Abstract
Background
Pitx2 is the homeobox gene located in proximity to the human 4q25 familial atrial fibrillation locus. When deleted in the mouse germline, Pitx2 haploinsufficiency predisposes to pacing induced atrial fibrillation indicating that reduced Pitx2 promotes an arrhythmogenic substrate. Previous work focused on Pitx2 developmental functions that predispose to atrial fibrillation. Although Pitx2 is expressed in postnatal left atrium, it is unknown whether Pitx2 has distinct postnatal and developmental functions.
Methods and Results
To investigate Pitx2 postnatal function, we conditionally inactivated Pitx2 in the postnatal atrium while leaving its developmental function intact. Unstressed adult Pitx2 homozygous mutant mice display variable R-R interval with diminished P-wave amplitude characteristic of sinus node dysfunction, an atrial fibrillation risk factor in human patients. An integrated genomics approach in the adult heart revealed Pitx2 target genes encoding cell junction proteins, ion channels, and critical transcriptional regulators. Importantly, many Pitx2 target genes have been implicated in human atrial fibrillation by genome wide association studies. Immunofluorescence and transmission electron microscopy studies in adult Pitx2 mutant mice revealed structural remodeling of the intercalated disc characteristic of human atrial fibrillation patients.
Conclusions
Our findings, revealing that Pitx2 has genetically separable postnatal and developmental functions, unveil direct Pitx2 target genes that include channel and calcium handling genes as well as genes that stabilize the intercalated disc in postnatal atrium.
Keywords: atrial fibrillation, sinus node dysfunction, genome-wide analysis, genetics, knockout models, Pitx2
Atrial fibrillation (AF), the most prevalent adult arrhythmia in humans, results in an increased risk of stroke, dementia and heart failure 1. Electrical impulses that are critical for a coordinated, physiologic heartbeat originate in the sinoatrial node (SAN) and are transduced from the pacemaker region into both atria. In AF, fibrillatory atrial impulses override normal conduction pathways with resulting irregular ventricular conduction.
Because of its clinical significance, great efforts have been expended to investigate the genetic underpinnings of AF using unbiased genome wide approaches such as genome wide association study (GWAS). Multiple studies have revealed a single nucleotide variant (SNV) on human 4q25 that was associated with familial AF (reviewed in 2). Patients with the 4q25 variant exhibited early onset AF that was independent from known AF risk factors. The 4q25 SNV also had prognostic value as patients with this variant are prone to AF recurrence after ablation therapy 3. The gene poor 4q25 region harbors the Pitx2 homeobox gene which has been implicated in AF predisposition using mouse models 4-7.
Atrial fibrillation and associated arrhythmias
AF may result from new, pathologic sources of electrical impulses. For example, many cases of ectopic electrical activity originate in the pulmonary vein 8. Other sites of ectopy include the left atrial posterior wall, superior vena cava, interatrial septum, crista terminalis, and coronary sinus myocardium 9, 10. In addition to ectopy, other causes of AF involve atrial myopathy that disrupts normal atrial conduction and promotes re-entrant circuits. One common example of AF secondary to myopathy is fibrosis that in some cases may be due to elevated Tgfβ signaling 11.
Work from the Framingham study has shown that patients with PR interval prolongation, also called first degree atrioventricular block (AVB), often develop AF 12. In addition to AVB, A sinus node dysfunction (SND) is also an AF risk factor in human patients 13. Notably, progression to higher grade arrhythmias with time is also common reflecting the importance of aging in arrhythmogenesis. Although the mechanistic connection between SND, PR interval prolongation, and AF are poorly understood, all three conditions may involve an atrial myopathy with defective atrial impulse conduction 14.
Predisposition to AF may result from a developmental defect that results in an adult heart with subclinical abnormalities that subsequently manifest as overt disease after environmental insults or aging. Alternatively, postnatal homeostatic genes may be required to maintain normal tissue structure and physiology. Small changes in homeostatic gene level may result in subclinical disease until an AF-inducing stress is encountered.
Pitx2 and predisposition to atrial fibrillation
Pitx2 encodes three isoforms, Pitx2a, Pitx2b, and Pitx2c that are generated by alternative splicing and dual promoter usage 4. Pitx2c is generated via an intergenic promoter while the Pitx2a and Pitx2b isoforms, generated by alternative splicing, use an upstream 5′ flanking promoter. The Pitx2c isoform is expressed on the left side of the embryo while the Pitx2a and Pitx2b isoforms are expressed symmetrically in the head within eyes and craniofacial strucutures4. In the mouse, Pitx2c expression continues in the postnatal atrium while human PITX2C is also the predominant isoform in left atrium 4, 6, 15. Studies in isoform-specific knock out mice in our lab also revealed that the Pitx2c isoform is the dominant isoform in determining left right asymmetry (LRA) during development 16. Pitx2 haploinsufficient (Pitx2null +/−) adult mice and Pitx2c +/− adult mice were prone to AF when challenged by programmed stimulation indicating that reduced Pitx2c levels during development creates an arrhythmogenic substrate 4, 6. Notably, Pitx2 levels are also decreased in the atria of human AF patients 5.
Previous experiments indicated that SAN genes were expanded in left superior caval vein and left atrium of Pitx2 null/+ and Pitx2 null/null mutant embryos indicating a developmental defect4. Optical mapping experiments showed Pitx2 conditional mutant embryos had a functional left-sided SAN that could override normal atrial cardiomyocyte depolarization 7. Furthermore, Pitx2c heterozygous adult mice had shortened action potential duration without fibrosis or structural defects suggesting an electrophysiologic mechanism for arrhythmogenesis in Pitx2c germline mutants 6.
It is unknown whether Pitx2 has a postnatal homeostatic function. To study Pitx2 postnatal function, we generated a Pitx2 conditional knock out (CKO) mouse line that deletes Pitx2 in postnatal atrium. The adult Pitx2 CKO mice had abnormal electrocardiography with irregular R-R interval and low voltage P waves indicating SND with impaired atrial conduction. A genome-wide search for Pitx2 targets by ChIP-Seq and microarray assays revealed genes encoding cell adhesion/cell junction proteins, ion channels, and transcription factors. Many Pitx2 target genes are AF risk genes identified in human GWAS 2. Using immunofluoresence and transmission electron microscopy (TEM) we obtained evidence for structural remodeling of the intercalated disc (ID) – the structure that mediates cardiomyocyte electro-mechanical coupling 17. Our data indicate a postnatal role for Pitx2 in AF predisposition and uncover novel Pitx2 target genes that provide new mechanistic etiologies for AF.
Materials and Methods
Mouse alleles and transgenic lines
The MCK-Cre transgenic line, R26R transgenic line, Pitx2 conditional null or floxed allele and Flag knock in allele have been described 4, 18. The Pitx2 conditional null allele contains LoxP sites flanking the exon encoding the N terminus of homeodomain and conditionally removes function of all Pitx2 isoforms.
Telemetry ECG
Implantation of the telemetry transmitter ETA-F10 (Data Sciences International) was done following the protocol of the manufacturer. Transmitters were placed intraperitoneally and ECG leads were at a lead II configuration. ECG data was collected by DSI Telemetric Physiological Monitor System and processed by Dataquest A.R.T. 3.1 software (Data Sciences International) without stress from restraint or anesthesia.
Microarray assay
Pitx2 control and mutant hearts were collected from 3-, 6- and 12-week-old mice. At each time point, three controls and three mutants were collected as biological replicates. cDNA microarray analysis was performed using Affymetrix GeneChip Mouse Genome 430 2.0 Array (Affymetrix, Santa Clara, CA).
ChIP-Sequencing
Chromatin immunoprecipitation was performed as described previously using EZ ChIP™ kit (Millipore) and Anti-FLAG M2 Affinity Gel (Sigma) 4. For library construction, 10-50 ng of purified ChIP DNA was end-repaired using Ion Plus Fragment Library Kit (Part no. 4471252, Life Technologies) and purified with two rounds of AMPure® XP beads (Beckman Coulter, Brea, CA) capture to size select fragments 100–250 bp in length. End-repaired DNA was ligated to Ion Torrent compatible adapters. Followed by 15 cycles of PCR amplification before downstream template preparation. DNA fragments of completed library ranged from approximately 170–220 bp in length. Sequencing was performed on Ion Torrent PGM system (Life Technologies).
Statistical analysis
Microarray raw data were generated using Affymetrix MAS5.0, global scaling was performed to set mean target intensity to 100. The normality of the microarray data was checked using quantile-quantile plot and Shapiro-Wilk test on R (3.0.1). Differential expressed genes were detected using R package limma (3.16.8) 19 with threshold p-value ≤ 0.05 and fold change ≥ 1.5. Benjamini-Hochberg correction was used to calculate false discovery rate. All the differential expressed genes have FDR < 0.35. Clusters were automatically detected using R package HOPACH (2.20.0) 20. The distance matrix was calculated using cosine angle method. Over-represented gene ontology terms were identified using GO-Elite 21 with Z score > 3, FDR < 0.10. FDR was calculated using Benjamini-Hochberg correction after permutation (n = 5000). qRT-PCR and luciferase reporter assay results were tested using two-sample Wilcoxon test in R. Difference of R-R interval between wild type and mutant mice was tested using two-tailed, unpaired Student’s t-test using R (n > 400). ChIP-Seq peaks were identified using Homer 22 under the assumption that the local density of the reads follow an Poisson distribution. The peaks were called using cutoff: read number enrichment ≥ 4 folds, FDR < 0.001, FDR effective Poisson p-value < 3.7e-8, and minimum read number 5.
Results
Generation of Pitx2 conditional knock out mice
To generate a Pitx2 conditional loss of function (Pitx2 CKO) allele that deletes all Pitx2 isoforms in postnatal atrium, mice with the muscle creatine kinase (MCK)-Cre driver were crossed to mice bearing the Pitx2 conditional null (Flox) allele (Fig. S1 A). MCK-Cre directs Cre activity in skeletal and cardiac muscle with a perinatal onset around birth 18, 23. We examined Cre activity using the R26R LacZ reporter. MCK-Cre activity initiates in atria after birth and within ventricles in a mosaic pattern after E15.5 (Fig. S1B). Since Pitx2 is predominantly expressed in left atrium at these fetal stages and in adult, MCK-Cre is a valuable tool to address Pitx2 function in postnatal left atrium 4, 16. Both immunostaining and realtime RT-PCR revealed that Pitx2 was efficiently deleted in neonatal left atrium (Fig. S1 C, D). Although Pitx2 is also inactivated in skeletal muscle, we did not detect skeletal muscle phenotypes in Pitx2 CKO mice perhaps due to overlapping function with Pitx3 24.
Pitx2 CKO mice have abnormal cardiac conduction
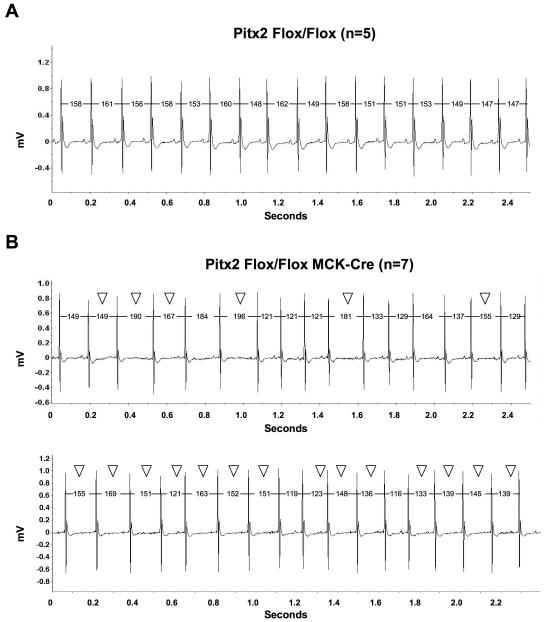
We implanted telemetry transmitters into control and Pitx2 CKO adult mice and collected resting electrocardiography (ECG) data in awake mice. Compared to normal sinus rhythm in controls (Fig. 1 A), ECG tracing for Pitx2 CKO mice show abnormal heart rate with irregular R-R intervals and low voltage P waves (Fig. 1 B, Table S1). This phenotype was observed in all Pitx2 CKO mice studied by resting telemetry ECG but none of the controls (Table S2). As sinus node sets heart rhythm and P wave represents atrial depolarization, these phenotypes indicate sinus node dysfunction (SND) with impaired atrial conduction. Importantly, SND is closely associated with AF in human patients 12.
Figure 1.
Pitx2 conditional knock out mice have abnormal heart function – sinus node dysfunction with impaired atrial conduction. By telemetry ECG tracing in awake mice, heart function of the adult control (Pitx2 Flox/Flox) and Pitx2 CKO (Pitx2 Flox/Flox MCK-Cre) mice were tested. (A) In control mice, normal sinus rhythm was observed. (B) In the Pitx2 CKO mice, mutant heart function including irregular R-R intervals and low voltage of P waves (open arrowheads) were observed. Tracings from one control and two Pitx2 CKO mice are shown. All mice shown were 3-month-old. The unit of R-R interval is millisecond (ms).
Unbiased discovery of Pitx2 regulated target genes
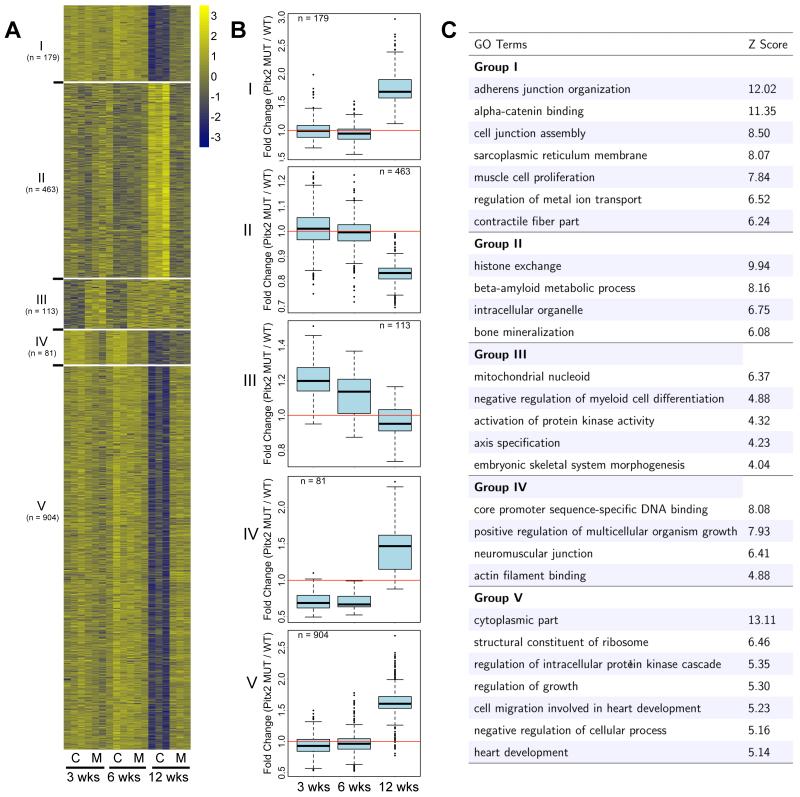
We performed microarray transcriptional profiling on 3-, 6-, and 12-week-old whole hearts from Pitx2 CKO and control mice(Fig. S2). Most genes were upregulated in Pitx2 CKO mutant hearts suggesting that Pitx2 is a transcriptional repressor in the adult heart (Fig. 2 A, S3). We performed clustering analysis on all differentially expressed genes and identified five gene expression categories in Pitx2 CKO hearts compared to controls. Groups I, IV, and V genes were significantly upregulated in the Pitx2 CKO at the 12 week stage. In contrast, group III genes were upregulated at the 3 and 6 week stages in Pitx2 CKO hearts. Group II genes, the only gene class that was downregulated, were reduced at 12 weeks in Pitx2 CKO hearts (Fig. 2 B). Genes in groups I,IV, and V were involved in cell junction assembly, as well as, proliferation and migration (Fig. 2 C).
Figure 2.
Age-related gene expression profiling in Pitx2 mutants. (A) Clustering analysis of gene expression profile across three time points in control and Pitx2 mutant hearts. Expression patterns of five representative groups are shown in heat map. The color bar represents relative expression level for each gene across different samples. (B) Differential expression between Pitx2 mutant and control across three time points. Mean value for each group was used to calculate fold change between mutants and controls. Box plots for selected clusters show median (middle line), third and first quartile (top and bottom hinges of the boxes), whiskers and outliers (black dots). (C) Gene ontology analysis of genes in each group. GO terms related to biological process and molecular function categories.
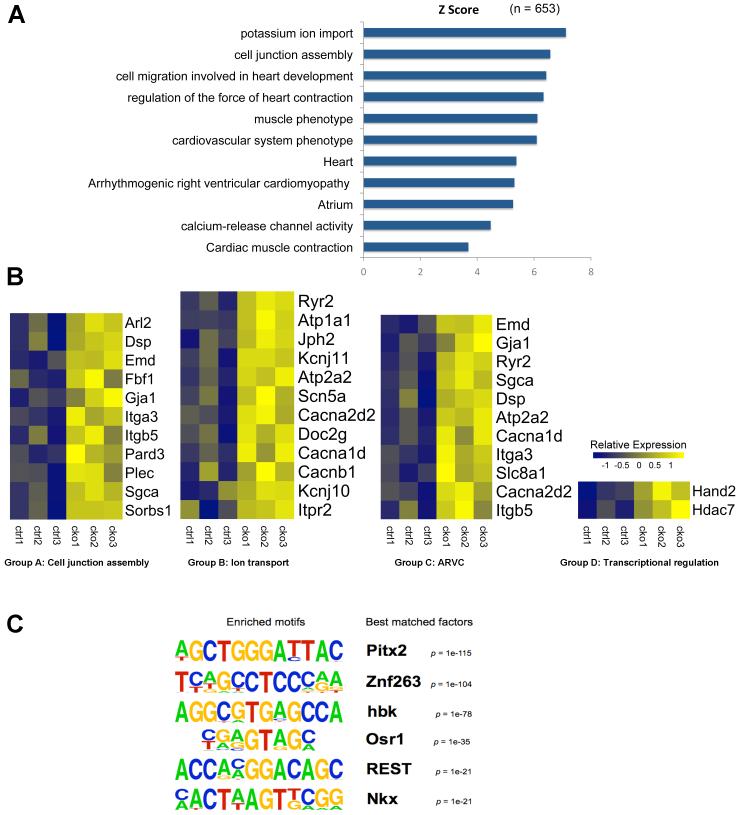
To investigate Pitx2 direct target genes, a ChIP-Seq assay was performed at the 12 week stage on heart tissue. Overlay of Pitx2 ChIP-Seq data and the 12-week expression profiling revealed genes that are likely direct Pitx2 target genes. Gene ontology (GO) analysis of the ChIP-Seq/microarray overlay gene-set indicated that most Pitx2 target genes are involved in cell-cell junction organization and ion channel physiology (Fig. 3 A, S4). Another important GO term providing insight into Pitx2 mediated arrhythmias was arrhythmogenic right ventricular cardiomyopathy (ARVC) thought to be a desmosome defect. We categorized Pitx2 direct target genes into four main groups based on gene function including cell junction assembly (Group A), ion transport (Group B), ARVC (Group C), and transcriptional regulation (Group D, Fig. 3 B). Enriched ChIP sequencing tags (peaks) were identified in potential regulatory chromosomal regions for these targets (Fig. 4 A, S5).
Figure 3.
Overlay of Pitx2 ChIP-Seq and gene expression profiling assays for Pitx2 CKO heart. (A) Gene ontology (GO) analysis for genes (n=653) from overlay of Pitx2 ChIP-Seq and Pitx2 CKO microarray assays (genes in close proximity to Pitx2 binding chromosomal regions identified by Pitx2 ChIP-Seq assay that also show significant change in microarray assay for Pitx2 CKO heart). Both assays performed on 3-month-old adult mouse hearts. (B) Heat map of gene expression profiling obtained by Pitx2 CKO microarray assay. The color bar represents relative expression level of log-transformed value for each gene across different samples. Ctrl1-Ctrl3: control mouse heart (Pitx2 Flox/Flox); CKO1-CKO3: Pitx2 CKO mouse heart (Pitx2 Flox/Flox MCK-Cre). (C) Motif analysis for peaks from Pitx2 ChIP-Seq (n=11,280). Eriched DNA binding motifs and best matched factors are shown.
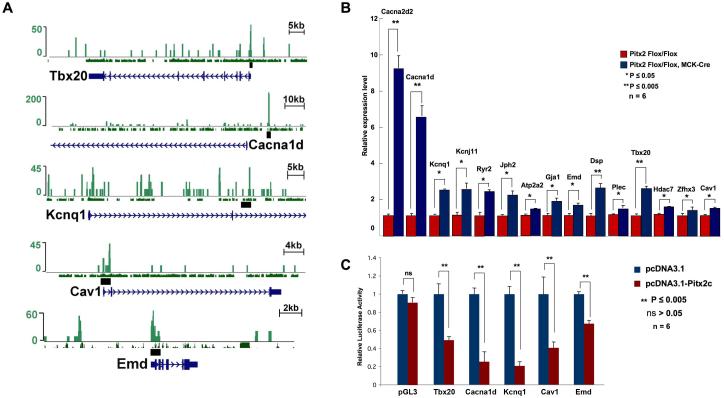
Figure 4.
Validation of potential Pitx2 targets from ChIP-Seq and microarray assays. (A) Genome browser tracks for potential targets of Pitx2 in adult heart identified by ChIP-Seq. Peaks from ChIP-Seq, conservation with human genome and the chromosomal regions used in reporter assay are shown. Normalized ChIP-Seq tag numbers are shown on the Y-axis. (B) Alteration in expression levels for the potential Pitx2 targets were detected by realtime RT-PCR in the left atrium of 3 to 4-month-old control and Pitx2 conditional knock out mice. (n=6) (C) Reporter assay for ChIP-Seq identified potential targets of Pitx2. Chromosomal regions were named after the genes in closest proximity. Values and error bars represent mean and standard deviation (n=6).
Motif analysis identified the Pitx2 binding element as the most highly enriched in the ChIP-Seq dataset validating our experiment 25 (Fig. 3 C). Other transcription factor binding sites were also enriched in the ChIP-Seq data set providing further insight into potential Pitx2 co-factors in the adult heart (Fig. 3 C). Among potential Pitx2 co-factors, Nkx2.5 is of great interest because it has been implicated in human AF in GWAS 26.
Our data sets indicated that other genes that have been implicated in human AF through GWAS or other genetic analysis are potential Pitx2 target genes such as Kcnq1, Cav1, and Zfhx327-33. Other Pitx2 target genes such as Cacna1d and Tbx20, have also been directly implicated in AF in mouse models or other human arrhythmias such as prolonged QRS (Table S3) 26, 34-36.
Pitx2 directly regulates genes involved in cell junction assembly, ion transport, and transcriptional regulation
We validated gene expression changes for genes in ChIP-Seq/microarray overlay by realtime RT-PCR. We also evaluated a number of candidate genes, previously implicated in human arrhythmias, that were unchanged on the microarray. Among these were Kcnq1, Cav1, Tbx20, and Zfhx3. In Pitx2 CKO hearts, we found increases in gene expression levels for ion channel and calcium handling genes such as Cacna1d, Cacna2d2, Kcnq1, Kcnj11, Ryr2, Jph2, and Atp2a2. We saw upregulated expression of genes encoding transcriptional regulators Hdac7, Tbx20, and Zfhx3 as well as Cav1, a component of calveolae. Significantly, elevated Hdac activity has been implicated in AF vulnerability, structural remodeling, and fibrosis 37. Tbx20 is a central component of a gene regulatory network that includes channel genes and other transcriptional regulators 38. Zfhx3, a homeodomain zinc finger transcription factor that was implicated in AF via GWAS, is also required for normal pituitary development as is Pitx239. Caveolins are known to associate with channel proteins and are thought to modify channel function 40. Our results also revealed higher mRNA expression levels for the genes Gja1, Emd, Dsp, and Plec, encoding ID components that are required for structural homeostasis and signaling between cardiomyocytes (Fig. 4B). Genes that were unchanged or reduced in RT-PCR assays are listed in Table S4.
Based on phylogenetic conservation and DNAase hypersensitivity, we cloned 1-3 kb DNA sequences flanking multiple Pitx2 ChIP-Seq peaks (Fig. 4A) into luciferase reporters and performed transactivation experiments in cultured cells. Luciferase activity for the ChIP-Seq regions that are in close proximity to Tbx20, Cacna1d, Kcnq1, Cav1, and Emd were significantly repressed upon Pitx2 co-transfection indicating Pitx2 directly represses gene expression through binding to these chromosomal regions (Fig. 4C).
Pitx2 mutants have an atrial cardiomyopathy with disrupted junctional complexes
Among the top Pitx2 target gene function categories are cell junction assembly and ARVC. To investigate ID status in Pitx2 CKO hearts, we used immunostaining with antibodies against β-catenin to label adherens junctions in the ID. β-catenin mediates cell adhesion between cardiomyocytes and also functions as a cell signaling molecule once activated by Wnt signaling. In controls, β-catenin clearly marked IDs while in Pitx2 CKO there were fewer distinct IDs and more β-catenin expression on lateral aspects of cardiomyocytes (Fig. 5 A-D).
Figure 5.
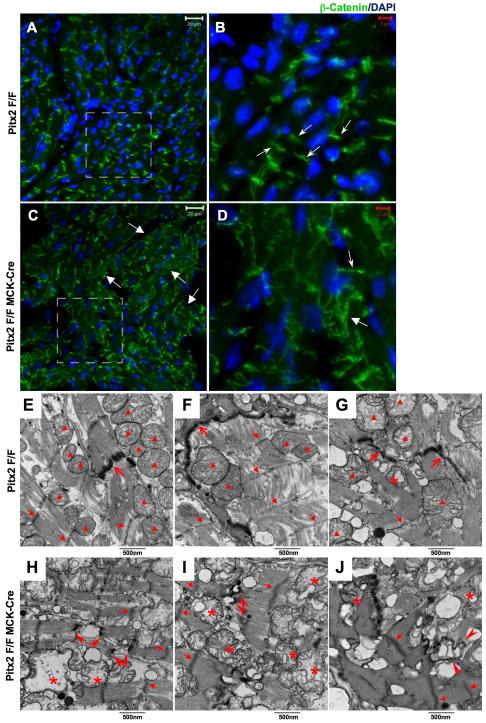
Pitx2 CKO mouse heart has altered expression pattern for β-catenin and disrupted intercalated discs (IDs). (A-D) β-catenin immunostaining on left atrium sections of 3-month-old control (Pitx2 Flox/Flox) (A-B) and Pitx2 CKO (C-D) mice shows enhanced expression of β-catenin on cardiomyocytes lateral aspect (big arrows in D) and in cytoplasm (big arrows in C). Small arrows point to intercalated discs. (E-J) Electron microscopy shows disrupted IDs in 1-year-old Pitx2 CKO heart. (E-G) in control left atrium, IDs (big arrows) and Mitochondia were intact (triangles). (H-J) in Pitx2 CKO left atrium, some IDs were disrupted (arrowheads). Large number of mitochondria were deteriorated (asterisks). Indistinct Z-lines were observed in Pitx2 CKO left atrium (small arrows).
We used transmission electron microscopy (TEM) to further investigate ID structure in Pitx2 mutants. In contrast to controls (Fig. 5 E-G), Pitx2 mutant IDs had widened spaces between junctions indicating ID deterioration (Fig. 5 H-J). We also found evidence for mitochondrial dysfunction with swollen and vacuolated mitochondria in Pitx2 CKO hearts (Fig. 5 H-J). Pitx2 CKO cardiomyocytes also had indistinct Z-lines indicating that Pitx2 is required for maintaining normal sarcomere cytoarchitecture (Fig. 5 H-J).
Discussion
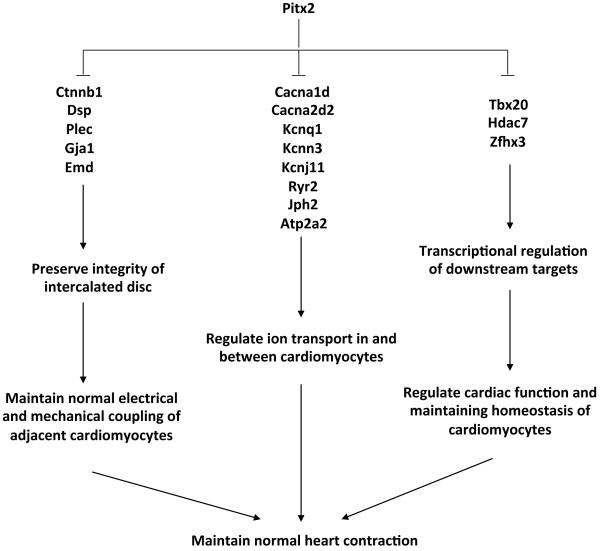
Pitx2 is the gene in proximity to the 4q25 familial AF locus. Previous work indicated that reduced Pitx2 levels result in AF predisposition. Prior to our experiments, it was unknown whether Pitx2 was important in postnatal heart since previously reported adult phenotypes could result from developmental defects. In addition to uncovering a Pitx2 postnatal function, our findings reveal novel Pitx2 target genes and that Pitx2 is required for ID homeostasis in postnatal atrium (Fig. 6).
Figure 6.
Model of Pitx2’s function in postnatal heart. In postnatal heart, Pitx2 binds to its targets through DNA binding domain and regulates expression of targets. Three groups of genes were identified to be targets of Pitx2. The first group contains genes regulating cell adhesion/cell junction assembly, which is critical for preserving normal structure of intercalated disc. The second group contains genes that regulate ion transportation in and between cardiomyocytes, which are important for normal conduction of electrical signal that induces heart contraction. The third group contains transcription regulators that regulate a large number of downstream targets and involved in regulating cardiac function and maintaining homeostasis of cardiomyocytes.
Pitx2 postnatal deletion results in sinus node dysfunction
Pitx2 deletion in postnatal atrium caused resting atrial arrhythmias indicating that in addition to its developmental function, Pitx2 has a genetically separable postnatal function in left atrial homeostasis. The predominant arrhythmia we observe with resting telemetry is SND that is known to be clinically linked to AF in human patients. SND, can result from defective impulse generation from the sinus node. In addition, SND similar to AF, is associated with diffuse atrial remodeling resulting in an arrhythmogenic substrate. Consistent with this, we observe anatomic disruption of cardiomyocyte junctions in Pitx2 CKO hearts 14.
The Pitx2c+/− mice that have a 37% reduction in Pitx2c levels have no structural abnormalities or obvious histologic defects but are still prone to AF 6. Molecular analysis indicates that Pitx2c+/− mice have electrical remodeling with changes in channel gene expression. The authors also noted changes in desmosomal genes, such as Dsg2, raising the possibility that Pitx2c+/− mice may also be prone to structural remodeling. Taken together with previous studies, our findings reveal that Pitx2 has both developmental and postnatal functions that predispose the heart to atrial arrhythmias.
Pitx2 regulates intercalated disc maturation and homeostasis
Our array data reveal that major changes in gene expression were detected at 12 weeks of age. Moreover, ChIP-Seq data support the model that Pitx2 directly regulates genes involved in ID homeostasis. Structural remodeling of intercellular connections and an increase in fibrosis represent known anatomic substrates for AF. Notably, Pitx2 mutants have upregulated expression of ID genes suggesting that the correct stiochiometry of ID component genes is important to maintain ID structure. The immunofluorescence showing β-catenin mislocalization and TEM revealing breakdown of intercellular junctions also strongly support the notion that Pitx2 is required for ID maturation and homeostasis.
In the developing and postnatal heart, ID components including desmosomes, gap junctions, and adherens junctions are initially expressed in a dispersed pattern. As the heart matures, ID components become localized into the ID at the cardiomyocyte termini 41. At the transcriptional level, expression of genes such as Emd and Gja1 is reduced as the heart matures42, 43. Intriguingly, Emd is known to function as a β-catenin interacting protein that also modulates β-catenin intracellular localization 44. It is conceivable that lateralized β-catenin that we observe in the Pitx2 mutant left atrium may be due to elevated Emd levels. Together, our data indicate that Pitx2 is required for this transcriptional downregulation of ID component genes in left atrium myocardium.
Our findings have implications for AF progression to chronic AF. The severe anatomic abnormalities observed in Pitx2 CKO hearts suggest that these are irreversible structural defects. Moreover, the TEM data suggest that the Pitx2 CKO hearts have deteriorating mitochondria suggesting cellular compromise further supporting the notion that Pitx2 CKO arrhythmias are irreversible. The TEM data also revealed that Pitx2 CKO atrial cardiomyocytes had Z-line defects suggesting that Pitx2 also regulates sarcomere homeostasis. Interestingly, we previously observed formation of M-lines and more mature sarcomeric structure in Pitx2 CKO extraocular muscle, that normally lacks M-lines, suggesting context dependent functions for Pitx2 in sarcomere regulation 45.
Pitx2 and ion current regulation
Our data support previous observations that multiple genes involved in calcium handling and potassium channels are changed in Pitx2 mutants 6. The ChIP-Seq data indicate that multiple genes that compose the calcium release unit such as Ryr2, Jph2, and Atp2a2 are directly regulated by Pitx2 46. Genes involved in calcium homeostasis have been implicated in both AF initiation and progression to chronic AF 47. For example, Ca+2/calmodulin dependent protein kinase II delta (CaMKII delta) has been shown to phosphorylate the Ryanodine Receptor 2 (RyR2) at S2814 resulting in incomplete RyR2 closure with calcium leak. Increased phosphorylation of RyR2 has been demonstrated in human patients with chronic AF, as well as, other AF models 48, 49. Our ChIP-Seq/microarray datasets indicate that RyR2 is upregulated in Pitx2 mutant adult hearts suggesting that in addition to phosphorylation transcriptional regulation may be a regulatory node for modulating calcium handling in atrial cardiomyocytes.
In Pitx2 CKO postnatal hearts the inward rectifier Kcnj11 was both directly bound by Pitx2 and upregulated at the mRNA level, a likely AF risk factor 50. This upregulation is consistent with the shortened action potential duration that has been previously described in Pitx2 mutants 5, 6.
Pitx2 and AF in the developing and postnatal heart
Our findings suggest that postnatal atrial cardiomyocytes require persistent Pitx2 expression for ID homeostasis and maturation. Pitx2 regulates separate mechanisms, a developmental mechanism and a postnatal homeostatic mechanism, that result in an arrhythmogenic substrate. This is also reflected in the expression dynamics of Pitx2 target genes in the developmental and postnatal contexts. Scn5a and Kcnj10 are downregulated in Pitx2 mutant atrium during development but upregulated in postnatal mutants 5. One explanation for this is that Pitx2 may have separate cofactors at developmental and postnatal stages that result in distinct transcriptional readouts.
Our findings also have implications for human AF. Previous GWAS implicated multiple genes as potential contributors to the pathogenesis of human AF and prolonged PR interval 2. Our Pitx2 ChIP-Seq data identified several of these genes as potential Pitx2 target genes in postnatal hearts (Table S2)2, 26. Since Pitx2 regulates many of these genes in the postnatal and adult heart, it is conceivable that drugs can be developed to modulate the molecular interaction between Pitx2 and its target genes. Since fetal therapies currently lack feasibility, our findings that many important Pitx2 regulated events occur postnatally strengthens the likelihood that Pitx2 mediated AF may be treatable in the future.
Supplementary Material
Acknowledgments
We thank Dr. AJ Marian for comments on the manuscript.
Funding Sources: This work was supported by NIH grants: 1F32HL105041 (YT); 5R01HL093484-04 (J.F.M.); R01 EY-015306 (HK); 5R01HD044157 (AMM).
Footnotes
Conflict of Interest Disclosures: None.
References
- 1.Benjamin EJ, Chen PS, Bild DE, Mascette AM, Albert CM, Alonso A, et al. Prevention of atrial fibrillation: Report from a national heart, lung, and blood institute workshop. Circulation. 2009;119:606–618. doi: 10.1161/CIRCULATIONAHA.108.825380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mahida S, Ellinor PT. New advances in the genetic basis of atrial fibrillation. J Cardiovasc Electrophysiol. 2012;23:1400–1406. doi: 10.1111/j.1540-8167.2012.02445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Husser D, Adams V, Piorkowski C, Hindricks G, Bollmann A. Chromosome 4q25 variants and atrial fibrillation recurrence after catheter ablation. J Am Coll Cardiol. 2010;55:747–753. doi: 10.1016/j.jacc.2009.11.041. [DOI] [PubMed] [Google Scholar]
- 4.Wang J, Klysik E, Sood S, Johnson RL, Wehrens XH, Martin JF. Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc Natl Acad Sci U S A. 2010;107:9753–9758. doi: 10.1073/pnas.0912585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chinchilla A, Daimi H, Lozano-Velasco E, Dominguez JN, Caballero R, Delpon E, et al. Pitx2 insufficiency leads to atrial electrical and structural remodeling linked to arrhythmogenesis. Circ Cardiovasc Genet. 2011;4:269–279. doi: 10.1161/CIRCGENETICS.110.958116. [DOI] [PubMed] [Google Scholar]
- 6.Kirchhof P, Kahr PC, Kaese S, Piccini I, Vokshi I, Scheld HH, et al. Pitx2c is expressed in the adult left atrium, and reducing pitx2c expression promotes atrial fibrillation inducibility and complex changes in gene expression. Circ Cardiovasc Genet. 2011;4:123–133. doi: 10.1161/CIRCGENETICS.110.958058. [DOI] [PubMed] [Google Scholar]
- 7.Ammirabile G, Tessari A, Pignataro V, Szumska D, Sutera Sardo F, Benes J, Jr., et al. Pitx2 confers left morphological, molecular, and functional identity to the sinus venosus myocardium. Cardiovasc Res. 2012;93:291–301. doi: 10.1093/cvr/cvr314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haissaguerre M, Jais P, Shah DC, Takahashi A, Hocini M, Quiniou G, et al. Spontaneous initiation of atrial fibrillation by ectopic beats originating in the pulmonary veins. N Engl J Med. 1998;339:659–666. doi: 10.1056/NEJM199809033391003. [DOI] [PubMed] [Google Scholar]
- 9.Katritsis DG, Giazitzoglou E, Korovesis S, Karvouni E, Anagnostopoulos CE, Camm AJ. Conduction patterns in the cardiac veins: Electrophysiologic characteristics of the connections between left atrial and coronary sinus musculature. J Interv Card Electrophysiol. 2004;10:51–58. doi: 10.1023/B:JICE.0000011485.98197.df. [DOI] [PubMed] [Google Scholar]
- 10.Lin WS, Tai CT, Hsieh MH, Tsai CF, Lin YK, Tsao HM, et al. Catheter ablation of paroxysmal atrial fibrillation initiated by non-pulmonary vein ectopy. Circulation. 2003;107:3176–3183. doi: 10.1161/01.CIR.0000074206.52056.2D. [DOI] [PubMed] [Google Scholar]
- 11.Everett THt, Olgin JE. Atrial fibrosis and the mechanisms of atrial fibrillation. Heart Rhythm. 2007;4:S24–27. doi: 10.1016/j.hrthm.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng S, Keyes MJ, Larson MG, McCabe EL, Newton-Cheh C, Levy D, et al. Long-term outcomes in individuals with prolonged pr interval or first-degree atrioventricular block. JAMA. 2009;301:2571–2577. doi: 10.1001/jama.2009.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts-Thomson KC, Sanders P, Kalman JM. Sinus node disease: An idiopathic right atrial myopathy. Trends Cardiovasc Med. 2007;17:211–214. doi: 10.1016/j.tcm.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 14.Lee JM, Kalman JM. Sinus node dysfunction and atrial fibrillation: Two sides of the same coin? Europace. 2013;15:161–162. doi: 10.1093/europace/eus223. [DOI] [PubMed] [Google Scholar]
- 15.Hsu J, Hanna P, Van Wagoner DR, Barnard J, Serre D, Chung MK, et al. Whole genome expression differences in human left and right atria ascertained by rna sequencing. Circ Cardiovasc Genet. 2012;5:327–335. doi: 10.1161/CIRCGENETICS.111.961631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu C, Liu W, Palie J, Lu MF, Brown NA, Martin JF. Pitx2c patterns anterior myocardium and aortic arch vessels and is required for local cell movement into atrioventricular cushions. Development. 2002;129:5081–5091. doi: 10.1242/dev.129.21.5081. [DOI] [PubMed] [Google Scholar]
- 17.Balse E, Steele DF, Abriel H, Coulombe A, Fedida D, Hatem SN. Dynamic of ion channel expression at the plasma membrane of cardiomyocytes. Physiol Rev. 2012;92:1317–1358. doi: 10.1152/physrev.00041.2011. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Y, Cheng G, Dieter L, Hjalt TA, Andrade FH, Stahl JS, et al. An altered phenotype in a conditional knockout of pitx2 in extraocular muscle. Invest Ophthalmol Vis Sci. 2009;50:4531–4541. doi: 10.1167/iovs.08-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 20.van der Laan MJ, Pollard KS. A new algorithm for hybrid hierarchical clustering with visualization and the bootstrap. Journal of Statistical Planning and Inference. 2003;117:275–303. [Google Scholar]
- 21.Zambon AC, Gaj S, Ho I, Hanspers K, Vranizan K, Evelo CT, et al. Go-elite: A flexible solution for pathway and ontology over-representation. Bioinformatics. 2012;28:2209–2210. doi: 10.1093/bioinformatics/bts366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and b cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bruning JC, Michael MD, Winnay JN, Hayashi T, Horsch D, Accili D, et al. A muscle-specific insulin receptor knockout exhibits features of the metabolic syndrome of niddm without altering glucose tolerance. Mol Cell. 1998;2:559–569. doi: 10.1016/s1097-2765(00)80155-0. [DOI] [PubMed] [Google Scholar]
- 24.L’Honore A, Coulon V, Marcil A, Lebel M, Lafrance-Vanasse J, Gage P, et al. Sequential expression and redundancy of pitx2 and pitx3 genes during muscle development. Dev Biol. 2007;307:421–433. doi: 10.1016/j.ydbio.2007.04.034. [DOI] [PubMed] [Google Scholar]
- 25.Amendt BA, Sutherland LB, Semina EV, Russo AF. The molecular basis of rieger syndrome. Analysis of pitx2 homeodomain protein activities. J Biol Chem. 1998;273:20066–20072. doi: 10.1074/jbc.273.32.20066. [DOI] [PubMed] [Google Scholar]
- 26.Pfeufer A, van Noord C, Marciante KD, Arking DE, Larson MG, Smith AV, et al. Genome-wide association study of pr interval. Nat Genet. 2010;42:153–159. doi: 10.1038/ng.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfeufer A, Sanna S, Arking DE, Muller M, Gateva V, Fuchsberger C, et al. Common variants at ten loci modulate the qt interval duration in the qtscd study. Nat Genet. 2009;41:407–414. doi: 10.1038/ng.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newton-Cheh C, Eijgelsheim M, Rice KM, de Bakker PI, Yin X, Estrada K, et al. Common variants at ten loci influence qt interval duration in the qtgen study. Nat Genet. 2009;41:399–406. doi: 10.1038/ng.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gudbjartsson DF, Holm H, Gretarsdottir S, Thorleifsson G, Walters GB, Thorgeirsson G, et al. A sequence variant in zfhx3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet. 2009;41:876–878. doi: 10.1038/ng.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holm H, Gudbjartsson DF, Arnar DO, Thorleifsson G, Thorgeirsson G, Stefansdottir H, et al. Several common variants modulate heart rate, pr interval and qrs duration. Nat Genet. 2010;42:117–122. doi: 10.1038/ng.511. [DOI] [PubMed] [Google Scholar]
- 31.Chen YH, Xu SJ, Bendahhou S, Wang XL, Wang Y, Xu WY, et al. Kcnq1 gain-of-function mutation in familial atrial fibrillation. Science. 2003;299:251–254. doi: 10.1126/science.1077771. [DOI] [PubMed] [Google Scholar]
- 32.Ellinor PT, Lunetta KL, Albert CM, Glazer NL, Ritchie MD, Smith AV, et al. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet. 2012;44:670–675. doi: 10.1038/ng.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benjamin EJ, Rice KM, Arking DE, Pfeufer A, van Noord C, Smith AV, et al. Variants in zfhx3 are associated with atrial fibrillation in individuals of european ancestry. Nat Genet. 2009;41:879–881. doi: 10.1038/ng.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Z, He Y, Tuteja D, Xu D, Timofeyev V, Zhang Q, et al. Functional roles of cav1.3(alpha1d) calcium channels in atria: Insights gained from gene-targeted null mutant mice. Circulation. 2005;112:1936–1944. doi: 10.1161/CIRCULATIONAHA.105.540070. [DOI] [PubMed] [Google Scholar]
- 35.Sotoodehnia N, Isaacs A, de Bakker PI, Dorr M, Newton-Cheh C, Nolte IM, et al. Common variants in 22 loci are associated with qrs duration and cardiac ventricular conduction. Nat Genet. 2010;42:1068–1076. doi: 10.1038/ng.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baig SM, Koschak A, Lieb A, Gebhart M, Dafinger C, Nurnberg G, et al. Loss of ca(v)1.3 (cacna1d) function in a human channelopathy with bradycardia and congenital deafness. Nat Neurosci. 2011;14:77–84. doi: 10.1038/nn.2694. [DOI] [PubMed] [Google Scholar]
- 37.Liu F, Levin MD, Petrenko NB, Lu MM, Wang T, Yuan LJ, et al. Histone-deacetylase inhibition reverses atrial arrhythmia inducibility and fibrosis in cardiac hypertrophy independent of angiotensin. J Mol Cell Cardiol. 2008;45:715–723. doi: 10.1016/j.yjmcc.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen T, Aneas I, Sakabe N, Dirschinger RJ, Wang G, Smemo S, et al. Tbx20 regulates a genetic program essential to adult mouse cardiomyocyte function. J Clin Invest. 2011;121:4640–4654. doi: 10.1172/JCI59472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qi Y, Ranish JA, Zhu X, Krones A, Zhang J, Aebersold R, et al. Atbf1 is required for the pit1 gene early activation. Proc Natl Acad Sci U S A. 2008;105:2481–2486. doi: 10.1073/pnas.0712196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilde AA, Brugada R. Phenotypical manifestations of mutations in the genes encoding subunits of the cardiac sodium channel. Circ Res. 2011;108:884–897. doi: 10.1161/CIRCRESAHA.110.238469. [DOI] [PubMed] [Google Scholar]
- 41.Angst BD, Khan LU, Severs NJ, Whitely K, Rothery S, Thompson RP, et al. Dissociated spatial patterning of gap junctions and cell adhesion junctions during postnatal differentiation of ventricular myocardium. Circ Res. 1997;80:88–94. doi: 10.1161/01.res.80.1.88. [DOI] [PubMed] [Google Scholar]
- 42.Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wheeler MA, Warley A, Roberts RG, Ehler E, Ellis JA. Identification of an emerin-beta-catenin complex in the heart important for intercalated disc architecture and beta-catenin localisation. Cell Mol Life Sci. 2010;67:781–796. doi: 10.1007/s00018-009-0219-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou Y, Gong B, Kaminski HJ. Genomic profiling reveals pitx2 controls expression of mature extraocular muscle contraction-related genes. Invest Ophthalmol Vis Sci. 2012;53:1821–1829. doi: 10.1167/iovs.12-9481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dobrev D, Voigt N, Wehrens XH. The ryanodine receptor channel as a molecular motif in atrial fibrillation: Pathophysiological and therapeutic implications. Cardiovasc Res. 2011;89:734–743. doi: 10.1093/cvr/cvq324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sood S, Chelu M, Van Oort R, Skapura D, Santonastasi M, Dobrev D, et al. Intracellular calcium leak due to fkbp12.6 deficiency in mice facilitates the inducibility of atrial fibrillation. Heart Rhythm. 2008;5:1047–1054. doi: 10.1016/j.hrthm.2008.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chelu MG, Sarma S, Sood S, Wang S, van Oort RJ, Skapura DG, et al. Calmodulin kinase ii-mediated sarcoplasmic reticulum ca2+ leak promotes atrial fibrillation in mice. J Clin Invest. 2009;119:1940–1951. doi: 10.1172/JCI37059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dobrev D, Wehrens XH. Calmodulin kinase ii, sarcoplasmic reticulum ca2+ leak, and atrial fibrillation. Trends Cardiovasc Med. 2010;20:30–34. doi: 10.1016/j.tcm.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kharche S, Garratt CJ, Boyett MR, Inada S, Holden AV, Hancox JC, et al. Atrial proarrhythmia due to increased inward rectifier current (i(k1)) arising from kcnj2 mutation--a simulation study. Prog Biophys Mol Biol. 2008;98:186–197. doi: 10.1016/j.pbiomolbio.2008.10.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.