Abstract
Mandible shape in the mouse is a complex trait that is influenced by many genetic factors. However, little is known about the action of single genes on adult mandible shape so far, since most developmentally relevant genes are already required during embryogenesis, i.e. knockouts lead to embryonic death or severe deformations, before the mandible is fully formed. We employ here a geometric morphometrics approach to identify subtle phenotypic differences caused by dosage effects of candidate genes. We use mouse strains with specific gene modifications (knock-outs and knock-ins) to compare heterozygous animals with controls from the same stock, which is expected to be equivalent to a change of gene expression of the respective locus. Such differences in expression level are also likely to occur as part of the natural variation. We focus on Bmp pathway genes (Bmp4, its antagonist Noggin and combinations of Bmp5–7 genotypes), but include also two other developmental control genes suspected to affect mandible development in some way (Egfr and Irf6). In addition, we study effects of Hoxd13, as well as an extracellular matrix constituent (Col2a1). We find that subtle, but significant shape differences are caused by differences in gene dosage of several of these genes. The changes seen for Bmp4 and Noggin are partially compatible with the action of these genes known from birds and fish. We find significant shape changes also for Hoxd13, although this gene has so far only been implicated in skeletal patterning processes of the limbs. Comparing the effect sizes of gene dosage changes to the variation found in natural populations of mice as well as QTL effects on mandible shape, we find that the effect sizes caused by gene dosage changes are at the lower end of the spectrum of natural variation, but larger than the average additive effects found in QTL studies. We conclude that studying gene dosage effects have the potential to provide new insights into aspects of craniofacial development, variation and evolution.
Keywords: shape analysis, morphometry, dosage effects, mandible development, Mus musculus
Introduction
The mouse mandible is an excellent model for the study of shape (Atchley 1991), since it has a complex outline that can readily be described with a set of landmarks and is therefore amenable to quantitative statistical comparisons based on geometric morphometrics approaches (Klingenberg 2010). This approach has been used in the context of QTL studies (e.g., Klingenberg et al. 2001, Leamy et al. 2008), chromosome substitution strains (Boell et al. 2011) and for studying the phenotype space of wildtype variation in natural populations of Mus musculus (Boell and Tautz 2011). We explore here the approach of using gene dosage differences for assessing the effects of single genes on mandible shape, along the lines suggested by Cooper and Albertson (2008) and exemplified in zebrafish by Albertson et al. (2007) and LeClair et al. (2009). The most obvious candidate genes for such an approach are Bmp4 and Noggin, which have been shown to play key roles in the development of bird beaks (Abzhanov et al. 2004, Wu et al. 2004) and fish jaws (Albertson et al. 2005, Parsons and Albertson 2009). In the mouse, Bmp4 knockouts are embryonic lethal (Winnier et al. 1995), but a role in mandible development has been inferred from tissue-specific inactivation and overexpression studies (Liu et al. 2005; Bonilla-Claudio et al. 2012). Other Bmp signalling genes are also of interest, of which we are looking here at Bmp5, Bmp6 and Bmp7. While Bmp5 and Bmp6 knockouts show only subtle phenotypes (Solloway et al. 1998, 1999), Bmp7 knockout mice have underdeveloped mandibles (Zouvelou et al. 1999). Further candidate genes that have been implicated in mandible development are Irf6, Egfr and Col2a1. Irf6 is a transcription factor involved in epidermal (keratinocyte) development and its inactivation causes craniofacial phenotypes in mice and humans (Ingraham et al. 2006). Similar phenotypes were found for knockouts of Egfr, a TGF-alpha signalling receptor that is expressed in the developing mandible (Miettinen et al. 1999). Col2a1 is a structural compound of the cartilaginous precursors of developing bone and animals homozygous for a Gly574Ser mutation have abnormal craniofacial structure, as well as a shortened mandible (Maddox et al. 1998). The only gene in our dataset for which neither mandibular phenotypes nor craniofacial expression have so far been reported is Hoxd13, a posteriorly expressed Hox gene which is involved in limb and digit patterning in mice and humans (Bruneau et al. 2001; Kuss et al. 2009).
Phenotypic variation can be associated with gene expression differences that can be the basis of evolutionary change (Schadt et al. 2003; Wittkopp et al. 2004; Veitia et al. 2008; Huang et al. 2010; Delker et al. 2011). In a heterozygous knockout animal, transcription from a knockout locus is expected to be 50% of the wildtype level, although there may be secondary effects that partially compensate for the loss of one allele (Malone et al. 2012). An up to 2-fold difference in expression appears to be a realistic emulation of natural expression variation. For example, in a recent eQTL study in Arabidopsis (Cubillos et al. 2012), the average fold change was found to be 1.4. Expression differences can lead to dosage imbalances and haplo-insufficiency with corresponding phenotypic effects (Veitia et al. 2008; Huang et al. 2010). Using geometric morphometrics approaches, one can trace even subtle phenotypic effects. Previous studies have employed this to assess changes in mouse skull morphology caused by the Brachyrrhine (Br) mutation (Willmore et al. 2006), the Papps2 gene (Hallgrimson 2006) as well as dosage effects caused by segmental aneuploidy (Hill et al. 2007). Comparable studies have also been done to study Fgf8 and Glypican4 in adult zebrafish (Albertson et al. 2007; LeClair et al. 2009). Studying heterozygous knockout animals may therefore provide a general approach to assess sensitivity of craniofacial shape with respect to expression differences that should be comparable to natural variation.
Materials and methods
Mouse strains
Since we expect that gene dosage effects on mandible shape are subtle, it is necessary to control for other confounding influences such as genetic background and breeding conditions. Although the lines used here are nominally in a C57BL/6J background (all were backcrossed to C57BL/6J for more than 10 generations), small differences between C57BL/6J animals coming from different laboratories or sub-strains are still possible. Hence, our approach is based on comparing heterozygous animals for the respective allele with wildtype control animals from the same breeding stock of the respective allele, brought up within the same time interval. This ensures that the animals were raised under the same conditions and with the same food, i.e. variance due to possible plasticity effects (Boell and Tautz 2011) is minimized. Shape differences between stocks are already established around week 2 and stabilize around week 8 (Boell and Tautz 2011), therefore all animals in the study were at least 8 weeks old (detailed below). Mice were genotyped for the segregating allele and their heads were transferred into ethanol and stored until scanned.
Alleles studied
Bmp4: represented by an allele with a modified cleavage site (Bmp4S2KHAMyc) that influences the long-range signalling ability of the ligand (Cui et al. 2001) that is expected to enhance the range of action. The allele Bmp4HAMyc represents a knockin into the endogenous Bmp4 locus to introduce an in frame HA epitope tag within the prodomain following amino acid 61 (FEATLYPYDVPDYALQMFG; HA epitope underlined) and an in frame myc tag within the mature domain, four amino acids downstream of the S1 cleavage site (RAKRSPKHEQKLISEEDLHPQR; S1 site italicized, myc epitope underlined). The modified allele Bmp4S2KHAMyc represents a knockin point mutation that introduces a serine to lysine amino acid change at the S2 cleavage site (RISR-RIKR) in addition to the HA and myc epitope tags described above. The animals were grown by Sylvia Nelsen and Jan Christian at Oregon Health and Science University, Portland, USA. The ages of the animals were between 8–16 weeks.
Other BMPs: Bmp5;Bmp7 and Bmp6;Bmp7 double mutants are described in Solloway and Robertson (1999) and Tillemann et al. (2010) and were raised by Claude Brodski and Orna Novikov at the Ben-Gurion-University of the Negev, Israel. Age differences between the Bmp5/7 and 6/7 mice were not larger than a week. In this case, wildtype animals were unfortunately unavailable. Therefore, our comparison was between homozygous Bmp5 and homozygous Bmp6 knockout mice, both in a Bmp7 heterozygous background.
Noggin: animals were derived from the knockout line described in McMahon et al. (1998) and were raised by YiPing Chen at Tulane University, USA. All animals were about 10 weeks old.
Irf6: animals were derived from the knockout line described in Ingraham et al. (2006) and were grown by Brian Schutte and Youssef A. Kousa at Michigan State University, USA. The ages of the animals ranged between 10–12 weeks.
Col2a1: represented by the B6 (Cg)-Col2a1sedc/J strain from the Jackson laboratory, a spontaneous mutation described in Maddox et al. (1998). Animals were grown by Louis Boell at the MPI for Evolutionary Biology in Plön, Germany. All animals were 8 weeks old.
Egfr: represented by the C57BL/6J-EgfrVel/J strain from the Jackson laboratory, derived from ENU mutagenesis. Animals were grown by Louis Boell at the MPI for Evolutionary Biology in Plön, Germany. Heterozygous mice were identified by their phenotype (secretions at eyelids, curly vibrissae, coat with velvet-like texture, according to Miettinen et al. 2009). All animals were 8 weeks old.
Hoxd13: animals were derived from the Hoxd13 knockout line described in Dollé et al. (1993) and were grown by Pia Kuss at the Max-Planck Institute für molekulare Genetik, Berlin, Germany. All animals were 12 weeks old.
Data collection
Specimens were scanned with a computer tomograph (microCT - VivaCT 40, Scanco, Bruettisellen, Switzerland). Two-dimensional x-ray images of right hemi-mandibles were obtained from micro-CT data as described in Boell and Tautz (2011). 14 Landmarks (Figure 1) were digitized on the images using tpsDig2 (Rohlf 2005a) and tpsUtil (Rohlf 2005b), producing a set of 28 two-dimensional raw coordinates for each specimen. In order to reduce measurement error, all specimens (except for the CSS) were digitized twice and the mean coordinates from both rounds of digitization were used for further analyses. Specimen position inside the scanner appears to produce no additional error (see Boell and Tautz 2011).
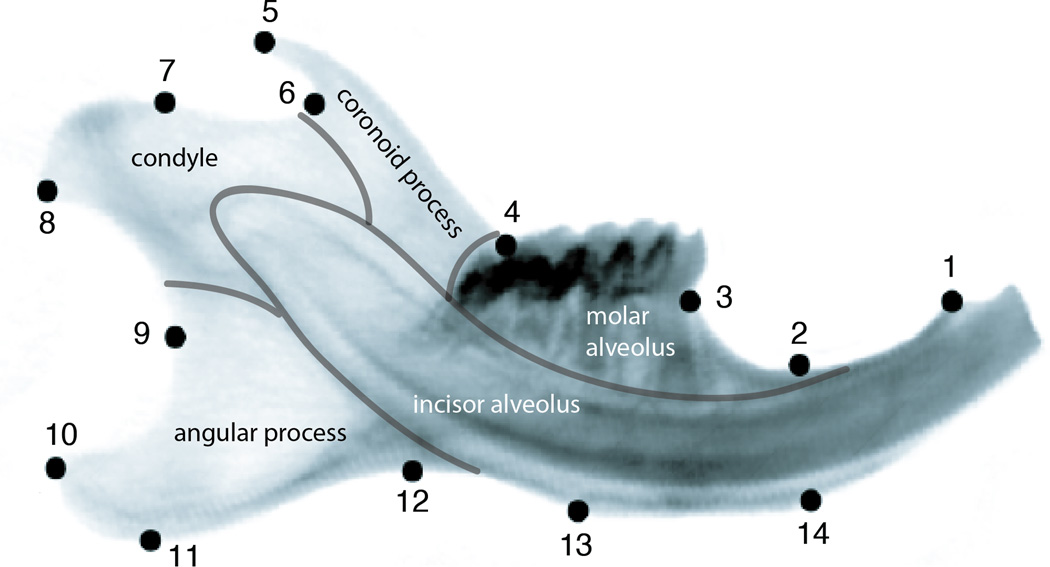
Figure 1.
Positions of the 14 landmarks used in this study on the outline of a mouse hemimandible radiograph. Morphological modules of the mandible are indicated by grey lines and the respective anatomical designations are indicated. The morphological positions of the landmarks are for LM1: Anterior terminus of bone dorsal of the incisor; LM2: Minimum of depression on dorsal side of incisor ramus; LM3: Bone/teeth transition anterior of M1; LM4: Intersection of ascending ramus with tooth row; LM5: Tip of processus coronoideus; LM6: Minimum of depression posterior to processus coronoideus; LM7: Anterior margin of condylar articular surface; LM8: Posteroventral tip of condyle; LM9: Minimum of depression formed by condyle and processus angularis; LM10: posterodorsal tip of processus angularis; LM11: Posteroventral tip of processus angularis; LM12: Minimum of depression formed by processus angularis and incisor ramus; LM13: Posterior margin of muscle insertion area on ventral side of incisor ramus; LM14: Anterior margin of muscle insertion area on ventral site of incisor ramus.
Statistical analyses
Geometric morphometrics analyses were done in MorphoJ (Klingenberg 2011). Pairwise comparisons were carried out between heterozygotes and controls for each gene (except Bmp5/6, as this comparison was between the respective homozygous knockouts) as indicated in Table 1. For two-dimensional landmark data, the number of degrees of freedom after Procrustes superimposition is 2×(number of landmarks) - 4. The number of degrees of freedom must not exceed the smallest within-group sample size in between-group comparisons (Zelditch et al. 2004). Therefore, we had to reduce the number of landmarks in each comparison according to the smallest sample involved. In order not to miss out significant shape differences due to an arbitrary selection of landmarks, we first identified the landmarks yielding the most pronounced differences between group mean shapes in each comparison. This was done by first Procrustes-superimposition of the specimens belonging to each pairwise comparison based on all 14 landmarks, then averaging the shape configurations within each of the two groups to be compared, then Procrustes-superimposition of the group mean shapes and calculating the shape difference vector between the group means. This vector consisted of 14 subvectors indicating the displacement of each landmark. Landmarks were prioritized according to the lengths of their displacement vectors. For significance testing, a number of landmarks with highest priority were chosen such that the resulting degrees of freedom did not exceed the sample size of the smallest group in the respective comparison (Table 1). The original datasets were then reduced to the chosen set of landmarks and again Procrustes-superimposed. The significance of Procrustes mean shape difference between groups in each comparison was assessed using the permutation procedure implemented in the discriminant function option in MorphoJ, using 1,000 rounds of permutation to generate an expected distribution of shape differences. We did not rely on Mahalanobis distances because the estimation of within-group variances on which these are based is more likely to be affected by the low sample sizes in our study than mean shape. In order to correct for multiple comparisons, significance levels were Holm-Bonferroni adjusted post hoc.
Table 1.
Genes, genotypes compared (sample size for each genotype shown in brackets), degrees of freedom (DF) available and landmarks used for the assessment of significant shape differences. Significant p-values after Bonferroni-Holm post hoc adjustment are marked with an asterisk. Note that Bmp4S2KHAmyc is an allele which is expected to enhance the range of action of the gene rather than diminishing it (see discussion).
| Gene | control Genotype (N) |
test Genotype (N) |
DF | Landmarks used | p (shape) | p (size) |
|---|---|---|---|---|---|---|
| Bmp4 | Bmp4HAmyc/+ (8) | Bmp4S2KHAmyc/+ (12) | 8 | 5,6,7,8,9,14 | 0.006* | 0.47 |
| Bmp5 vs 6 | Bmp 5−/−;Bmp7+/− (7) | Bmp 6−/−;Bmp7+/− (6) | 6 | 3,7,8,11,12 | <0.0001* | 0.59 |
| Bmp7 | Bmp 5−/−;Bmp7+/− (7) | Bmp 5−/−;Bmp7+/+ (8) | 6 | 5,6,7,9,10 | 0.012 | 0.8 |
| Noggin | Noggin+/+ (9) | Noggin+/− (10) | 8 | 4,7,11,12,13 | <0.0001* | <0.0001* |
| Irf6 | Irf6+/+ (19) | Irf6+/− (14) | 14 | 1,4,5,7,8,10,11,12,14 | 0.15 | 0.05 |
| Col2a1 | Col2a1+/+ (15) | Col2a1+/− (15) | 14 | 4,5,7,8,9,10,11,12,14 | 0.016 | 0.44 |
| Egfr | Egfr+/+ (29) | Egfr+/− (23) | 22 | all except 4 | 0.006* | 0.75 |
| Hoxd13 | Hoxd13+/+ (14) | Hoxd13+/− (9) | 8 | 5,6,8,9,10,11 | 0.007* | 0.39 |
Results
To assess the impact of gene dosage of candidate genes on mandible shape, we compared the landmark configurations between animals carrying mutant or knockout alleles and control animals on the same genetic background and reared in the same animal facility. Our candidate genes were Bmp4, Bmp5, Bmp6, Bmp7, Noggin, Irf6, Col2a1, Egfr and Hoxd13. Sample numbers and results of the pairwise comparisons are shown in Table 1. We found significant shape differences in five out of the eight comparisons and a significant size difference in one case. All significant shape differences are depicted in Figure 2.
Figure 2.
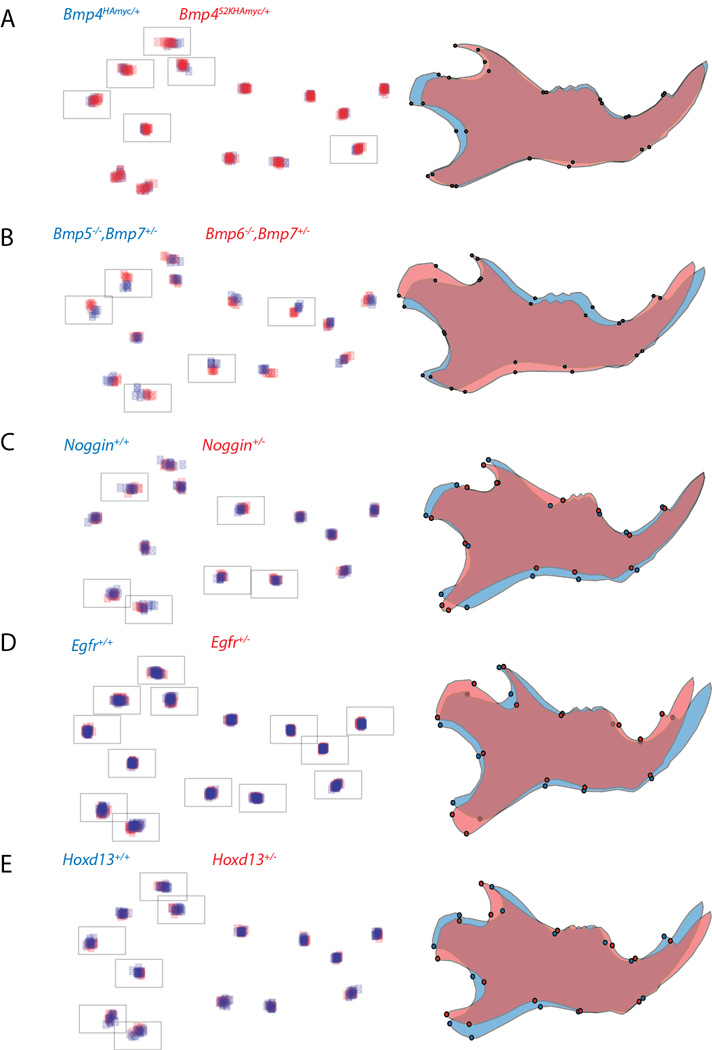
Procrustes coordinates (left) and overall shape differences (right) between pairs of genotypes for which significant shape differences were found. The procrustes coordinates are superimposed for all individuals, blue represents the reference shapes, red the mutant shapes. The framed coordinates (landmarks) were used for significance testing after separate Procrustes superimposition. The visualization of shape differences was done by using the “warped outline drawing” function in MorphoJ (Klingenberg 2011), the differences are exaggerated to unit Procrustes distance of 0.1.
The Bmp4S2KHAMyc allele shows an elongated coronoid process and a shortened condyle (Figure 2A). The comparison of mice carrying Bmp5 versus Bmp6 knockout alleles in a Bmp7 heterozygous background (Bmp5−/−;Bmp7+/− vs Bmp6−/−;Bmp7+/−) showed pronounced shape differences with respect to an elongated and upward shifted condyle. Furthermore, the molar alveolus was anteriorly shallower (Figure 2B). On the other hand, Bmp7 heterozygous mice on a Bmp5 knockout background (Bmp 5−/−;Bmp7+/− vs. Bmp 5−/−;Bmp7+/+) did not show significant shape differences (Table 1).
Mandibles from heterozygous Noggin mice showed a shortened condyle, a narrower distal angular process and the base of their coronoid process is anteriorly protracted (Figure 2C). Furthermore, Noggin+/− mandibles are 7% larger than Noggin+/+ mandibles, which is a significant size difference (Table 1).
Heterozygous Egfr animals exhibit shape changes throughout the mandible including extended and shifted condyle and angular process, a more slender coronoid process and a shifted incisor (Figure 2D). In mandibles of Hoxd13 heterozygous mice, the coronoid process is shifted posteriorly and the angular process is narrower and shifted dorsally (Figure 2E).
Comparison of effect sizes
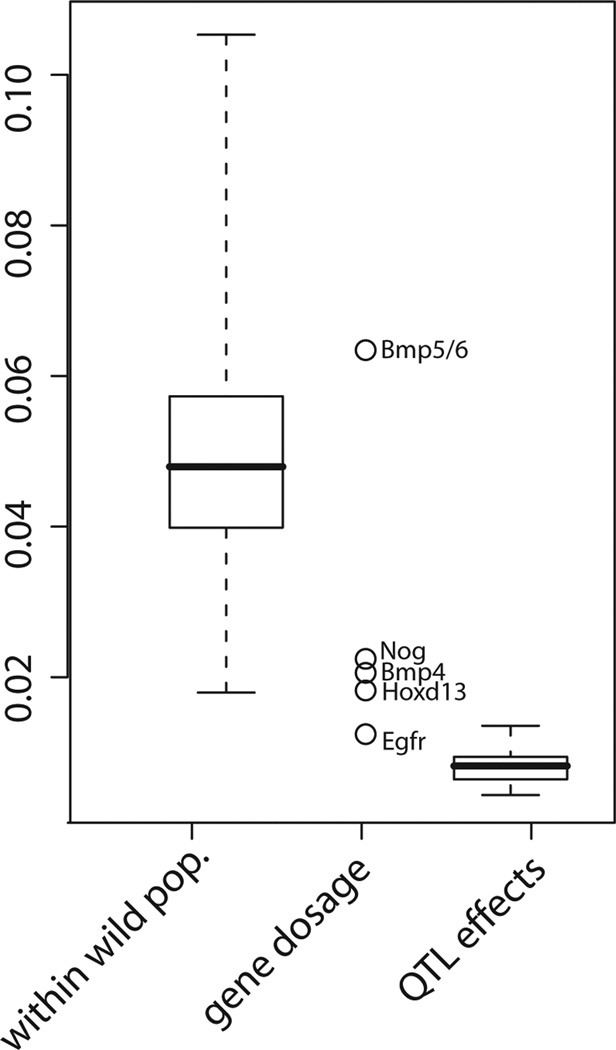
The above described effects of gene dosage on mouse mandible shape are subtle and would not easily be identified without the statistical approach used here. In order to put such differences into a wider quantitative genetic and evolutionary context, we compared the shape distances corresponding to significant dosage effects found in this study with shape distances found between individuals of wild populations of Mus musculus domesticus, as described in Boell and Tautz (2011) and with QTL effects from crosses between two inbred strains as described in Leamy et al. (2008) (Figure 3). All Procrustes distances in the comparison are based on the same set of 14 landmarks. The largest difference is seen for the comparison between Bmp5 nulls and Bmp6 nulls, which may be because knockout homozygotes at two different loci instead of knockout heterozygotes at one locus are compared. The effects of the other genes are smaller than the average difference between two wild mice caught from the same wild population. They are, however, still considerably larger than the majority of additive genetic effects in the Leamy et al. (2008) study.
Figure 3.
Effect sizes of gene dosage compared with phenotypic distances within populations among wild caught individuals of M. m. domesticus (data taken from Boell and Tautz 2011) and with QTL effect sizes taken from Leamy et al. (2008). All shape differences/distances are in units of Procrustes distance and based on the same set of landmarks.
Discussion
Our results show that knockout or mutant alleles of candidate genes in heterozygous condition can cause subtle, but significant shape differences that can be analysed in a geometric morphometrics framework. Hence, this approach can complement the phenotypic analysis of such alleles and may at the same time provide insights into possible degrees of natural variation due to differences in expression levels between alleles.
From an evolutionary perspective, Bmp4 is the best studied candidate with respect to changes in craniofacial shape. In Darwin´s finches, the amount and onset of Bmp4 expression is correlated with species-specific beak shapes, although this applies only to the upper beak, not to the lower one (Abzhanov et al, 2004). Injection of Bmp4 into the facial ectoderm leads to smaller beaks, whereas more Bmp4 in the mesenchyme leads to increased beak width and depth (Abzhanov et al. 2004). In cichlids, increased Bmp4 expression in the developing jaw leads to shorter, deeper jaws with increased mechanical advantage, and variants of Bmp4 co-segregate with corresponding adaptive phenotypic variation in natural populations (Albertson et al. 2005). These similarities have led Albertson et al. (2005) to postulate Bmp4 as “a key player in vertebrate diversity”. A role for Bmp4 in facial skeletal development of the mouse was inferred by studying conditional overexpression and cell-specific knockouts (Liu et al. 2005; Bonilla-Claudio et al. 2012). We have used here an allele of Bmp4 that interferes with Bmp4 signalling by enhancing the susceptibility of a proteolytic processing site, which makes the protein more active in long-range signalling in overexpression assays (Cui et al. 2001). The mandible shape phenotype which we observe for this allele shows a shortened condyle and an elongated coronoid process. This shape change is comparable with overexpression phenotypes in birds and fish, because the shortening of the condyle implies a shortening of the proximo-distal axis, whereas the elongation of the coronoid implies an increase in depth of the posterior mandible. Given that the Bmp4S2KHAMyc allele is expected to enhance the range of action of Bmp4 in the mandible, this would suggest an effect at least roughly comparable to the Bmp4 dosage effect across vertebrates.
Noggin is an extracellular signalling molecule antagonizing Bmp4. Its function is partially redundant to that of Chordin, i.e. knockout strains for either gene show only subtle effects on the mandible, while the double knockout leads to severe truncation of the mandible (Stottmann et al. 2001). Wu et al. (2006) showed for birds that an increase of Bmp4 amount locally stimulates growth in the developing beak, whereas increasing the amount of Noggin has the reverse effect. Given that the Bmp4S2KHAMyc allele that we have studied here is predicted to enhance Bmp4 signalling range, one would expect that a reduction of its antagonist Noggin in the heterozygous knockout mice should result in a similar phenotype. The shortening of the condyle is indeed comparable between the two phenotypes (Figure 2A, C) and would therefore support this notion. Other shape changes, on the other hand, such as the shifted angular process (Figure 2 C) are only found for Noggin. Hence, the interactions are likely to be more complex and may in fact involve the other Bmp genes. It is also unclear why Noggin heterozygotes show an increased size.
Bmp5 and Bmp6 knockouts have no major overt phenotype or skeletal changes, but ossification processes are affected (Solloway et al. 1998, Solloway and Robertson 1999). Bmp5 is expressed in the 1st branchial arch ectoderm at embryonic day E8.5 (Solloway and Robertson 1999), in the lower jaw molar epithelium at E13.0–13.5 (Vaahkotari et al. 1996), and in the mandible at E14.5 (Diez-Roux et al. 2011). Bmp6 is expressed in the lower jaw molar epithelium at E13.0–13.5 (Vaahtokari et al. 1996). Given these expression data, it appears plausible that either gene may affect mandible development in some ways. In fact, the shape difference seen between Bmp5 and Bmp6 homozygous knockout mice is by far the largest observed in our study (Figure 3). This may be for two reasons: first, the comparison involves in this case homozygous knockout animals instead of heterozygous knockouts; second, the observed shape difference may reflect cumulative effects of both Bmp5 and Bmp6. In any case, our results suggest that at least one of the two genes are indeed in some way involved in generating the mandible shape, although this had previously not been suspected.
Bmp7 is expressed in many tissues during mouse development, including the precartilaginous mesenchyme of the developing mandible (Lyons et al. 1995). However, we observe no significant effect on mandible shape in the heterozygous mice, even though they are on a Bmp5 homozygous mutant background (Table 1). Hence, we do not expect that the Bmp5 vs. Bmp6 comparisons discussed above are affected by the heterozygous state of Bmp7 in the respective animals. On the other hand, Bmp7 knockout affects the development of teeth and other ectodermal appendages of the orofacial complex and a function in epithelial-mesenchymal interactions was suggested (Zouvelou et al. 2009).
The shape changes seen for the Egfr heterozygous mice appear also at least partially to correspond to phenotypic effects seen in the knockout mice. Among various other major effects, Miettinen et al. (1995) showed that knockout mice develop foreshortened mandibles and we observe in the heterozygote mice a shortened and somewhat tilted incisor (Figure 2D).
We found no significant shape changes for Col2a1 and Irf6, although direct and indirect effects on mandibular development in homozygous knockout mice were described for these genes (Maddox et al. 1998, Ingraham et al. 2006). However, one has to keep in mind that very subtle effects would require larger sample sizes to achieve statistical significance.
An influence of Hoxd13 on mandible shape was so far not described. Hoxd13 belongs to the posterior Hox genes and is strongly expressed in the developing limbs of the mouse (Bruneau et al. 2001). Accordingly, it was so far mostly studied in the context of limb and digit development in humans and mice (Bruneau et al. 2001; Kuss et al. 2009), where it has specific roles in skeletal patterning (Salsi et al. 2008). However, given that Hoxd13 is known to have a more complex expression including various tissues and organs throughout the body (Cantile et al. 2009), it seems possible that there is also an as yet undetected expression in the tissues leading to the formation of the mandible. This will need to be further studied.
The comparison of effect sizes shown in Figure 3 suggests that the gene dosage effects described in our study are smaller than the shape differences between individual wild-caught mice, but larger than most QTL effects. Shape differences between wild-caught individuals are due to environmental differences, differences in age and probably the cumulative effects of many genes, so it is not surprising that they are comparatively large. On the other hand, the comparatively small size of QTL effects may be due to two reasons: first, the mutations studied here may be more drastic in effect than segregating natural variants in the strains used for the QTL experiments, and second, QTL effects represent average effects of genomic regions over multiple genetic backgrounds, whereas the mutations are observed on uniform backgrounds, some of which may enhance the apparent effect sizes.
Conclusions
We show that subtle effects of gene dosage on an adult trait, such as mandible shape, can be detected using geometric morphometrics. The Bmp4 and Noggin effects found in this approach appear to be partially comparable with developmental and evolutionary effects of the Bmp4 pathway in birds and fish. This would suggest that the approach is suitable to identify basic gene functions on shape and may also be extended to 3D shape analyses (Schunke et al. 2012). On the other hand, the results are complex, as multiple parts of the mandible appear to be affected at the same time. Such pleiotropic effects would be expected for genes involved in general signalling processes. This raises the question whether there are any specific genes that are regulating specific aspects of shape components, or whether the whole shape is a result of the action of a developmental network, involving epistasis and developmental buffering. We expect that the analysis of gene dosage effects on shape differences may be a way to approach the role of specific genes versus general effects.
Acknowledgements
The authors are indebted to Christine Pfeifle and Heike Harre for help with mouse breeding. The work was funded by institutional resources of the Max-Planck Society to DT. Financial support to BCS (#DE13513) and YAK (#1F31DE022696-01) came from the NIH National Institute of Dental and Craniofacial Research. Financial support to CB came from the Israel Science Foundation (grant No. 1391/11).
References
- Abzhanov A, Protas M, Grant BR, Grant PR, Tabin CJ. Bmp4 and morphological variation of beaks in Darwin´s finches. Science. 2004;305:1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- Albertson RC, Streelmann JT, Kocher TD, Yelick PC. Integration and evolution of the Cichlid mandible: the molecular basis of alternate feeding strategies. PNAS. 2005;102:16287–16292. doi: 10.1073/pnas.0506649102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson RC, Yelick PC. Fgf8 haploinsufficiency results in distinct craniofacial defects in adult zebrafish. Dev Biol. 2007;15:505–515. doi: 10.1016/j.ydbio.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchley WR. A model for development and evolution of complex morphological structures. Biol Rev. 1991;66:101–157. doi: 10.1111/j.1469-185x.1991.tb01138.x. [DOI] [PubMed] [Google Scholar]
- Boell L, Tautz D. Micro-evolutionary divergence patterns of mandible shapes in wild house mouse (Mus musculus) populations. BMC Evol Biol. 2011;11:306. doi: 10.1186/1471-2148-11-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boell L, Gregorova S, Forejt J, Tautz D. A comparative assessment of mandible shape in a consomic strain panel of the house mouse (Mus musculus) - implications for epistasis and evolvability of quantitative traits. BMC Evol Biol. 2011;11:309. doi: 10.1186/1471-2148-11-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla-Claudio M, Wang J, Bai1 Y, Klysik E, Selever J, Martin JF. Bmp signaling regulates a dose-dependent transcriptional program to control facial skeletal development. Development. 2012;139:709–719. doi: 10.1242/dev.073197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneau S, Johnson KR, Yamamoto M, Kuroiwa A, Duboule D. The mouse Hoxd13(spdh) mutation, a polyalanine expansion similar to human type II synpolydactyly (SPD), disrupts the function but not the expression of other Hoxd genes. Dev Biol. 2001;237(2):345–353. doi: 10.1006/dbio.2001.0382. [DOI] [PubMed] [Google Scholar]
- Cantile M, Franco R, Tschan A, Baumhoer D, Zlobec I, Schiavo G, Forte I, Bihl M, Liguori G, Botti G, Tornillo L, Karamitopoulou-Diamantis E, Terracciano L, Cillo C. HOX D13 expression across 79 tumor tissue types. Int J Cancer. 2009 Oct 1;125(7):1532–1541. doi: 10.1002/ijc.24438. [DOI] [PubMed] [Google Scholar]
- Cooper WJ, Albertson RC. Quantification and variation in experimental studies of morphogenesis. Dev Biol. 2008;15:295–302. doi: 10.1016/j.ydbio.2008.06.025. [DOI] [PubMed] [Google Scholar]
- Cubillos FA, Yansouni J, Khalili H, Balzergue S, Elftieh S, Martin-Magniette M-L, Serrand Y, Lepiniec L, Baud S, Dubreucq B, Renou J-P, Camilleri C, Loudet O. Expression variation in connected recombinant populations of Arabidopsis thaliana highlights distinct transcriptome architectures. BMC Genomics. 2012;13:117. doi: 10.1186/1471-2164-13-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Hackenmiller R, Berg L, Jean F, Nakayama T, Thomas G, Christian JL. The activity and signaling range of mature BMP- is regulated by sequential cleavage at two sites within the prodomain of the precursor. Gen Dev. 2001;15:2797–2802. doi: 10.1101/gad.940001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delker C, Quint M. Expression level polymorphisms: heritable traits shaping natural variation. Trends Plant Sci. 2011;16:481–488. doi: 10.1016/j.tplants.2011.05.009. [DOI] [PubMed] [Google Scholar]
- Diez-Roux G, Banfi S, Sultan M, Geffers L, Anand S, et al. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1000582. e1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollé P, Dierich A, LeMeur M, Schimmang T, Schuhbaur B, Chambon P, Duboule D. Disruption of the Hoxd-13 gene induces localized heterochrony leading to mice with neotenic limbs. Cell. 1993 Nov 5;75(3):431–441. doi: 10.1016/0092-8674(93)90378-4. [DOI] [PubMed] [Google Scholar]
- Hallgrímsson B, Brown JJY, Ford-Hutchinson AF, Sheets HD, Zelditch ML, Jirik FR. The brachymorph mouse and the developmental basis for canalization and morphological integration. Evol Dev. 2006;8:61–73. doi: 10.1111/j.1525-142X.2006.05075.x. [DOI] [PubMed] [Google Scholar]
- Hill C, Reeves RH, Richtsmeier JT. Effects of aneuploidy on skull growth in a mouse model of Down syndrome. J Anat. 2007;210:394–405. doi: 10.1111/j.1469-7580.2007.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N, Lee I, Marcotte EM, Hurles ME. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001154. e1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingraham CR, Kinoshita A, Kondo S, Yang B, Sajan S, Trout KJ, Malik MI, Dunnwald M, Goudy SL, Lovett M, Murray JC, Schutte BC. Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6. Irf6 Nat Genet. 2006;38:1335–1340. doi: 10.1083/ng1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KR, Sweet HO, Donahue LR, Ward-Bailey P, Bronson RT, Davisson MT. A new spontaneous mouse mutation of Hoxd13 with a polyalanine expansion and phenotype similar to human synpolydactyly. Hum Mol Genet. 1998 Jun;7(6):1033–1038. doi: 10.1093/hmg/7.6.1033. [DOI] [PubMed] [Google Scholar]
- Klingenberg CP, Leamy LJ, Routman EJ, Cheverud JM. Genetic architecture of mandible shape in mice: effects of quantitative trait loci analyzed by geometric morphometrics. Genetics. 2001;157:785–802. doi: 10.1093/genetics/157.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg CP. Evolution and development of shape: integrating quantitative approaches. Nat Rev Gen. 2010;11:623–635. doi: 10.1038/nrg2829. [DOI] [PubMed] [Google Scholar]
- Klingenberg CP. MorphoJ: an integrated sowftware package for geometric morphometrics. Mol Ecol Resour. 2011;11:353–357. doi: 10.1111/j.1755-0998.2010.02924.x. [DOI] [PubMed] [Google Scholar]
- Kuss P, Villavicencio-Lorini P, Witte F, Klose J, Albrecht AN, Seemann P, Hecht J, Mundlos S. Mutant Hoxd13 induces extra digits in a mouse model of synpolydactyly directly and by decreasing retinoic acid synthesis. J Clin Invest. 2009 Jan;119(1):146–156. doi: 10.1172/JCI36851. Epub 2008 Dec 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leamy LJ, Klingenberg CP, Sherratt E, Wolf JB, Cheverud JM. A search for quantitative trait loci exhibiting imprinting effects on mouse mandible size and shape. Heredity. 2008;101:518–526. doi: 10.1038/hdy.2008.79. [DOI] [PubMed] [Google Scholar]
- LeClair EE, Mui SR, Huang A, Topczewska JM, Topczewski J. Craniofacial skeletal defects of adult zebrafish Glypican 4 (knypek) mutants. Dev Dyn. 2009;238:2550–2563. doi: 10.1002/dvdy.22086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Selever J, Murali D, Sun X, Brugger SM, Ma L, Schwartz RJ, Maxson R, Furuta Y, Martin JF. Threshold-specific requirements for Bmp4 in mandibular development. Dev Biol. 2005 Jul 15;283(2):282–293. doi: 10.1016/j.ydbio.2005.04.019. [DOI] [PubMed] [Google Scholar]
- Lyons KM, Hogan B, Robertson EJ. Colocalization of Bmp7 and Bmp2 RNAs suggests that these factors cooperatively mediate tissue interactions during murine development. MOD. 1995;50:71–83. doi: 10.1016/0925-4773(94)00326-i. [DOI] [PubMed] [Google Scholar]
- Maddox BK, Garofalo S, Horton WA, Richardson MD, Trune DR. Craniofacial and otic capsule abnormalities in a transgenic mouse strain with a Col2a1 mutation. J Craniofac Genet Dev Biol. 1998;18:195–201. [PubMed] [Google Scholar]
- Malone JH, Cho DE-Y, Mattiuzzo NR, Artieri GC, Jiang L, Dale RK, Smith HE, McDaniel J, Munro S, Salit M, Andrews J, Przytycka TM, Oliver B. Mediation of Drosophila autosomal dosage effects and compensation by network interactions. Genome Biol. 2012;13:r28. doi: 10.1186/gb-2012-13-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon JA, Takada S, Zimmerman LB, Fan CM, Harland RM, McMahon AP. Noggin-mediated antagonism of BMP signaling is required for growth and patterning of the neural tube and somite. Genes Dev. 1998;12:1438–1252. doi: 10.1101/gad.12.10.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen PJ, Chin JR, Shum L, Slavkin HC, Shuler CF, Derynck R, Werb Z. Epidermal growth factor receptor function is necessary for normal craniofacial development and palate closure. Nat Gen. 1999;22:69–73. doi: 10.1038/8773. [DOI] [PubMed] [Google Scholar]
- Parsons KJ, Albertson RC. Roles for Bmp4 and CaM1in shaping the jaw: evo-devo and beyond. Annu Rev Genet. 2009;43:369–388. doi: 10.1146/annurev-genet-102808-114917. [DOI] [PubMed] [Google Scholar]
- Rohlf FJ. tpsDig, digitize landmarks and outlines, version 2.05. Department of Ecology and Evolution, State University of New York at Stony Brook, Software; 2005a. [Google Scholar]
- Rohlf FJ. tpsUtil, file utility program, version 1.26. Department of Ecology and Evolution, State University of New York at Stony Brook, Software; 2005b. [Google Scholar]
- Salsi V, Vigano MA, Cocchiarella F, Mantovani R, Zappavigna V. Hoxd13 binds in vivo and regulates the expression of genes acting in key pathways for early limb and skeletal patterning. Dev Biol. 2008 May 15;317(2):497–507. doi: 10.1016/j.ydbio.2008.02.048. Epub 2008 Mar 8. [DOI] [PubMed] [Google Scholar]
- Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, Ruff TG, Milligan SB, Lamb JR, Cavet G. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- Schunke AC, Bromiley PA, Tautz D, Thacker NA. TINA manual landmarking tool: software for the precise digitization of 3D landmarks. Front Zool. 2012;9:6. doi: 10.1186/1742-9994-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solloway MJ, Robertson EJ. Early embryonic lethality in Bmp5;Bmp7 double mutant mice suggests functional redundancy within the 60A subgroup. Development. 1999;126:1753–1768. doi: 10.1242/dev.126.8.1753. [DOI] [PubMed] [Google Scholar]
- Solloway MJ, Dudley AT, Bikoff EK, Lyons KM, Hogan BL, Robertson EJ. Mice lacking Bmp6 function. Dev. Genet. 1998;22:321–339. doi: 10.1002/(SICI)1520-6408(1998)22:4<321::AID-DVG3>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Stottmann RW, Anderson RM, Klingensmith J. The BMP antagonists Chordin and Noggin have essential but redundant roles in mouse mandibular outgrowth. Dev Biol. 2001;240:457–473. doi: 10.1006/dbio.2001.0479. [DOI] [PubMed] [Google Scholar]
- Tilleman H, Hakim V, Novikov O, Liser K, Nashelsky L, Di Salvio M, Krauthammer M, Scheffner O, Maor I, Mayseless O, Meir I, Kayam G, Sela-Donenfeld D, Simeone A, Brodski C. Bmp5/7 in concert with the mid-hindbrain organizer control development of noradrenergic locus coeruleus neurons. Mol Cell Neurosci. 2010;45:1–11. doi: 10.1016/j.mcn.2010.05.003. [DOI] [PubMed] [Google Scholar]
- Vaahtokari A, Åberg T, Jernvall J, Keränen S, Thesleff I. The enamel knot as a signalling center in the developing mouse tooth. MOD. 1996;54:39–43. doi: 10.1016/0925-4773(95)00459-9. [DOI] [PubMed] [Google Scholar]
- Veitia RA, Bottani S, Birchler JA. Cellular reactions to gene dosage imbalance: genomic, transcriptomic and proteomic effects. Trends Genet. 2008;24:390–397. doi: 10.1016/j.tig.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Willmore KE, Zelditch ML, Young N, Ah-Seng A, Lozanoff S, Hallgrímsson B. Canalization and developmental stability in the brachyrrhine mouse. J Anat. 2006;208:361–372. doi: 10.1111/j.1469-7580.2006.00527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnier G, Blessing M, Labosky PA, Hogan BL. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995 Sep 1;9(17):2105–2116. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG. Evolutionary changes in cis and trans gene regulation. Nature. 2004;430(6995):85–88. doi: 10.1038/nature02698. [DOI] [PubMed] [Google Scholar]
- Wu P, Jiang T-X, Shen J-Y, Widelitz RB, Chuong C-M. Morphoregulation of avian beaks: comparative mapping of growth zone activities and morphological evolution. Dev Dyn. 2006;235:1400–1412. doi: 10.1002/dvdy.20825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Jiang T-X, Suksaweang S, Widelitz RB, Chuong C-M. Molecular shaping of the beak. Science. 2004;305:1465–1466. doi: 10.1126/science.1098109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelditch M, Swiderski D, Sheets DH, Fink W. Geometric morphometrics for biologists. New York, London: Elsevier Academic Press; 2004. [Google Scholar]
- Zouvelou V, Luder H-U, Mitsiadis TA, Graf D. Deletion of BMP7 affects the development of bones, teeth, and other ectodermal appendages of the orofacial complex. J Exp Zool (Mol Dev Evol) 2009;312B:361–374. doi: 10.1002/jez.b.21262. [DOI] [PubMed] [Google Scholar]