitivity in patients with recurrent non–small-cell lung cancer [J] J Clin Oncol. 2005;23(28):6829–6837. doi: 10.1200/JCO.2005.01.0793. [DOI] [PubMed] [Google Scholar]
- 10.Sandier A, Gray R, Perry MC, et al. Paclitaxel-carboplatin alone or with bevacizumab for non–small-cell lung cancer [J] N Engl J Med. 2006;355(24):2542–2550. doi: 10.1056/NEJMoa061884. [DOI] [PubMed] [Google Scholar]
- 11.Reck M, von Pawel J, Zatloukal P, et al. Phase III trial of cisplatin plus gemcitabine with either placebo or bevacizumab as first-line therapy for nonsquamous non–small-cell lung cancer: AVAil [J] J Clin Oncol. 2009;27(8):1227–1234. doi: 10.1200/JCO.2007.14.5466. [DOI] [PubMed] [Google Scholar]
- 12.Massarelli E, Varella-Garcia M, Tang X, et al. KRAS mutation is an important predictor of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer [J] Clin Cancer Res. 2007;13(10):2890–2896. doi: 10.1158/1078-0432.CCR-06-3043. [DOI] [PubMed] [Google Scholar]
- 13.Pao W, Wang TV, Riely GJ, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib [J] PLoS Med. 2005;2(1):e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stefanou D, Batistatou A, Arkoumani E, et al. Expression of vascular endothelial growth factor (VEGF) and association with microvessel density in small-cell and non–small-cell lung carcinomas [J] Histol Histopathol. 2004;19(1):37–42. doi: 10.14670/HH-19.37. [DOI] [PubMed] [Google Scholar]
- 15.Herbst RS, Sandier AB. Non–small cell lung cancer and antiangiogenic therapy: what can be expected of bevacizumab? [J] Oncologist. 2004;9:19–26. doi: 10.1634/theoncologist.9-suppl_1-19. Suppl. 1. [DOI] [PubMed] [Google Scholar]
- 16.Dowlati A, Gray R, Sandier AB, et al. Cell adhesion molecules, vascular endothelial growth factor, and basic fibroblast growth factor in patients with non–small cell lung cancer treated with chemotherapy with or without bevacizumab—an Eastern Cooperative Oncology Group Study [J] Clin Cancer Res. 2008;14(5):1407–1412. doi: 10.1158/1078-0432.CCR-07-1154. [DOI] [PubMed] [Google Scholar]
- 17.Eberhard DA, Johnson BE, Amler LC, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non–small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib [J] J Clin Oncol. 2005;23(25):5900–5909. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 18.Lee KH, Min HS, Han SW, et al. ERCC1 expression by immunohistochemistry and EGFR mutations in resected non–small cell lung cancer [J] Lung Cancer. 2008;60(3):401–407. doi: 10.1016/j.lungcan.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 19.Kamiyoshihara M, Hirai T, Kawashima O, et al. Primary large cell carcinoma of the lung in the patients undergoing pulmonary resection: a comparison between pre- and postoperative diagnosis [J] Kyobu Geka. 1998;51(6):464–468. [PubMed] [Google Scholar]
- 20.Meert AP, Martin B, Delmotte P, et al. The role of EGF-R expression on patient survival in lung cancer: a systematic review with meta-analysis [J] Eur Respir J. 2002;20(4):975–981. doi: 10.1183/09031936.02.00296502. [DOI] [PubMed] [Google Scholar]
- 21.Jeon YK, Sung SW, Chung JH, et al. Clinicopathologic features and prognostic implications of epidermal growth factor receptor (EGFR) gene copy number and protein expression in non–small cell lung cancer [J] Lung Cancer. 2006;54(3):387–398. doi: 10.1016/j.lungcan.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 22.Dacic S, Flanagan M, Cieply K, et al. Significance of EGFR protein expression and gene amplification in non–small cell lung carcinoma [J] Am J Clin Pathol. 2006;125(6):860–865. doi: 10.1309/H5UW-6CPC-WWC9-2241. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki S, Dobashi Y, Sakurai H, et al. Protein overexpression and gene amplification of epidermal growth factor receptor in nonsmall cell lung carcinomas. An immunohistochemical and fluorescence in situ hybridization study [J] Cancer. 2005;103(6):1265–1273. doi: 10.1002/cncr.20909. [DOI] [PubMed] [Google Scholar]
- 24.Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers [J] J Natl Cancer Inst. 2005;97(5):339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 25.Hirsch FR, Varella-Garcia M, Cappuzzo F, et al. Combination of EGFR gene copy number and protein expression predicts outcome for advanced non–small-cell lung cancer patients treated with gefitinib [J] Ann Oncol. 2007;18(4):752–760. doi: 10.1093/annonc/mdm003. [DOI] [PubMed] [Google Scholar]
- 26.Huang SF, Liu HP, Li LH, et al. High frequency of epidermal growth factor receptor mutations with complex patterns in non–small cell lung cancers related to gefitinib responsiveness in Taiwan [J] Clin Cancer Res. 2004;10(24):8195–8203. doi: 10.1158/1078-0432.CCR-04-1245. [DOI] [PubMed] [Google Scholar]
- 27.Mitsudomi T, Yatabe Y. Mutations of the epidermal growth factor receptor gene and related genes as determinants of epidermal growth factor receptor tyrosine kinase inhibitors sensitivity in lung cancer [J] Cancer Sci. 2007;98(12):1817–1824. doi: 10.1111/j.1349-7006.2007.00607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirsch FR, Varella-Garcia M, Bunn PA, Jr, et al. Epidermal growth factor receptor in non–small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis [J] J Clin Oncol. 2003;21(20):3798–3807. doi: 10.1200/JCO.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 29.Parra HS, Cavina R, Latteri F, et al. Analysis of epidermal growth factor receptor expression as a predictive factor for response to gefitinib (“Iressa”, ZD1839) in non–small-cell lung cancer [J] Br J Cancer. 2004;91(2):208–212. doi: 10.1038/sj.bjc.6601923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perez-Soler R, Chachoua A, Hammond LA, et al. Determinants of tumor response and survival with erlotinib in patients with non–small-cell lung cancer [J] J Clin Oncol. 2004;22(16):3238–47. doi: 10.1200/JCO.2004.11.057. [DOI] [PubMed] [Google Scholar]
- 31.Andrieux LO, Fautrel A, Bessard A, et al. GATA-1 is essential in EGF-mediated induction of nucleotide excision repair activity and ERCC1 expression through ERK2 in human hepatoma cells [J] Cancer Res. 2007;67(5):2114–2123. doi: 10.1158/0008-5472.CAN-06-3821. [DOI] [PubMed] [Google Scholar]
- 32.Kim JS, Kim MA, Kim TM, et al. Biomarker analysis in stage III–IV (M0) gastric cancer patients who received curative surgery followed by adjuvant 5-fluorouracil and cisplatin chemotherapy: epidermal growth factor receptor (EGFR) associated with favourable survival [J] Br J Cancer. 2009;100(5):732–738. doi: 10.1038/sj.bjc.6604936. [DOI] [PMC free article] [PubMed] [Google Scholar]