Abstract
The molecular basis of CNS myelin regeneration (remyelination) is poorly understood. We generated a comprehensive transcriptional profile of the separate stages of spontaneous remyelination that follow focal demyelination in the rat CNS and found that transcripts that encode the retinoid acid receptor RXR-γ were differentially expressed during remyelination. Cells of the oligodendrocyte lineage expressed RXR-γ in rat tissues that were undergoing remyelination and in active and remyelinated multiple sclerosis lesions. Knockdown of RXR-γ by RNA interference or RXR-specific antagonists severely inhibited oligodendrocyte differentiation in culture. In mice that lacked RXR-γ, adult oligodendrocyte precursor cells efficiently repopulated lesions after demyelination, but showed delayed differentiation into mature oligodendrocytes. Administration of the RXR agonist 9-cis-retinoic acid to demyelinated cerebellar slice cultures and to aged rats after demyelination caused an increase in remyelinated axons. Our results indicate that RXR-γ is a positive regulator of endogenous oligodendrocyte precursor cell differentiation and remyelination and might be a pharmacological target for regenerative therapy in the CNS.
Introduction
Following acute demyelination in the CNS, adult oligodendrocyte precursor cells (OPCs) can migrate to the area of injury, differentiate into oligodendrocytes and restore myelin sheaths1, 2, 3. However, this natural regenerative process, or spontaneous remyelination, is limited in demyelinating diseases such as multiple sclerosis4, 5, owing in part to the failure of adult OPCs to differentiate into myelinating oligodendrocytes6, 7, 8. The failure to restore CNS myelin after injury compromises the integrity of axons and leaves them vulnerable to degeneration9. Although the genes that regulate the proliferation and differentiation of OPCs during development have been intensively studied, relatively little is known about the molecular signals that regulate the function of adult OPCs after demyelination. The identification of key signaling networks associated with remyelination would improve our understanding of how OPCs respond to injury, and help researchers to identify pharmacological targets for the development of regenerative therapeutics that could encourage myelin regeneration10, 11.
We have used a well-established and highly tractable toxin-induced demyelination method in rats12, combined with laser capture microdissection (LCM) and microarray analysis of selectively isolated lesions, to generate a complete transcriptome of the separate stages of spontaneous CNS remyelination. We found that the transcript that encodes RXR-γ was substantially upregulated during the regenerative phase of remyelination and we detected RXR-γ expression in oligodendrocyte lineage cells in remyelinating lesions in the rat CNS and in tissue from individuals with multiple sclerosis. By using pharmacological and genetic manipulation methods, we found that activation of RXR stimulated oligodendrocyte differentiation to enhance remyelination. RXR signaling therefore represents a regenerative therapeutic target for promoting CNS remyelination in the demyelinated brain.
Results
Increased Rxrg transcripts in CNS remyelinating lesions
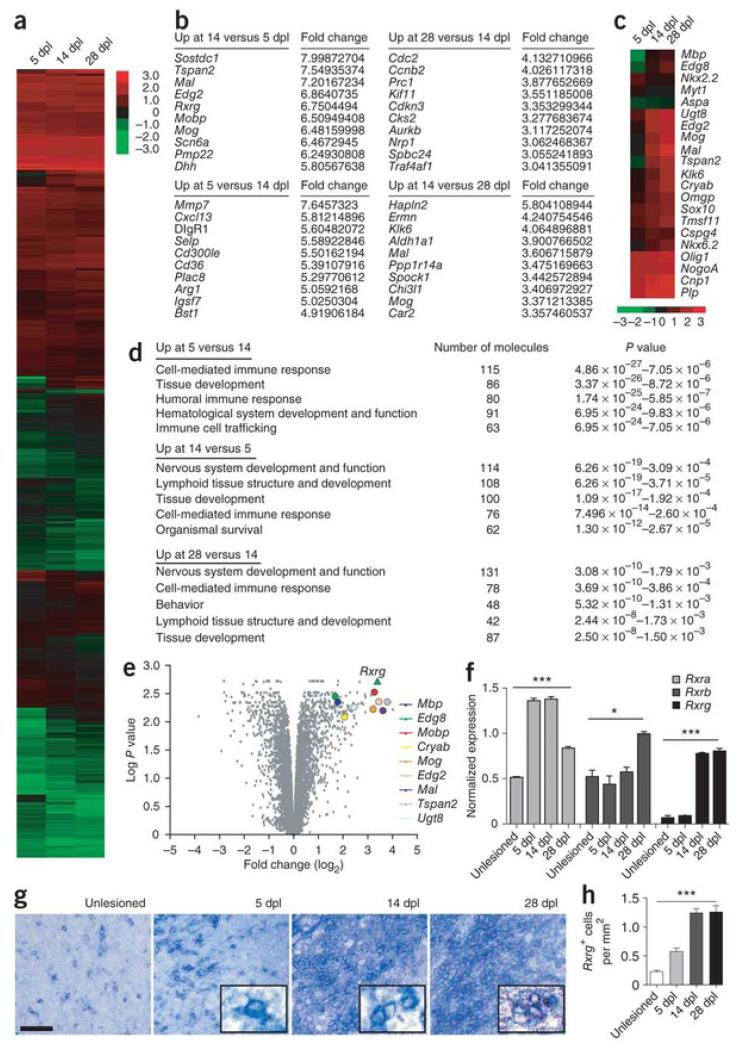
We induced focal demyelinations in the rat caudal (inferior) cerebellar peduncle (CCP)12 and isolated lesioned tissues at 5, 14 and 28 days post-lesion (dpl) using LCM. For microarray analysis, we used three independently lesioned rats per time point to provide three biological replicates. We hybridized labeled RNAs onto the Illumina Rat RefSeq chip, which contains more than 22,000 genes, and analyzed them using the Illumina BeadStudio and R statistical tools (lumi, limma and fspma packages). We identified 8,754 genes that were differentially expressed (3,197 genes with P < 0.05) over the three post-lesion time points (Fig. 1a and Supplementary Table 1). The genes that were most highly expressed at 5 dpl compared with 14 or 28 dpl were associated with inflammation, including Mmp7, Cxcl13 and Arg1, whereas the genes that were most highly expressed at 14 dpl compared with 5 dpl were associated with myelination, including Tspan2, Mal, Lpar1 (also called Edg2), Mobp and Mog (P < 0.05; Fig. 1b). Indeed, an analysis of known genes involved in myelination revealed that most showed increased expression at 14 or 28 dpl compared with 5 dpl (Fig. 1c). We also found that genes that are specific to the OPC lineage, such as Nkx2-2 and Myt1, showed decreased expression at 14 dpl compared with 5 dpl, which suggests that the OPC population had differentiated into oligodendrocytes by 14 dpl.
Figure 1. Differential expression of Rxrg in CNS remyelination transcriptome.
(a) Hierarchical clustering and graphical analysis of differentially expressed genes at 5, 14 and 28 dpl (P < 0.05). (b) Ten most upregulated genes at each time point relative to the other time points. (c) Graphical analysis showing the differential expressions of known genes associated with myelination (P < 0.05). (d) Top five overall physiological functions in lesions at 5, 14 and 28 dpl using Ingenuity pathway analysis of upregulated genes from each time point. (e) Volcano plot (x axis = log2 FC at 14 dpl compared to 5 dpl; y axis = log2 P) showing highly differentially expressed genes associated with myelination genes. Rxrg (green triangle; x, y = 3.3752, 2.7084) is shown as a highly expressed transcript at 14 dpl compared to 5 dpl. (f) Real-time qPCR detection of Rxra, Rxrb and Rxrg expression from laser-captured lesions during remyelination (n = 3). Rxrg is barely detectable in non-lesioned CCPs and at 5 dpl, and highly expressed at 14 and 28 dpl. (g) In situ hybridization shows significant increase of Rxrg+ cells in the CCP at 14 dpl and 28 dpl compared to non-lesioned and 5 dpl CCP. Scale bar, 50 μm. (h) Quantification of Rxrg+ cells in lesioned CCPs at 5, 14 and 28 dpl (n = 3 per time point). Mean ± s.e.m. are shown. *P < 0.05, ***P < 0.001, one-way ANOVA.
We next performed ingenuity pathway analysis (IPA) by submitting the list of genes that were differentially expressed between 5 and 14 dpl and between 14 and 28 dpl to elucidate the overall physiology of remyelination and active signaling pathways associated with each regeneration time point. The top physiological systems networks at 5 dpl, based on genes that showed higher expression at 5 dpl than at 14 dpl, involved the immune response, which indicates the presence of active inflammation (Fig. 1d and Supplementary Table 2). Moreover, the top networks at 14 and at 28 dpl, based on genes that were upregulated at 14 dpl compared with 5 dpl and at 28 dpl compared with 14 dpl, involved nervous system development and function, with a decreased immune response, which indicates that remyelination activity was increasing. The most significant pathways (P < 0.001) that were detected at 5 dpl were associated with macrophage and inflammation activities, such as Fcγ receptor–mediated phagocytosis in macrophages or monocytes and interleukin signaling, whereas the most significant signaling pathways (P < 0.001) at 14 dpl were related to cell metabolism and proliferation or differentiation; these included inositol phosphate metabolism and Notch signaling pathways (Supplementary Table 3). These results show that the overall molecular signature of CNS remyelination involves distinct and temporally regulated signaling pathways that are characterized by active inflammation at 5 dpl and by the initiation of remyelination at 14 dpl.
To identify genes that are potentially involved in CNS remyelination, we performed a volcano plot analysis by plotting the fold change (log2 FC) of genes that were differentially expressed between 5 and 14 dpl against their significance (log P). We hypothesize, on the basis of our transcriptome analysis, that gene pathways that are upregulated or enriched at 14 dpl stimulate remyelination (that is, OPC differentiation). We found that retinoid X receptor gamma (Rxrg) was one of the most significantly upregulated genes at 14 dpl (log2 FC = 3.375; log P = 2.708) and clustered with many genes that are involved in myelination (Fig. 1e and Supplementary Table 4). RXR-γ and the other RXR members, RXR-α and RXR-β, are nuclear receptors that work through heterodimeric association with other nuclear receptors, such as retinoic acid receptors (RARs), thyroid hormone receptors, vitamin D receptors (VDRs), peroxisome proliferator activator receptors (PPARs) and liver X receptors (LXRs) to regulate cell proliferation, differentiation and apoptosis13. Rxra and Rxrb were also differentially expressed over the three post-lesion time points in our remyelination transcriptome, although the difference in expression was smaller than for Rxrg. We also detected the differential expression of genes that heterodimerize with RXR14, including Thra, Thrb, Nr1h3 (LXRα), Nr2f1 (COUP-TFI) and Nr4a2 (Nurr1), which supports the idea that RXR signaling is involved in remyelination (Supplementary Table 5). Moreover, IPA on the full list of differentially expressed genes from all three post-lesion time points showed that pathways associated with RXR signaling were enriched (Table 1, Supplementary Fig. 1 and Supplementary Table 6), which suggests that RXR signaling is highly active in lesions and might contribute to remyelination.
Table 1. Genes associated with RXR signaling during remyelination.
| RXR pathways | −Log(P) | Ratio | Genes |
|---|---|---|---|
|
| |||
| LXR/RXR activation | 1.12 × 10−10 | 4.49 × 10−1 | Il1a, Lcat, Ccl7, Tnfrsf11b, Rela, Hadh, Lpl, Rxra, Cd14, Cd36, Srebf1, Echs1, Ngfr, Hmgcr, Lbp,Il1r2, Tnfrsf1a, Rxrb, Ncor1, Nr1h2, Mmp9, Tlr4, Il18, Ccl2, Apoc1, Abcg 1, Apoe, Irf3, Rxrg, Il1b,Nr1h3 |
| VDR/RXR activation | 1.22 × 10−1 | 4.81 × 10−1 | Tgfb2, Prkch, Gadd45a, Prkcd, Cebpa, Rxra, Hes1, Il12a, Gtf2b, Cd14, Prkcz, Prkca , Prkce, Prkcq,Trpv6, Klk6, Igfbp6, Calb1, Pdgfa, Prkcb, Hr, Psmc5, Cxcl10, Yy1, Rx rb, Foxo1, Spp1, Ncor1, Ccl5,Cebpb, Cdkn1b, Csnk2a1, Klf4, Cyp24a1, Ccnc, Rxrg, Igfbp3 |
| LPS/IL-1–mediated inhibition of RXR function | 1.42 × 10−1 | 3.32 × 10−1 | Aldh1a2, Tnfrsf11b, Sod3, Abcb1, Rxra, Cpt1a, Gstt1, Gstm1, Aldh3b1, Ngfr, Acox1, Il1r2, Ces2,Myd88, Nr1h2, Tlr4, Slc27a1, Apoc1, Apoe, Map3k1, Fabp7, Xpo1, Scar b1, Acsl4, Acsl3, Fmo4,Rara, Ndst1, Maoa, Il1b, Nr1h3, Cpt2, Maob, Abcb9, Hmgcs 2, Fabp3, Sult1a1, Cd14, Aldh3a2,Srebf1, Gstk1, Mgmt, Cat, Gsto1, Aldh1a1, Lbp, Abcc3, Acsl5, Acsl6, Tnfrsf1a, Hs3st1, Chst10,Gstm3, Gstp1, Aldh9a1, Jun, Abcg1, Mgst1, Acsl1, Gstt2, Fabp4, Hmgcs1 |
| PPARα/RXR activation | 1.74 × 10−1 | 3.86 × 10−1 | Tgfb2, Adcy3, Gpd2, Gna11, Smad4, Prkar1a, Cand1, Adcy4, Rxra, Prkca, Tgfb1, Pl cb3, Nfkbia,Tgfbr2, Mapk14, Nras, Acox1, Il1r2, Prkcb, Prkag1, Prkaca, Plcb1, Mapk 1, Stat5b, Map2k6,Prkar2b, Raf1, Tgfbr1, Slc27a1, Nfkbib, Acvr2b, Pdia3, Acadl, Plc g2, Il1b, Mapk3, Rela, Lpl, Adcy5,Hsp90aa1, Map2k2, Smad2, Adcy2, Prkag2, Cd36 , Plcd4, Plcg1, Got2, Prkaa1, Mras, Shc1, Kras,Adcy8, Acaa1, Ikbkb, Plcl1, Ncor1, G pd1, Jak2, Jun, Ncoa6, Map2k1, Plcd1, Plcb2 |
| CAR/RXR activation | 2.16 | 3.1 × 10−1 | Sult1a1, Abcc3, Rxrb, Mcl1, Ccnd1, Abcb1, Gadd45b, Aldh1a1, Rxra |
| FXR/RXR activation | 2.69 | 2.41 × 10−1 | Il1a, Foxo1, Mapk12, Sdc1, Vldlr, Rxra, Il18, Akt1, Cyp27a1, Srebf1, Slc4a2, Apoe, Cyp8b1, Scarb1,Pon1, Rara, G6pc3, Abcb4, Il1b, Nr1h3 |
| PXR/RXR activation | 3.74 | 2.79 × 10−1 | Abcc3, Ces2, Prkag1, Prkaca, Abcb9, Hmgcs2, Foxo1, Prkar1a, Rela, Abcb1, Prkar 2b, Rxra, Akt1,Cpt1a, Prkag2, Gstm1, Aldh3a2, Nr3c1, Aldh1a1 |
| TR/RXR activation | 6.19 | 3.49 × 10−1 | Klf9, Me1, Thrsp, Frap1, Hif1a, Pfkp, Rxra, Pik3c3, Eno1, Srebf1, Rcan2, Trh, Strbp, Ucp2, F10,Rxrb, Slc16a3, Pik3ca, Ncor1, Thrb, Thra, Akt1, Pik3cb, Nrgn, Hdac3, Nc oa6, Pik3r2, Scarb1, Rxrg |
To validate RXR expression in remyelination, we performed real-time quantitative polymerase chain reaction (qPCR) on reverse-transcribed mRNAs isolated from non-lesioned CCPs and lesioned CCPs at 5 dpl, 14 dpl and 28 dpl by LCM (Fig. 1f). The expression of Rxra, Rxrb and Rxrg was consistent with their differential expression in silico. In particular, Rxrg was barely expressed at 5 dpl, but showed markedly increased expression (by nearly eightfold) at 14 dpl and remained highly expressed at 28 dpl. Moreover, in situ hybridization analysis of focally demyelinated rat brains showed that the density of Rxrg-expressing cells in lesions was at least 3 times greater at 14 and 28 dpl than at 5 dpl (Fig. 1g,h), supporting the idea that it was actively expressed at the onset of remyelination.
RXR-γ expression in oligodendrocyte lineage cells
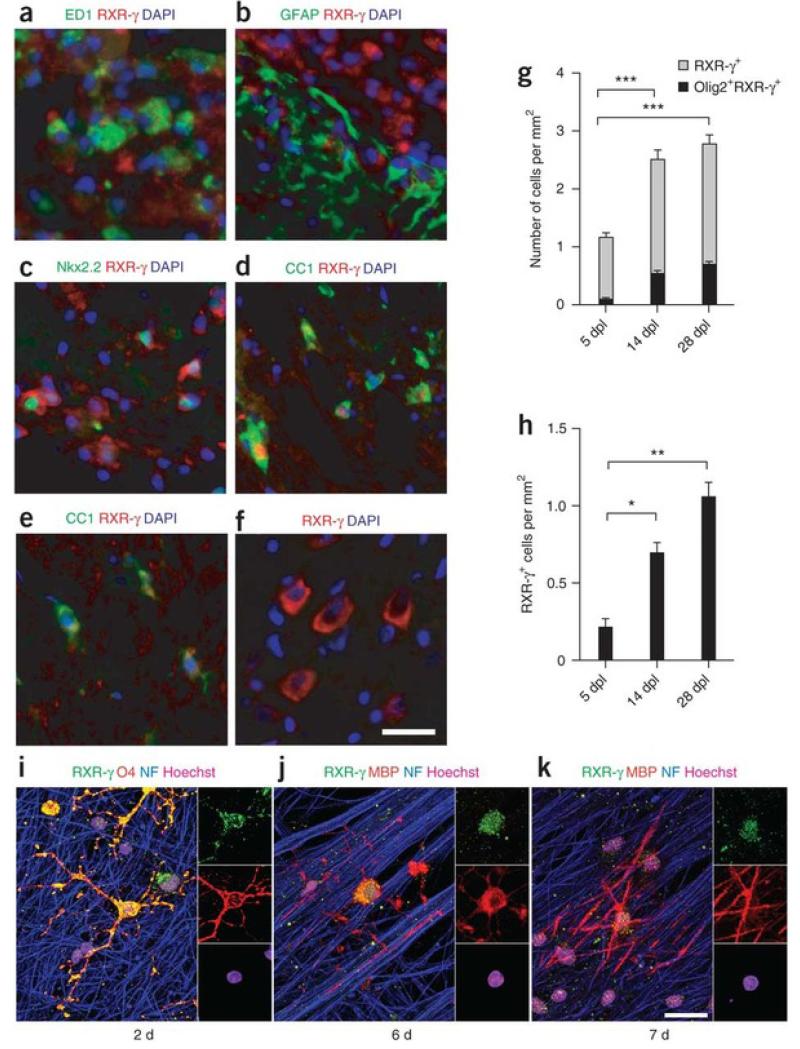
The remyelination environment is composed of demyelinated axons, activated adult OPCs, regenerated oligodendrocytes, microglia or macrophages and reactive astrocytes15. To determine which cells expressed RXR-γ, we performed immunfluorescence analysis on focally demyelinated rat brains. RXR-γ was detected predominantly in the cytosol of macrophages expressing the marker ED1+(Fig. 2a) and rarely in reactive astrocytes (Fig. 2b). We also found RXR-γ in the cytosol of Nkx2.2+ OPCs (Fig. 2c) and in the nuclei of CC1+ oligodendrocytes (Fig. 2d), suggesting that it might be translocated to influence OPC differentiation. In non-lesioned white matter, RXR-γ was faintly detectable in CC1+ oligodendrocytes (Fig. 2e). In non-lesioned gray matter, RXR-γ was most strongly expressed in neurons, such as in the striatum (Fig. 2f). These results confirm that RXR-γ is highly expressed in the injured environment. To determine what percentage of RXR-γ+ cells in lesions are oligodendrocyte lineage cells, we performed in situ hybridization against Rxrg followed by immunoperoxidase staining using antibodies to the oligodendrocyte lineage marker Olig2 (Supplementary Fig. 2). Oligodendrocyte lineage cells (Olig2+ RXR-γ+) represented about 8.7% of RXR-γ+cells at 5 dpl, 21.5% at 14 dpl and 25.5% at 28 dpl (Fig. 2g), suggesting that most RXR-γ+ cells might be macrophages. When we quantified RXR-γ+ oligodendrocytes in lesions, we found that the number of RXR-γ+ CC1+ cells increased significantly from 5 dpl to 14 dpl and 28 dpl (P = 0.0022), suggesting that OPC differentiation increased in the lesions over time (Fig. 2h).
Figure 2. RXR-γ expression by oligodendrocyte lineage cells.
There are several lines of evidence that RXR-γ might be involved in remyelination. After spinal cord contusions, all three RXR members become actively expressed in the cytosol of reactive microglia, neurons, astrocytes and oligodendrocytes, which suggests that RXR signaling is involved in the injury response of the damaged CNS16. Moreover, only RXR-γ was found to translocate from the oligodendrocyte cytosol to the nucleus after injury16. We also performed a database search through the recent detailed microarray analysis of purified oligodendrocyte lineage cells and found that RXR-γ is substantially enriched in purified OPCs17. To confirm that nuclear expression of RXR-γ correlates with OPC differentiation and myelination, we carried out immunostaining analysis on purified co-cultures of OPCs and dorsal root ganglion (DRG) neurons. After 2 d in co-culture, we detected RXR-γ predominantly in the cytosol of oligodendrocyte precursor cell bodies and processes (Fig. 2i). However, as these cells differentiated and began to myelinate, RXR-γ was no longer detected in the processes and became restricted to the cell bodies and nuclei (Fig. 2j,k), suggesting that RXR-γ redistributes to the nucleus during OPC differentiation.
RXR-γ expression in multiple sclerosis lesions
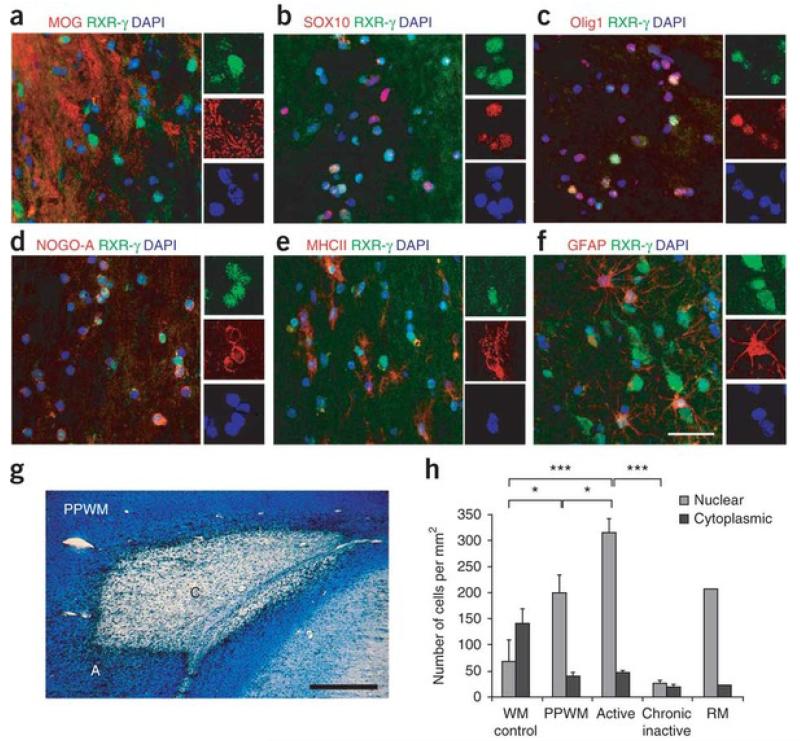
To examine RXR-γ expression in multiple sclerosis lesions, we performed immunostaining analyses on snap-frozen post-mortem brain sections from randomly chosen individuals with multiple sclerosis, including secondary progressive (two cases) and relapsing remitting (one case) and from three non-neurological controls (Supplementary Table 7). In the active borders of multiple sclerosis lesions, we found RXR-γ expression in either the nucleus or the cytosol of oligodendrocyte lineage cells (Fig. 3a–d). Quantification of RXR-γ+ cells in these lesions showed that 84.5 ± 5.7% of these cells expressed Olig1 in the nucleus, indicating that RXR-γ is expressed by activated OPCs that have probably migrated to lesions in response to demyelination in multiple sclerosis. We also found RXR-γin activated microglia or macrophages (Fig. 3e) and reactive astrocytes (Fig. 3f).
Figure 3. Expression of RXR-γ in multiple sclerosis lesions.
(a–f) Co-immunolabeling for RXR-γ (green) and (in red) MOG (a), Sox10 (b), Olig1 (c), NOG0-A (d), MHCII (e) or GFAP (f) in active multiple sclerosis lesion areas. Nuclei were visualized with Hoechst (blue). (g) Luxol fast blue staining followed by anti-MHCII immunoperoxidase labeling showing a typical chronic active multiple sclerosis lesion with active border (A) and chronic inactive core (C), as well as peri-plaque white matter (PPWM). (h) Quantification of nuclear and cytoplasmic RXR-γ+ cells in multiple sclerosis lesions reveals significantly more nuclear RXR-γ+ cells in active lesions, PPWM and remyelinated shadow plaques (RM) compared to chronic inactive lesions and normal appearing white matter (WM) from non-neurological cases. Scale bars: a–f, 50 μm; g, 2 mm. Mean ± s.e.m. are shown. *P < 0.05, ***P < 0.001; one-way ANOVA.
To determine the density of cells that expressed RXR-γ in active and inactive areas of damage, we used Luxol fast blue (LFB) staining of brain sections from individuals with multiple sclerosis followed by immunoperoxidase staining with the macrophage or monocyte marker MHCII to differentiate the active border (LFB+ MHCII+) from the chronic inactive core (LFB− MHC+) of lesions (Fig. 3g). We also quantified the number of RXR-γ+ cells in the peri-plaque white matter (PPWM) around the lesion, in a remyelinated shadow plaque lesion, and in control normal-appearing white matter from non-neurological cases. Active lesions and PPWM contained a significantly greater density (active lesions versus control: P < 0.001; PPWM vs control: P < 0.05) of RXR-γ+ cells than did control white matter (Fig. 3h). By contrast, chronic inactive core lesions contained significantly fewer RXR-γ+ cells (P < 0.05) than did normal-appearing white matter. We also detected substantially more nuclear than cytoplasmic RXR-γ expression in active lesions, PPWM and the remyelinated shadow plaque (>4-fold) than in chronic inactive cores. In contrast to the multiple sclerosis lesions, we detected more cytoplasmic than nuclear RXR-γ expression (>2-fold) in the control white matter, suggesting that RXR-γ may be sequestered in the cytoplasm of terminally differentiated cells in the normal adult CNS. The detection of nuclear RXR-γ in active lesions, shadow plaque and PPWM suggests these are areas of high cellular activity that might be associated with repair. Moreover, the diminished expression of RXR-γ in chronic inactive lesions correlates well with the impairment of remyelination in the progressive stages of multiple sclerosis, and therefore supports the idea that RXR signaling is involved in repairing demyelinated CNS axons.
RXR-γ loss-of-function impairs OPC differentiation
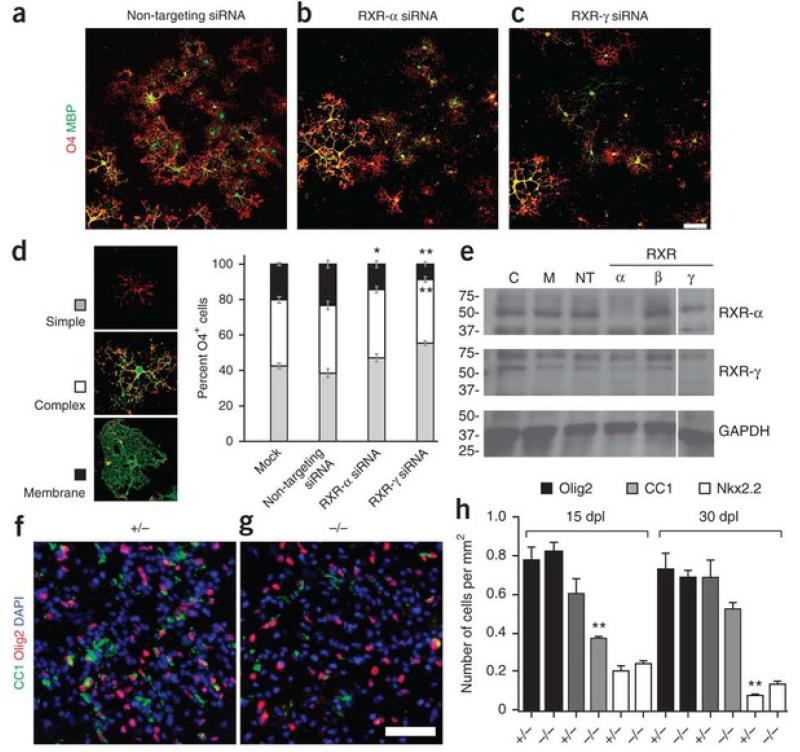
To determine whether RXR-γ regulates the differentiation of OPCs, we transfected cultured OPCs with non-targeting small inhibitory RNAs (siRNAs) as control (Fig. 4a), siRNAs against Rxra (Fig. 4b) or siRNAs against Rxrg (Fig. 4c). After 72 h in differentiation medium, oligodendrocytes that had been transfected with RXR-γsiRNA were less morphologically differentiated than controls. We immunostained oligodendrocyte lineage cells using the O4 monoclonal antibody, which recognizes both immature and differentiated oligodendrocytes, and an antibody to myelin basic protein (MBP), which marks only differentiated oligodendrocytes. We then determined the differentiation state of oligodendrocyte lineage cells on the basis of their morphologies as defined by multiple process outgrowth (simple), extensive process outgrowth and branching (complex) and terminal membrane expansion (membrane; Fig. 4d). Control non-targeting siRNAs did not influence OPC differentiation, as the percentages of O4+ oligodendrocytes with simple, complex or membrane morphologies appeared similar to those in mock, non-transfected oligodendrocytes (Fig. 4d). By contrast, cells transfected with RXR-γ siRNAs showed a significant increase in oligodendrocytes displaying simple morphologies (P < 0.001) and a decrease in those with membrane morphologies (P < 0.001), indicating a failure to achieve efficient differentiation. We also found that significantly fewer cells transfected with RXR-α siRNAs had MBP+ membrane sheets (P < 0.05), although not as few as cells transfected with RXR-γ siRNAs. Moreover, the percentage of MBP+ cells decreased by more than 30% in cells transfected with RXR-α or RXR-γ siRNA compared to controls (Supplementary Fig. 3). Immunostaining analysis revealed that oligodendrocyte lineage cells expressed both RXR-α and RXR-γ but hardly any RXR-β in culture (Supplementary Fig. 3). Western blot analysis confirmed that the siRNAs specifically knocked down expression of RXR-α and RXR-γ, respectively, and did not interfere with the expression of other RXR members (Fig. 4e and Supplementary Fig. 4). These results suggest that both RXR-α and RXR-γ are involved in stimulating oligodendrocyte differentiation.
Figure 4. Loss of RXR-γ function impairs oligodendrocyte differentiation.
(a–c) Purified OPCs transfected with non-targeting siRNAs (a), RXRα siRNAs (b) and RXR-γ siRNAs (c) and visualized with O4 (red) and antibodies to MBP (green) after 72 h in differentiation medium. Scale bar, 25 μm. (d) Morphological criteria for the maturation state of differentiating oligodendrocyte defined as simple, complex or membrane morphologies. Cells transfected with RXR-γ siRNAs resulted in increased percentage of O4+ oligodendrocytes with simple morphologies and decreased percentage of complex membrane morphologies compared to mock-treated and non-targeting siRNA–transfected cells. (e) Western blot shows the specificity of RXR-α or RXR-γknockdowns. The position of molecular weight standards (in kilodaltons) is shown on the left. Full-length blot presented in Supplementary Figure 4. C, untransfected control; M, mock-transfected; NT, non-targeting siRNA; RXRα, β, γ, RXRα, β or γ siRNA. (f,g) Ventral spinal cord lesions of Rxrg+/− (f) and Rxrg−/− (g) mice stained with antibodies to CC1 (green) and Olig2 (red) 15 d after demyelination. Nuclei visualized with Hoechst (blue). (h) Quantification of oligodendrocyte lineage cells at 15 and 30 dpl shows no difference in the density of total Olig2+ cells between homozygous and heterozygous mutant mice, but a reduction of CC1+ cells and increased Nkx2.2+ cells in lesions of homozygous compared to heterozygous mutant mice. Scale bar, 50 μm. Mean ± s.e.m. are shown. *P < 0.05, **P < 0.01; Student’s t test.
Previous studies have shown that mice that lack RXR-γ show no gross abnormalities during development and are viable after birth18. However, as adults these mutants show thyroid hormone resistance, changes in metabolic activity and depressive behaviors mediated by reduced dopaminergic signaling19, 20, 21, which suggests that RXR-γ regulates homeostatic functions in the adult CNS. To determine whether CNS remyelination requires RXR-γ, we performed focal demyelination on Rxrg−/− mice by injecting lysolecithin into the spinal cord ventral funiculus of adult mice and analyzed spinal cord lesions at 15 and 30 dpl. At 15 dpl, the density of ED1+ macrophages or activated microglia, GFAP+ reactive astrocytes and Olig2+ oligodendrocyte lineage cells were not substantially different in the lesions of Rxrg−/− mice compared with controls (Fig. 4f–h and Supplementary Fig. 5), which suggested that RXR-γ was not required for the recruitment of these cells into lesions. Moreover, there was no obvious difference between the two groups regarding oligodendrocyte lineage cell survival in lesions (Supplementary Fig. 5). By contrast, we found a significant reduction (P = 0.0094) in the number of CC1+ oligodendrocytes in the lesions of Rxrg−/− mice compared with controls (Fig. 4f–h). However, this decrease appeared to be transient as by 30 dpl the number of CC1+ cells in lesions in Rxrg−/− mice had increased almost to the level seen in control mice (Fig. 4h). We also detected more Nkx2.2+ OPCs inRxrg−/− mice than in controls at 15 and 30 dpl, suggesting that OPCs were less efficient at differentiating into oligodendrocytes in the absence of RXR-γ. We next performed semi-thin resin section analysis and found no obvious difference in remyelination between Rxrg−/− and control mice at 30 dpl (Supplementary Fig. 5). These results suggest that the loss of RXR-γ impairs OPC differentiation after demyelination, but also that compensatory mechanisms might eventually overcome the absence of RXR-γ signaling to regenerate myelinating oligodendrocytes.
RXR antagonists inhibit OPC differentiation
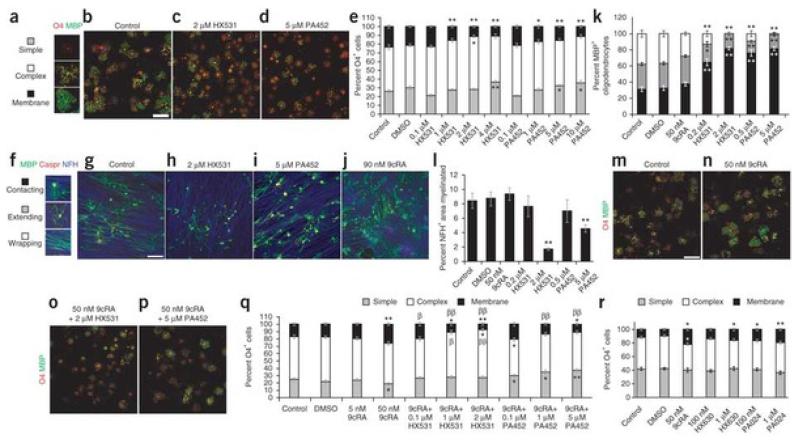
To abolish all RXR activity, we treated OPC cultures with a synthetic RXR-selective antagonist (HX531 or PA452)22 (Fig. 5). After 72 h, we immunostained oligodendrocyte lineage cells with O4 antibody and antibody to MBP, and determined their differentiation states from their morphologies (Fig. 5a). Compared to untreated or control cultures (Fig. 5b), increasing concentrations of HX531 (Fig. 5c) or PA452 (Fig. 5d) resulted in oligodendrocyte lineage cells displaying more simple morphologies and fewer membrane morphologies (Fig. 5e). Moreover, the percentage of total MBP+ oligodendrocytes decreased in cells treated with either antagonist compared to controls (Supplementary Fig. 6), suggesting that RXR antagonists inhibited oligodendrocyte differentiation. We did not find a substantial difference in the number of oligodendrocyte lineage cells undergoing apoptosis between antagonist-treated and control cultures (Supplementary Fig. 7), indicating that RXR antagonists do not influence the survival of these cells and were not toxic at the concentrations analyzed.
Figure 5. Rexinoids influence oligodendrocyte differentiation and myelination.
(a–d) OPC cultures immunolabeled with antibodies to O4 (red) and MBP (green) after RXR antagonist treatment for 72 h. Compared to non-treated cells (b), treatment with HX531 (c; 2 μM) or PA452 (d; 5 μM) resulted in fewer mature oligodendrocytes. Scale bar, 25 μm. (e) Increasing antagonist concentration resulted in decreasing number of membrane sheet-bearing oligodendrocytes. (f–j) Oligodendrocyte-DRG co-cultures maintained for 10 d after addition of OPCs immunolabeled with anti-MBP (green), anti-Caspr (red) and anti-NFH (blue). (g) Control co-culture; (h) HX531 (2 μM); (i) PA452 (5 μM); (j) 9cRA (50 nM). Scale bar, 100 μm. (k,l) Increasing antagonist concentration resulted in decreased MBP+ oligodendrocytes (k) and less myelination (l). (m–p) OPC cultures labeled with O4 (red) and anti-MBP (green). (m) Untreated; (n) 9cRA alone; (o) 9cRA and HX531; (p) 9cRA and PA452. Scale bar, 25 μm. (q) Quantification showing that 50 nM 9cRA increased mature oligodendrocyte membranes, and low concentrations of HX531 and PA452 were sufficient to abrogate 9cRA-mediated differentiation. (r) Treatment of cultured OPCs with 9cRA, HX630 or PA024 resulted in increased oligodendrocyte membrane sheets. Mean ± s.e.m. are shown. *P < 0.05 versus control,**P < 0.005 versus control, †P < 0.05 versus 50 nM 9cRA, ††P < 0.005 versus 50 nM 9cRA; Student’s t test.
The observation that RXR signaling is required for the differentiation of oligodendrocyte lineage cells in purified OPC cultures raises the question of whether it also promotes myelin formation. We addressed this question by manipulating RXR signaling in co-cultures of OPCs and DRG neurons. We quantified myelination by MBP+ oligodendrocyte-axon contact (contacting), oligodendrocyte membrane extension on axons (extending) or myelin compaction based on elongated oligodendrocyte membranes over Caspr+ paranodal clusters (wrapping; Fig. 5f). Compared to control (Fig. 5g), administration of HX531 (Fig. 5h) or PA452 (Fig. 5i) to the co-cultures resulted in a concentration-dependent increase in the number of contacting oligodendrocytes and a marked reduction in the percentage of myelinating oligodendrocytes (Fig. 5k). To determine whether decreased myelination was caused by failed oligodendrocyte differentiation or a potential decrease in the number of axons, we calculated the percentage of neurofilament-labeled (NFH+) axons that were myelinated. The percentage of myelinated axons with respect to total NFH+ axons in the culture decreased substantially when either antagonist was added to the culture medium (Fig. 5l), suggesting that oligodendrocytes were stalled at the premyelinating stage. These results indicate that RXR signaling in oligodendrocytes is necessary for efficient myelination.
9-cis-retinoic acid improves CNS remyelination
9-cis-retinoic acid (9cRA), an isomer of the vitamin A-derived all-trans retinoic acid, is a known ligand for RXR activation23. It has been shown to activate transcription of the gene that encodes MBP24, which suggests that RXR signaling may promote OPC differentiation. To assess the role of 9cRA in OPC differentiation, we treated OPC cultures with 50 nM 9cRA for 48 h (Fig. 5). As thyroid hormone can influence RXR signaling, we omitted triidothyronine and thyroxine from the culture medium. Compared to control or untreated cultures (Fig. 5m), cultures treated with 9cRA had a higher percentage of MBP+ membrane sheets (Fig. 5n,q), which suggests that 9cRA promotes OPC differentiation. However, 9cRA can activate both RXRs and retinoic acid receptors (RARs)23. To confirm that 9cRA promoted OPC differentiation through RXRs, we administered 9cRA with the RXR antagonists HX531 (Fig. 5o) or PA452 (Fig. 5p) to OPC cultures. A low concentration of antagonist (0.1 μm HX531 or 0.1 μm PA452) was sufficient to abrogate 9cRA-induced OPC differentiation, and increasing concentrations of either antagonist in the presence of 9cRA further decreased the percentage of mature oligodendrocyte membranes (Fig. 5q), consistent with the idea that 9cRA stimulates OPC differentiation through RXR signaling. To confirm that RXR activation stimulates OPC differentiation, we treated OPC cultures with the selective RXR agonists HX630 or PA024 (refs. 25,26). Both agonists increased the elaboration of membrane sheets by cultured oligodendrocytes, consistent with the response of these cells to 9cRA treatment (Fig. 5r). We also examined the effect of 9cRA on myelinating co-cultures but did not observe a significant increase in myelination (P> 0.2; Fig. 5j,l), which indicates that either endogenous activation of RXR signaling was sufficient to achieve maximal oligodendrocyte differentiation by 10 d in co-culture, or 50 nM 9cRA was insufficient to increase differentiation from OPCs.
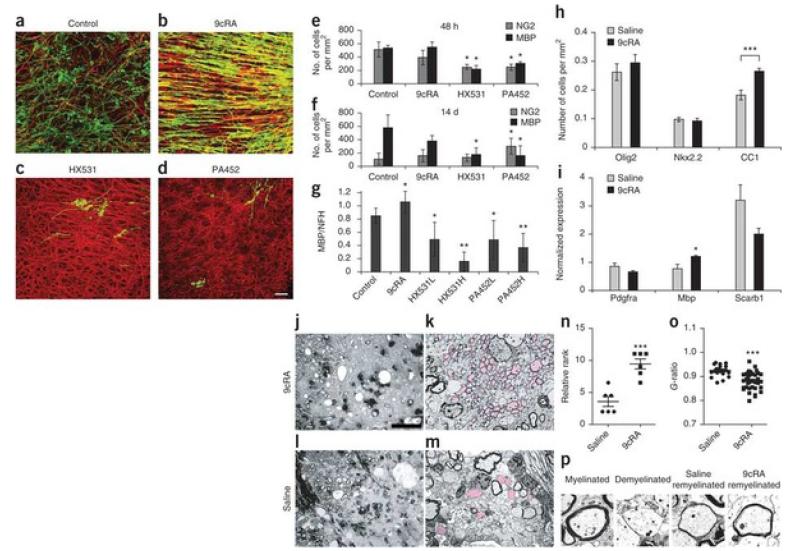
To find out whether activation of RXR signaling would promote CNS remyelination, we tested the effects of 9cRA on ex vivo demyelinated cerebellar slice cultures (Fig. 6). Mouse cerebellar slice cultures were treated overnight with lysolecithin after 14 d in vitro as described in rats to induce demyelination27. Cultures were then maintained in medium alone (Fig. 6a), 9cRA (Fig. 6b), the antagonists HX531 (Fig. 6c) or PA452 (Fig. 6d). We found that 9cRA treatment for 48 h or 14 d did not result in an increase in the number of NG2+ OPCs or MBP+ oligodendrocytes, whereas HX531 or PA452 resulted in a significant reduction (P < 0.05) in MBP+oligodendrocytes at both time points. However, an analysis of the percentage of myelin ensheathment revealed that 9cRA significantly increased (P < 0.05) the percentage of MBP+ membranes on NFH+ axons relative to control slices (Fig. 6a–d,g). Moreover, cell proliferation analysis using BrdU labeling revealed no difference in the density of BrdU+ cells between 9cRA–treated and control cultures (Supplementary Fig. 7); therefore, 9cRA does not seem to affect cell proliferation.
Figure 6. CNS remyelination is enhanced by 9 cis-retinoic acid.
(a) Control cerebellar slices fixed 10 d after demyelination with lysolecithin and immunolabeled with antibodies to NFH (red) and MBP (green). (b–d) Remyelination was increased by 9cRA (b), and decreased by HX531 (c) or PA452 (d). Scale bar, 20 μm. (e,f) Quantification of NG2+ and MBP+ cells, 48 h (e) or 14 d (f) after treatment. *P < 0.05; Student’s t test. (g) Quantification of remyelination after addition of 9cRA, or HX531 or PA452 at high (H, 2 or 5 μM, respectively) or low (L, 0.2 or 0.5 μM) concentrations. *P< 0.05, **P < 0.001; one-way ANOVA. (h) Treatment with 9cRA increased CC1+ cells in lesions in rats. Student’s t test. (i) Real-time qPCR analysis shows increased Mbpexpression in 9cRA-treated mice. Student’s t test. (j) Semi-thin section of a lesioned CCP 27 dpl after 9cRA treatment. Upper left corner shows normal myelinated axons. To the right is a large area of lesion showing axons outlined by thinly remyelinating membranes and dark macrophages. Scale bar, 50 μm. (k) Ultrastructural microscopy (1,500×) shows many remyelinated axons (pink) compared to axons that remained demyelinated. (l,m) Control animal (images as in j and k, respectively) shows few visible remyelinated axons. (n) Ranking analysis. Highest rank represents most remyelination. Mann-Whitney U test. (o) G-ratio is lower in 9cRA-treated mice compared to control mice. Student’s t test. (p) Representative images of myelinated, demyelinated, control remyelinated and 9cRA remyelinated axons. Mean ± s.e.m. are shown. ***P < 0.001.
We next asked whether exogenous 9cRA could similarly enhance remyelination in vivo. As retinoids can influence the adaptive immune response28, it would be difficult to distinguish direct effects of 9cRA on remyelination from those that were primarily immunomodulatory. We therefore used the focal toxin-induced model of demyelination for our experiment rather than a variant of experimental autoimmune encephalomyelitis (EAE), which is a widely used model for studying the immunological aspects of multiple sclerosis. The toxin model also provides a clear distinction between acute demyelination, whose induction is independent of the adaptive immune system, and subsequent remyelination, allowing the effects of 9cRA on remyelination to be specifically addressed. Furthermore, we performed our studies in aged adult rats, in which remyelination occurs less efficiently than in young adult rats and which therefore provide a more clinically relevant context29. Following focal demyelination, we injected 10 mg per kg per day of 9cRA or saline intraperitoneally for 14 d from 7 to 21 dpl, and then analyzed the extent of remyelination at 27 dpl. The densities of Olig2+ oligodendrocyte lineage cells and Nkx2.2+ OPCs at lesions in the 9cRA-treated group were similar to those in the saline-treated group (Fig. 6h). However, we detected an increase in CC1+differentiated oligodendrocytes in the 9cRA-treated group compared with the control group. Moreover, real-time qPCR on laser capture–microdissected lesions at 27 dpl showed a roughly 30% increase in Mbp expression, which indicates myelin regeneration, in 9cRA-treated mice compared with control mice (Fig. 6i). We did not observe a significant difference between 9cRA-treated and control groups in the expression of Pdgfra (P = 0.1513) or Scarb1 (P = 0.0864), which correlates with OPC or macrophage activities, respectively. We used semi-thin resin and ultrastructural analyses of CCP lesions at 27 dpl, and found that the 9cRA-treated group (Fig. 6j,k) had more remyelinated axons than the control group (Fig. 6l,m). The improvement in CNS remyelination after 9cRA treatment was confirmed by ranking analysis of the semi-thin resin sections (Fig. 6n). We also assessed the efficiency of remyelination by performing a G-ratio analysis, which describes the ratio of axon diameter to myelinated axon. We found that the 9cRA-treated group had a lower G-ratio than the saline-treated group owing to the presence of thicker remyelinated sheaths around axons (Fig. 6o,p), consistent with an acceleration of CNS remyelination.
Discussion
We have generated a transcriptional database of genes that are differentially expressed in association with spontaneous CNS remyelination after focal, toxin-induced demyelination in rat white matter. The remyelination transcriptome will be a useful resource that should allow the neuroscience and regenerative medicine communities to better understand the signaling networks and factors that are required when endogenous precursor cells repair the injured brain, as well as how normally expressed genes and signaling pathways in the white matter might be affected in pathologies of CNS demyelination or failed myelin regeneration.
The prominence of RXR signaling pathways in the remyelination transcriptome showed that RXR signaling was associated with the CNS regenerative response, and opens a new area of research on the role of RXRs in regenerative medicine. In the uninjured mammalian CNS, RXR-γ is generally expressed at low levels in all glial cells30, but it becomes actively expressed by activated microglia or macrophages, reactive astrocytes and oligodendrocytes after CNS injury16. We found that RXR-γ was also expressed after focal demyelination and in active multiple sclerosis lesions, which suggests that RXR-γ is a physiological signal of injury in the acutely lesioned brain. Functional analyses in cultured OPCs using siRNAs against RXR-γ or RXR-specific antagonists, and in RXR-γ null mice, showed inefficient oligodendrocyte differentiation, indicating that RXR-γ is an important regulator of remyelination. In purified human adult OPCs, microarray profiling did not detect RXRG transcripts31. We found that OPCs from normal-appearing white matter of multiple sclerosis brains expressed more RXR-γ in the cytoplasm than in the nucleus, which suggests that RXRG might not be actively transcribed in ‘resting’ adult OPCs. Therefore, as in rodents, RXR-γ is probably expressed and activated in response to CNS injury in humans.
The ability for RXR to heterodimerize with a number of nuclear receptors suggests that RXR can modulate the expression of different genes, depending on when and with which receptor it heterodimerizes. RXR can form permissive or non-permissive heterodimers with other nuclear receptors32. As 9cRA stimulated differentiation and enhanced CNS remyelination, RXR-γ probably acts though permissive heterodimerization. Candidate permissive heterodimers, including LXR and PPAR, have been described in oligodendrocyte lineage cells33, 34, 35. In our microarray data, we did not detect the differential expression of any members of the PPAR family, but we detected RXR-α, RXR-β, LXRα, COUP-TFI and Nurr1 as possible partners in permissive RXR-γ heterodimers. However, it remains to be determined which nuclear receptor(s) heterodimerize with RXR-γ in oligodendrocyte lineage cells after demyelination, and what genes are transcribed in response to RXR-γactivation to promote the differentiation of OPCs.
RXR agonists or rexinoids are widely available and show therapeutic promise for cancer cell differentiation therapy, as well as for the treatment of metabolic diseases32. In EAE, 9cRA can modulate inflammation36, suggesting that rexinoids might be useful for the treatment of inflammatory diseases of the nervous system such as multiple sclerosis. We have shown here that rexinoids can also stimulate oligodendrocyte differentiation and remyelination in the injured CNS, thereby illustrating an additional role of rexinoids as potential drugs for regenerative therapy in demyelinating disorders.
Methods
Focal demyelination
Female Sprague-Dawley rats aged 3 months (180–210 g) were used for LCM, in situ hybridization and immunostaining. Rats aged from 9 months to one year (250–300 g) were used for the 9-cis-retinoic acid experiment. All experiments were performed in compliance with UK Home Office regulations (Project License: 80/8718) Focal demyelination was induced bilaterally by stereotactically injecting 4.0 μl of 0.01% (ethidium bromide (vol/vol) in saline into caudal cerebellar peduncles (CCP). For analysis of Rxrg+/− and Rxrg−/− mutants, adult mice (~6 months old) received 1 μl of 1.0% lysolecithin (vol/vol) injection in the ventral funiculus, and mice were killed at 15 dpl and 30 dpl for analysis.
Laser capture microdissections and microarrays
Mice (n = 4 per group) were killed at 5, 14 and 28 dpl, and brains isolated and snap frozen. Cryosections (15 μm) were collected on PEN-membrane slides (P.A.L.M. Cat. No. 1400-1000), fixed in 70% ethanol (vol/vol), stained with 1% toluidine blue (wt/vol) and then dehydrated with ethanol and xylene. LCM was performed on PALM MicroBeam. Total RNA was isolated using RNAqueous-Micro (Ambion. Cat. No. AM1931) and used for microarray analysis and qPCR. For microarrays, total RNA from brains (n = 3) isolated from each post-lesion time point were amplified once and quantity checked. RNAs were hybridized onto Illumina Rat RefSeq slide, giving three biological replicates per time point, and differentially expressed genes were detected simultaneously with the Illumina BeadStation. Microarray analysis was performed at Cambridge Genomic Services (University of Cambridge).
Microarray and bioinformatics
The Ratref-12 expression Beadchip was used for the microarray study. The quality of the assay was assessed using the BeadStudio control panel. Raw data were loaded into R using lumi37, and were filtered using the Illumina detection value. This value is a P value that results from a statistical test between the beads representing the probes and the negative controls on the array; the lumi default filter counts any probes with a P value below 0.01 as present. The filtering was performed using the following criteria: any given probe needs to be present on at least one of the replicates at any of the time points. After the data were filtered, they were transformed using variance stabilization and then normalized using quantile normalization in lumi. The data were then analyzed using the R package fspma. This algorithm is designed to perform mixed-model ANOVA analysis. The model chosen had two main effects, the time points and samples. The latter was set as a random effect and ranking was performed on the time points. Additional comparisons were done using the R package limma. This package uses linear models to compare groups, and was used to perform pair-wise comparisons to compare the different time points between them. For both sets of results, the ANOVA and the pairwise comparisons, the P values were corrected using false discovery rate. The cut off used for the analysis was P < 0.05. Genes that were differentially expressed over three post-lesion time points from the ANOVA analysis (P < 0.05) were submitted to Cluster 3.0 for hierarchical clustering analysis (euclidian distance, centroid linkage clustering) and visualized using Java TreeView. IPA was performed using the Ingenuity Pathways Analysis software.
Antibodies and reagents
The primary antibodies for rodent experiments were mouse antibody to A2B5 (Millipore), rabbit antibody to Caspr (Abcam), mouse antibody to ED1 (Serotec), mouse antibody to GFAP (DAKO), rat antibody to MBP (Serotec), mouse antibody to Nkx2.2 (DHSB, University of Iowa), mouse O4 (Millipore), rabbit antibody to Olig2 (Millipore), chicken antibody to NFH (Encor Biotechnologies) and rabbit antibody to RXR-γ (Abcam). For multiple sclerosis tissues, they were rabbit antibody to RXR-γ(Abcam), rabbit antibody to RXRα (Santa Cruz Biotechnology), rabbit antibody to RXRβ (Santa Cruz Biotechnology), mouse monoclonal antibody to Olig1 (R&D Systems), polyclonal antibody to Sox10 (R&D Systems), polyclonal antibody to NOGO-A (Santa Cruz Biotechnology), antibody to MHC class II (mouse IgG1, DAKO) and antibody to GFAP (mouse IgG1, Millipore).
Immunohistochemistry
Rats were anaesthetized and perfused with 4% paraformaldehyde at 5 dpl, 14 dpl and 28 dpl before brains were isolated, postfixed and cryosectioned. 12-μm sections containing lesions were detected by rapid toluidine blue staining and light microscopy examination before collection and storage at −80 °C. Immunoperoxidase staining was performed using the Vectastain ABC Kit (Vector Laboratories). Non-radioactive in situ hybridization for RXR-γ was performed as described29. For the generation of the Rxrg probe, a 669-bp rat Rxrg cDNA fragment was isolated as described38 from cultured OPC cell extracts and subcloned into the CS2+ vector. Antisense Rxrg was generated with T7 polymerase after BamH1 linearization. Images were photographed and labeled cells were assessed using Axiovision 4.7.1 software (Zeiss). Statistical analysis was performed using Excel and Prism Graph Pad.
Purified rat OPC cultures
OPC cultures were prepared from neonatal Sprague-Dawley rats as described39. OPCs were seeded on PDL-coated 13-mm glass coverslips, and maintained in SATO medium without thyroxine and thiiodothyronine at 37 °C in 7.5% CO2. For differentiation experiments, OPCs were cultured in the presence of 1:1,000 DMSO, RXR agonists HX630 or PA024 (at 100 nM or 1 μM), 5 or 50 nM 9-cis-retinoic acid and/or synthetic RXR antagonists HX531 (at 0.1 μM, 1 μM, 2 μM or 4 μM) or PA452 (at 0.1 μM, 1 μM, 5 μM or 10 μM). Cultures were fixed for immunofluorescence analysis. Confocal z-stacks were acquired using Leica SPE confocal microscope, and analyzed using Image Pro Plus software (Media Cybernetics). Differentiation was analyzed by scoring cells as having ‘simple’ (short, non-interdigitating processes), ‘complex’ (longer, interdigitating processes, but not membrane sheets) or ‘membrane’ morphology (processes containing MBP-positive membrane sheets). Three 20× fields from each of four coverslips were analyzed per condition.
Myelinating rat OPC-DRG neuron co-cultures
Rat OPC and OPC-DRG co-cultures were prepared as described40, with the following modifications: DRG neuron cultures were prepared from embryonic day 15 (E15) Sprague-Dawley rats and cultured at a density of 2 × 105 cells per 22-mm coverslip. DRGs were maintained in DMEM with 10% fetal calf serum (FCS, vol/vol) and 100 ng ml−1 NGF (Serotec). DRG cultures were treated with fluorodeoxyuridine to remove contaminating cells. After 21 d, 2 × 105 OPCs were added per coverslip to DRGs, and co-cultures were maintained for a further 10 d in myelination medium (BME with ITS and Glutamax I supplements, 36 μg ml−1glucose and 0.5% FCS (all Invitrogen)). In myelination experiments, 1:1,000 DMSO, 50 nM 9-cis-retinoic acid, HX531 (0.2 or 2 μM) or PA452 (0.5 or 5 μM) were added. Cultures were fixed for immunocytochemistry. Oligodendrocytes were scored for their morphology as ‘contacting’ (processes touching but not aligning with axons), ‘extending’ (processes aligned with, but not surrounding axons) or ‘wrapping’ (MBP and Caspr-positive internodes clearly visible). The percentage of axon area myelinated was quantified using Image Pro Plus. For each field analyzed, percentage myelinated area was measured by extracting a mask image representing MBP-NFH colocalization from each layer of a confocal stack and carrying out an ‘extended depth of field’ projection of these mask images to form a single image representing the total myelinated area throughout the stack, the value of which was obtained using the software. Total myelinated area was then divided by the NFH-immunopositive area measured in that field, multiplied by 100. Three 20× fields from each of four coverslips were analyzed per condition.
Mouse cerebellar slice cultures
Remyelinating mouse cerebellar slice cultures were prepared based on previously published methods used for rats. Slices were exposed to control medium, 50 nM 9cRA, or low (0.2 or 0.5 μM) or high (2 or 5 μM) doses of the RXR antagonists HX531 and PA452, maintained for a further 14 d, and then processed for immunolabeling. Imaging was carried out as described above for oligodendrocyte cultures and myelinating co-cultures. Myelin was quantified using Image Pro Plus as described for the co-cultures. Two experiments were analyzed in duplicate.
siRNA transfections
After shake-off, OPCs were maintained in SATO medium with pen-strep (Invitrogen), 0.5% fetal bovine serum (FBS, vol/vol) and 10 nM PDGF and FGF overnight. Medium was removed and replaced with SATO with 0.5% FBS, and cells were transfected with 20 μM siRNA (or mock transfected) using 1% lipofectamine RNAiMAX transfection reagent (Invitrogen) in OPTI-MEM. siRNA sequences used were obtained from Dharmacon/Thermo Scientific (RXRα: L-089934-01; RXR-γ: L-083061-08; non-targeting: D-001810-10). Medium was replaced with SATO (without T3 or T4) with 0.5% FBS and pen-strep after 6 h. Cells were maintained in culture for 72 h, and then fixed with 4% PFA for 20 min at 4 °C or lysed for 10 min on ice with TEN buffer with 1% Triton X-100 (vol/vol) and 1× protease and phosphatase inhibitor cocktails (Calbiochem). Lysates were subjected to SDS-PAGE and western blotting.
9-cis retinoic acid analysis on focal demyelination
Aged rats (n = 10 per group) with focal demyelinating lesions were injected intraperitoneally with 10 mg per kg 9-cis-retinoic acid (BIOMOL International) or saline by from 7 to 21 dpl and killed at 27 dpl for remyelination analysis by microscopy and PCR. For semithin resin sections and electron microscopy, mice (n= 6 per group) were perfused with 4% glutaraldehyde and their brains removed. We obtained 1-mm transverse cerebellum/brain stem sections, processed them through osmium tetroxide, dehydrated them in ethanol, and embedded them in resin (TAAB Laboratories). We collected 1-μm resin sections on glass slides and stained them with 1% toluidine blue. The extent of remyelination was then assessed by light microscopy. For ultrastructural analysis, resin-embedded tissues were cut and examined with a Hitachi H-600 Electron Microscope. Ranking analysis (n = 4 per group) was performed using a 2-tailed Mann Whitney test and analyzed using Excel and Prism Graph Pad. G-ratio analysis was performed using NIH ImageJ. The remaining mice (n = 4 per group) were killed and their brains removed for LCM and qPCR analysis.
Quantitative PCR
PCR primers for rat Mbp, Pdgfra, Scarb1, B2M, Rxra, Rxrb, Rxrg and Actin were purchased from Gene Globe (Qiagen). Real-time qPCR was performed with Quanti-Tect SYBR green PCR Kit (Qiagen) and analyzed by Rotor Gene 6000 PCR analyzer (Corbett Research). Results were normalized against Actin or B2Mand expressed as mean ± s.e.m. Statistical analysis was performed using Prism Graph Pad.
Multiple sclerosis tissue samples and immunohistochemistry
Snap-frozen post-mortem multiple sclerosis brain samples were obtained from the French GIE NeuroCEB brain bank (D. Seilhean, Pitié-Salpêtrière Hospital) and the UK Multiple Sclerosis tissue Bank (R. Reynolds, Imperial College). Control brain samples from individuals who had died from non-neurological diseases were also obtained from the same sources. Tissues were collected with the donors’ fully informed consent through a prospective donor scheme following ethical approval. Three randomly chosen multiple sclerosis cases (Supplementary Table 6) were studied, including secondary progressive (two cases) and relapsing remitting (one case). For these multiple sclerosis cases, the mean age was 66.3 years (range: 65–74). The death-tissue preservation delay varied between 20 and 45 h. Histological assessment of the lesions was performed using Luxol fast blue/Cresyl violet and Oil-red-O (macrophages filled with myelin debris) histological staining. multiple sclerosis lesions were classified according to their inflammatory activity (KP1 immunolabeling) and on the basis of histological criteria of acute lesions (active demyelination, myelin vacuolation, inflammation or edema, minor gliosis and vague margin), chronic lesions (no myelin vacuolation, absence of inflammation, gliosis, axonal loss and sharp margin) and shadow plaques (myelin pallor). The expression of RXR-γ was studied in six multiple sclerosis lesions with active border (n = 3), chronic silent core (n = 5), and shadow plaques (n = 1, partially remyelinated).
Snap-frozen multiple sclerosis tissue sections were hydrated in PBS and microwaved in Vector unmasking solution, according to the manufacturer’s protocol. Briefly, sections were pre-incubated in blocking buffer (10% normal goat serum (vol/vol), 0.1% Triton-X 100 in PBS) for 1 h and incubated overnight with primary antibodies at 4 °C. After overnight incubation, slides were extensively washed in PBS/0.1% Triton X-100 and incubated with appropriate secondary antibodies. Quantification of RXR-γ+ cells was performed on ImageJ software of at least three serial multiple sclerosis sections (100 μm apart) in active borders, chronic silent core and PPWM from three distinct cases and in the normal-appearing white matter from non-neurological controls. Data are expressed as mean ± s.e.m. Non-parametric statistical tests were performed (one-way ANOVA) and the results were considered significant at P < 0.05.
Supplementary Material
Acknowledgments
We thank D. Seilhean (Service d’Anatomopathologie Neurologique, G-H Pitié-Salpêtrière, Paris) for classification of multiple sclerosis lesions, the French Brain Bank GIE NeuroCEB (Hôpital Pitié-Salpêtrière, Paris, France) and the United Kingdom Multiple Sclerosis Society Brain Bank (R. Reynolds, Imperial College, London) for multiple sclerosis tissue. All tissues were collected with the approval of the French and London Multicentre Research Ethics committees. Animal procedures were performed under a UK Home Office Project License. This work was supported by grants from the United Kingdom Multiple Sclerosis Society (R.J.M.F., C.ff.-C.), the Wellcome Trust (C.ff.-C.), the French Multiple Sclerosis foundation ARSEP (B.N.O.), the Biotechnology and Biological Sciences Research Council of the United Kingdom (C.ff.-C., J.B.), National Multiple Sclerosis Society (C.ff.-C., R.J.M.F., A.B.-V.E., B.N.O.), AP-HP Hôpital Pitié-Salpêtrière, Service d’Anatomopathologie Neurologique (B.N.O.) et des Maladies du Système Nerveux (A.B.-V.E.). A.W. holds a Wellcome Trust Intermediate Fellowship. A.A.J. holds a Fellowship from Multiple Sclerosis Society of Canada.
Footnotes
Accession codes: A full list of genes was deposited in NCBI GEO (accession code GSE24821). http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE24821
Competing financial interests: The authors declare no competing financial interests.
Contributor Information
Jeffrey K Huang, MRC Centre for Stem Cell Biology and Regenerative Medicine and Department of Veterinary Medicine, University of Cambridge, Cambridge, UK..
Andrew A Jarjour, MRC Centre for Regenerative Medicine and Multiple Sclerosis Society/University of Edinburgh Centre for Translational Research, Centre for Inflammation Research, The Queen’s Medical Research Institute, Edinburgh, UK..
Brahim Nait Oumesmar, Centre de Recherche de l’Institut du Cerveau et de la Moelle Epinière, Inserm U.975; Université Pierre et Marie Curie-Paris 6 UMR-S975; Cnrs UMR 7225; and AP-HP Groupe Hospitalier Pitié-Salpêtrière, Fédération de Neurologie, Paris cedex 13, France..
Christophe Kerninon, Centre de Recherche de l’Institut du Cerveau et de la Moelle Epinière, Inserm U.975; Université Pierre et Marie Curie-Paris 6 UMR-S975; Cnrs UMR 7225; and AP-HP Groupe Hospitalier Pitié-Salpêtrière, Fédération de Neurologie, Paris cedex 13, France..
Anna Williams, MRC Centre for Regenerative Medicine and Multiple Sclerosis Society/University of Edinburgh Centre for Translational Research, Centre for Inflammation Research, The Queen’s Medical Research Institute, Edinburgh, UK..
Wojciech Krezel, Institut de Génétique et de Biologie Moléculaire et Cellulaire, Department of Cell Biology and Development, Illkirch, France..
Hiroyuki Kagechika, Graduate School of Biomedical Science, Institute of Biomaterials and Bioengineering, Tokyo Medical and Dental University, Chiyoda-ku, Tokyo, Japan..
Julien Bauer, Department of Pathology, University of Cambridge, Cambridge, UK..
Chao Zhao, MRC Centre for Stem Cell Biology and Regenerative Medicine and Department of Veterinary Medicine, University of Cambridge, Cambridge, UK..
Anne Baron-Van Evercooren, Centre de Recherche de l’Institut du Cerveau et de la Moelle Epinière, Inserm U.975; Université Pierre et Marie Curie-Paris 6 UMR-S975; Cnrs UMR 7225; and AP-HP Groupe Hospitalier Pitié-Salpêtrière, Fédération de Neurologie, Paris cedex 13, France..
Pierre Chambon, Institut de Génétique et de Biologie Moléculaire et Cellulaire, Department of Cell Biology and Development, Illkirch, France..
Charles ffrench-Constant, MRC Centre for Regenerative Medicine and Multiple Sclerosis Society/University of Edinburgh Centre for Translational Research, Centre for Inflammation Research, The Queen’s Medical Research Institute, Edinburgh, UK..
Robin J M Franklin, MRC Centre for Stem Cell Biology and Regenerative Medicine and Department of Veterinary Medicine, University of Cambridge, Cambridge, UK..
References
- 1.Prineas JW, Connell F. Remyelination in multiple sclerosis. Ann. Neurol. 1979;5:22–31. doi: 10.1002/ana.410050105. [DOI] [PubMed] [Google Scholar]
- 2.Patani R, Balaratnam M, Vora A, Reynolds R. Remyelination can be extensive in multiple sclerosis despite a long disease course. Neuropathol. Appl. Neurobiol. 2007;33:277–287. doi: 10.1111/j.1365-2990.2007.00805.x. [DOI] [PubMed] [Google Scholar]
- 3.Raine CS, Wu E. Multiple sclerosis: remyelination in acute lesions. J. Neuropathol. Exp. Neurol. 1993;52:199–204. [PubMed] [Google Scholar]
- 4.Goldschmidt T, Antel J, König FB, Brück W, Kuhlmann T. Remyelination capacity of the MS brain decreases with disease chronicity. Neurology. 2009;72:1914–1921. doi: 10.1212/WNL.0b013e3181a8260a. [DOI] [PubMed] [Google Scholar]
- 5.Franklin RJM. Why does remyelination fail in multiple sclerosis? Nat. Rev. Neurosci. 2002;3:705–714. doi: 10.1038/nrn917. [DOI] [PubMed] [Google Scholar]
- 6.Chang A, Tourtellotte WW, Rudick R, Trapp BD. Premyelinating oligodendrocytes in chronic lesions of multiple sclerosis. N. Engl. J. Med. 2002;346:165–173. doi: 10.1056/NEJMoa010994. [DOI] [PubMed] [Google Scholar]
- 7.Kuhlmann T, et al. Differentiation block of oligodendroglial progenitor cells as a cause for remyelination failure in chronic multiple sclerosis. Brain. 2008;131:1749–1758. doi: 10.1093/brain/awn096. [DOI] [PubMed] [Google Scholar]
- 8.Wolswijk G. Chronic stage multiple sclerosis lesions contain a relatively quiescent population of oligodendrocyte precursor cells. J. Neurosci. 1998;18:601–609. doi: 10.1523/JNEUROSCI.18-02-00601.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nave KA, Trapp BD. Axon-glial signaling and the glial support of axon function. Annu. Rev. Neurosci. 2008;31:535–561. doi: 10.1146/annurev.neuro.30.051606.094309. [DOI] [PubMed] [Google Scholar]
- 10.Fancy SPJ, et al. Dysregulation of the Wnt pathway inhibits timely myelination and remyelination in the mammalian CNS. Genes Dev. 2009;23:1571–85. doi: 10.1101/gad.1806309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller RH, Mi S. Dissecting demyelination. Nat Neurosci. 2007;10:1351–4. doi: 10.1038/nn1995. [DOI] [PubMed] [Google Scholar]
- 12.Woodruff RH, Franklin RJM. Demyelination and remyelination of the caudal cerebellar peduncle of adult rats following stereotaxic injections of lysolecithin, ethidium bromide, and complement/anti-galactocerebroside: a comparative study. Glia. 1999;25:216–228. doi: 10.1002/(sici)1098-1136(19990201)25:3<216::aid-glia2>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 13.Germain P, et al. International Union of Pharmacology. LXIII. Retinoid X receptors. Pharmacol. Rev. 2006;58:760–772. doi: 10.1124/pr.58.4.7. [DOI] [PubMed] [Google Scholar]
- 14.Lefebvre P, Benomar Y, Staels B. Retinoid X receptors: common heterodimerization partners with distinct functions. Trends Endocrinol. Metab. 2010;21:676–683. doi: 10.1016/j.tem.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 15.Franklin RJM, ffrench-Constant C. Remyelination in the CNS: from biology to therapy. Nat. Rev. Neurosci. 2008;9:839–855. doi: 10.1038/nrn2480. [DOI] [PubMed] [Google Scholar]
- 16.Schrage K, Koopmans G, Joosten EAJ, Mey J. Macrophages and neurons are targets of retinoic acid signaling after spinal cord contusion injury. Eur. J. Neurosci. 2006;23:285–295. doi: 10.1111/j.1460-9568.2005.04534.x. [DOI] [PubMed] [Google Scholar]
- 17.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krezel W, et al. RXR gamma null mice are apparently normal and compound RXR alpha+/−/RXR beta−/−/RXR gamma−/− mutant mice are viable. Proc. Natl. Acad. Sci. USA. 1996;93:9010–9014. doi: 10.1073/pnas.93.17.9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haugen BR, et al. Retinoid X receptor gamma-deficient mice have increased skeletal muscle lipoprotein lipase activity and less weight gain when fed a high-fat diet. Endocrinology. 2004;145:3679–3685. doi: 10.1210/en.2003-1401. [DOI] [PubMed] [Google Scholar]
- 20.Brown NS, et al. Thyroid hormone resistance and increased metabolic rate in the RXR-gamma–deficient mouse. J. Clin. Invest. 2000;106:73–79. doi: 10.1172/JCI9422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krzyżosiak A, et al. Retinoid X receptor gamma control of affective behaviors involves dopaminergic signaling in mice. Neuron. 2010;66:908–920. doi: 10.1016/j.neuron.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi B, et al. Novel retinoid X receptor antagonists: specific inhibition of retinoid synergism in RXR-RAR heterodimer actions. J. Med. Chem. 2002;45:3327–3330. doi: 10.1021/jm0255320. [DOI] [PubMed] [Google Scholar]
- 23.Heyman RA, et al. 9-cis retinoic acid is a high affinity ligand for the retinoid X receptor. Cell. 1992;68:397–406. doi: 10.1016/0092-8674(92)90479-v. [DOI] [PubMed] [Google Scholar]
- 24.Pombo PM, Barettino D, Ibarrola N, Vega S, Rodríguez-Peña A. Stimulation of the myelin basic protein gene expression by 9-cis-retinoic acid and thyroid hormone: activation in the context of its native promoter. Brain Res. Mol. Brain Res. 1999;64:92–100. doi: 10.1016/s0169-328x(98)00311-8. [DOI] [PubMed] [Google Scholar]
- 25.Kagechika H, Shudo K. Synthetic retinoids: recent developments concerning structure and clinical utility. J. Med. Chem. 2005;48:5875–5883. doi: 10.1021/jm0581821. [DOI] [PubMed] [Google Scholar]
- 26.Nishimaki-Mogami T, et al. The RXR agonists PA024 and HX630 have different abilities to activate LXR/RXR and to induce ABCA1 expression in macrophage cell lines. Biochem. Pharmacol. 2008;76:1006–1013. doi: 10.1016/j.bcp.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 27.Birgbauer E, Rao TS, Webb M. Lysolecithin induces demyelination in vitro in a cerebellar slice culture system. J. Neurosci. Res. 2004;78:157–166. doi: 10.1002/jnr.20248. [DOI] [PubMed] [Google Scholar]
- 28.Klemann C, et al. Synthetic retinoid AM80 inhibits Th17 cells and ameliorates experimental autoimmune encephalomyelitis. Am. J. Pathol. 2009;174:2234–2245. doi: 10.2353/ajpath.2009.081084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sim FJ, Zhao C, Penderis J, Franklin RJM. The age-related decrease in CNS remyelination efficiency is attributable to an impairment of both oligodendrocyte progenitor recruitment and differentiation. J. Neurosci. 2002;22:2451–2459. doi: 10.1523/JNEUROSCI.22-07-02451.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moreno S, Farioli-Vecchioli S, Cerù MP. Immunolocalization of peroxisome proliferator-activated receptors and retinoid X receptors in the adult rat CNS. Neuroscience. 2004;123:131–145. doi: 10.1016/j.neuroscience.2003.08.064. [DOI] [PubMed] [Google Scholar]
- 31.Sim FJ, et al. Complementary patterns of gene expression by human oligodendrocyte progenitors and their environment predict determinants of progenitor maintenance and differentiation. Ann. Neurol. 2006;59:763–779. doi: 10.1002/ana.20812. [DOI] [PubMed] [Google Scholar]
- 32.Altucci L, Leibowitz MD, Ogilvie KM, de Lera AR, Gronemeyer H. RAR and RXR modulation in cancer and metabolic disease. Nat. Rev. Drug Discov. 2007;6:793–810. doi: 10.1038/nrd2397. [DOI] [PubMed] [Google Scholar]
- 33.Trousson A, et al. 25-hydroxycholesterol provokes oligodendrocyte cell line apoptosis and stimulates the secreted phospholipase A2 type IIA via LXR beta and PXR. J. Neurochem. 2009;109:945–958. doi: 10.1111/j.1471-4159.2009.06009.x. [DOI] [PubMed] [Google Scholar]
- 34.Granneman J, Skoff R, Yang X. Member of the peroxisome proliferator-activated receptor family of transcription factors is differentially expressed by oligodendrocytes. J. Neurosci. Res. 1998;51:563–573. doi: 10.1002/(SICI)1097-4547(19980301)51:5<563::AID-JNR3>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 35.Almad A, McTigue DM. Chronic expression of PPAR-delta by oligodendrocyte lineage cells in the injured rat spinal cord. J. Comp. Neurol. 2010;518:785–799. doi: 10.1002/cne.22242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diab A, et al. Ligands for the peroxisome proliferator-activated receptor-gamma and the retinoid X receptor exert additive anti-inflammatory effects on experimental autoimmune encephalomyelitis. J. Neuroimmunol. 2004;148:116–126. doi: 10.1016/j.jneuroim.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 37.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 38.Georgiades P, Brickell PM. Differential expression of the rat retinoid X receptor gamma gene during skeletal muscle differentiation suggests a role in myogenesis. Dev. Dyn. 1997;210:227–235. doi: 10.1002/(SICI)1097-0177(199711)210:3<227::AID-AJA4>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 39.McCarthy KD, de Vellis J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J. Cell Biol. 1980;85:890–902. doi: 10.1083/jcb.85.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laursen LS, Chan CW, ffrench-Constant C. An integrin-contactin complex regulates CNS myelination by differential Fyn phosphorylation. J. Neurosci. 2009;29:9174–9185. doi: 10.1523/JNEUROSCI.5942-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.