Abstract
The growth and development of plants are sensitive to their surroundings. Although numerous studies have analyzed plant transcriptomic variation, few have quantified the effect of combinations of factors or identified factor-specific effects. In this study, we performed RNA sequencing (RNA-seq) analysis on tobacco leaves derived from 10 treatment combinations of three groups of ecological factors, i.e., climate factors (CFs), soil factors (SFs), and tillage factors (TFs). We detected 4980, 2916, and 1605 differentially expressed genes (DEGs) that were affected by CFs, SFs, and TFs, which included 2703, 768, and 507 specific and 703 common DEGs (simultaneously regulated by CFs, SFs, and TFs), respectively. GO and KEGG enrichment analyses showed that genes involved in abiotic stress responses and secondary metabolic pathways were overrepresented in the common and CF-specific DEGs. In addition, we noted enrichment in CF-specific DEGs related to the circadian rhythm, SF-specific DEGs involved in mineral nutrient absorption and transport, and SF- and TF-specific DEGs associated with photosynthesis. Based on these results, we propose a model that explains how plants adapt to various ecological factors at the transcriptomic level. Additionally, the identified DEGs lay the foundation for future investigations of stress resistance, circadian rhythm and photosynthesis in tobacco.
Keywords: climate factors, soil factors, tillage factors, tobacco leaves, transcriptome, RNA-seq
1. Introduction
Given that plants root at the same spot throughout their life and that they have large surface areas in contact with the environment, environmental changes have a greater impact on the growth and survival of plants than they do on animals. Several environmental factors, including terrain, climate, soil properties, and soil water have been recognized as affecting plant growth and development and these factors are typically considered when selecting cultivation sites [1]. Among these environmental factors, climate factors (CFs: including light, temperature, air, rainfall, and wind) have the greatest impact on the spatial distribution of vegetation and the yield performance of crops [2], and account for much of the regional variation in crop production. SFs also have a critical effect on plant growth and crop yield, as they determine the physical environment of crop roots and are the major source of nutrients [3]. Soil fertility, texture, organic-matter content, and mineralogy greatly affect the quality and yield of crops. Because SFs can be carefully managed to promote the production of high-yielding and high-quality crops, soil fertility and plant nutrition supplies have been the focus of much research [4,5]. Over 65% of all cropland is supplemented with commercial fertilizer, lime, and soil conditioners, and almost all maize (Zea may) and over 80% of wheat (Triticum aestivum) and cotton (Gossypium hirsutum) are supplemented with commercial nutritional products to improve soil fertility and, ultimately, crop yield in the United States [1]. In addition to environmental factors, TFs also influence the yield and quality of food crops by affecting soil moisture, nutrient availability, temperature, and aeration [6]. Although no-tillage (NT) management has become more popular in North America, as this approach reduces soil erosion and cost and improves soil health, conventional tillage (CT) is still the major farming practice in Asia, South America, and Africa, since the NT approach tends to decrease yield and protein content of crops [7,8]. It is well known that various environmental factors and TFs mutually affect each other [9]. While the effect of each of the above-mentioned factors on crop production has been widely investigated, only a few studies have analyzed the influence of combinations of these factors or identified the specific impact of each factor at the transcriptomic level. Both in tobacco and Arabidopsis thaliana, the molecular and metabolic response of plants to a combination of drought and heat stress is distinct from that of plants subjected to each of these stresses applied individually, 454 transcripts in Arabidopsis were observed specifically expressed in cells during a combination of drought and heat stress [10,11]. Investigation on phenotypic plasticity of grapevine (Vitis vinifera) by comparing the berry transcriptome also revealed the relationships among differential gene expression profiles, environments, growing conditions and ripening parameters and identified several putative candidate genes for the definition of berry quality traits [12].
Recent advances in high-throughput sequencing technology have led to a dramatic increase in the production of sequencing data for RNA-seq and ChIP-seq analyses [13], providing rapid and cost-effective tools to monitor transcriptomic changes. Several studies have assessed global gene expression in different tissues, at different developmental stages, and in response to various environmental stimuli [14,15]. However, the emphasis has always been placed on transcriptomic variation in response to an individual treatment, and the effects of various combinations of treatments have been largely neglected. A transcriptome comparison of fertilized ovary and basal leaf meristem tissue of drought-treated and well-watered maize revealed that more drought-responsive genes were activated in the ovary than in the leaf meristem upon exposure to drought stress [14]. Whole-genome expression profile analysis of lupin (Lupinus spp.) identified 2128 genes that were differentially expressed in response to phosphate (Pi) deficiency stress, suggesting that novel mechanisms of Pi deficiency-induced metabolism and cytokinin and gibberellic acid signaling exist [16]. Transcriptome assembly from RNA-seq data identified 28,335 unique genes from sorghum (Sorghum bicolor L. Moench) root and shoot tissues challenged with polyethylene glycol (PEG)-induced osmotic stress and exogenous abscisic acid (ABA) [16]. Recent transcriptomic data from the leaves of field-grown rice (Oryza sativa) plants at various seasonal, diurnal, and developmental time points along with the corresponding meteorological data were successfully used to develop a statistical model that predicts the influence of variable environmental conditions on transcriptome dynamics, which was found to be predominantly governed by endogenous diurnal rhythms, ambient temperature, plant age, and solar radiation [17]. Model testing on the following year’s rice plants proved that this model was highly accurate at associating gene expression changes with environmental influences, thus suggesting a promising means of translating large amounts of laboratory-learnt knowledge into practical solutions for problems encountered in agricultural production. Despite numerous studies of plant genomes and transcriptomes, few studies have addressed the influence of combinations of environmental and other factors on gene expression profiles. Moreover, few extensive studies have quantified the transcriptome-wide effect of a specific factor although some research has focused on the transcriptomic variation caused by single factors.
Tobacco (Nicotiana tabacum) belongs to the agriculturally important Solanaceae family and is a valuable industrial and commercial crop in many countries, although consumption has steadily declined due to increased public awareness of smoking-related health risks and government regulations [18]. It is also an important model organism in plant genetics research, and is ideal for studies in phenotypic diversity, hybridization and ploidy manipulations, and functional characterization. Growth, flowering, and metabolism of tobacco plants are remarkably sensitive to environmental changes, especially to changes in the physical and chemical properties of the soil. Nitrogen and potassium are the nutrient elements with the greatest impact on tobacco growth and development. Optimal levels of nitrogen in the soil increase crop yield, while potassium improves tobacco quality [19]. Compared with CT, minimum tillage (MT) did not have a pronounced effect on tobacco yield, but significantly prolonged the vegetative growth stage [20]. To investigate the transcriptomic variation caused by CFs, SFs, and/or TFs, we analyzed the leaves of tobacco plants subjected to various treatment combinations. Our study provides novel insight into the molecular mechanisms whereby plants adapt to ecological changes and the relationship between various ecological factors at the transcriptomic level.
2. Results and Discussion
2.1. RNA-Seq Data Analyses
To compare transcriptomic variations in the leaves of tobacco plants exposed to different CFs, SFs, and TFs, ten samples cultivated in Kaiyang County (KY), Weining County (WN), and Tianzhu County (TZ) and exposed to different treatments were collected and used for RNA-seq analysis.
After removal of low quality and contaminated reads, a total of 58,466,453 50 bp raw reads were acquired, ranging from 5.2 to 6.2 M reads per sample, containing 8.28 gigabases (Gb) of sequence data (Table 1). We aligned the sequence reads against the tobacco SGN Unigene database (containing 84,602 unique ESTs) in the Solanaceae Genomics Network (SGN) [21], using TopHat with default parameters [22]. Seventy percent of the total reads were successfully mapped to the reference sequence (Table 1), resulting in about a 50-fold average coverage of the tobacco SGN Unigenes. Previous study demonstrated that 10 and 30 M (75 bp) reads could detect about 80% and all annotated chicken genes, respectively [23]. But, another study also indicated that RNA-seq density generated by about 6 M (36 bp) reads showed a strong congruence with expression metrics from array intensities (Pearson’s r = 0.90–0.91) [24]. Considering tobacco is a tetraploid, the sequencing depth in our study would be enough to identify highly expressed DEGs, but might be insufficient for detection of extremely low expression transcripts.
Table 1.
Summary of RNA-seq reads mapping to reference genes.
| Sample | Total reads | Mapped reads | % Mapped reads | Number of transcripts |
|---|---|---|---|---|
| KYC | 6,010,205 | 4,164,387 | 69.29% | 26,864 |
| KYP | 6,127,551 | 4,276,322 | 69.79% | 25,823 |
| KYsWN | 5,566,935 | 3,894,388 | 69.96% | 25,001 |
| KYsTZ | 6,020,409 | 4,162,982 | 69.15% | 25,550 |
| WNC | 5,252,135 | 3,663,806 | 69.76% | 23,971 |
| WNP | 5,913,266 | 4,259,827 | 72.04% | 24,185 |
| WNsKY | 5,218,808 | 3,704,089 | 70.98% | 24,261 |
| TZC | 6,091,837 | 4,254,123 | 69.83% | 25,128 |
| TZP | 6,005,103 | 4,222,155 | 70.31% | 25,540 |
| TZsKY | 6,260,204 | 4,455,821 | 71.18% | 26,100 |
| Overall | 58,466,453 | 41,057,900 | 70.22% | 31,057 |
KYC, TZC, and WNC represent samples came from KY, TZ, and WN without soil exchange and tillage treatment. KYP, TZP, and WNP represent samples harvested from the corresponding cultivated regions with tillage treatment. KYsTZ and KYsWN represent samples harvested from KY grown in soil from TZ and WN, respectively; TZsKY and WNsKY indicate samples collected from TZ and WN, respectively, and grown on KY soil. Total reads corresponds to the initial output of sequencing reads. Mapped reads refers to the number of reads mapped to the tobacco SGN Unigene reference sequence.
To quantify transcriptomic variations in our samples, Cufflinks was used to assemble all reads into transcript models [22]. Subsequently, expression levels for all transcripts were calculated in fragments per kilobase of exon model per million mapped reads (FPKM), a length-normalized measure of exonic read density that allows expression levels to be compared within or between different samples [25]. Using a threshold of mean FPKM higher than 10, an aggregate of 23,442 to 26,935 mRNA transcripts for each sample was observed. A total of 30,688 unique transcripts were expressed in ten samples (Table S1). The transcripts generated in our study most likely represent almost the complete transcriptome of tobacco leaves. Our results are in accordance with those from a previous tobacco transcriptome analysis in which the transcripts were assembled into a set of 40,642 high-quality unigenes [26]. The transcript number in our study was also much lower than that of the tobacco SGN Unigenes (84,602), indicating that more than half of the SGN unigenes are not expressed in tobacco leaves.
Expression distribution and box plot analysis revealed that ten samples in our study possessed similar expression patterns, with more than 80% of the genes being expressed between 10 and 100 FPKM and around 15% between 100 and 1000 FPKM (Figures S1 and S2). After normalization, box plot and gene expression level distributions both showed that the transcripts of ten samples had similar expression patterns and variation ranges and a normal distribution, suggesting that the ten sets of sequencing data are comparable and suitable for downstream transcriptomic variation analysis.
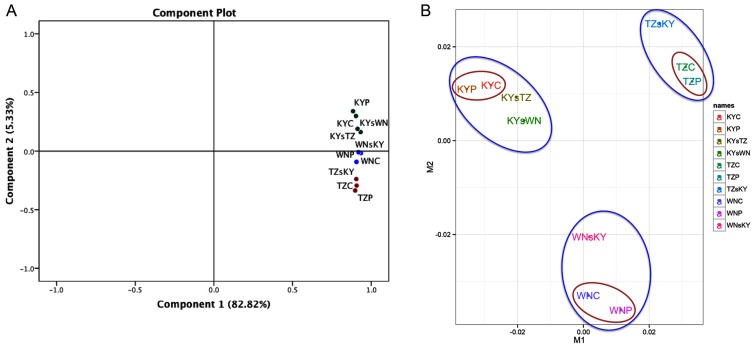
To confirm our RNA-seq data and to conduct a preliminarily comparison of the effects of three different ecological factors on tobacco leaf transcriptomes, we implemented a principal component analysis (PCA) of the sample correlation matrix calculated from log2-transfromed FPKM values (Figure 1A). The first principal component accounted for 82.82% of the total variability, which in this case corresponds to the reference sequence-specific variance, and the subsequent principal components accounted for 5.33% and 3.35% of overall variance, highlighting the difference between the samples affected by CFs and SFs, respectively (Figure 1 and Table S2), as the values in the component matrix between WN, TZ, and KY were quite different, while those within the same cultivated region varied only slightly (Table 2). Based on the PCA and MDC plots, the largest variance was caused by CFs, and the smallest by TFs (Figure 1). Although it is challenging to make direct comparisons between factors in the above-mentioned two plots, transcriptomic variation caused by SFs is markedly higher than that caused by TFs.
Figure 1.
PCA and MDC plots of log2-normalized FPKM of ten RNA-seq samples. In the PCA plot (A); green, blue, and brown discs represent samples from KY, WN, and TZ, respectively; In the MDS plot (B), the brown and blue ellipses indicate transcriptomic variation affected by TFs and CFs, respectively.
Table 2.
The number of DEGs in tobacco leaves affected by different CFs, SFs, and/or TFs.
| Group | No. | Combination | Number of DR genes | Number of UR genes | Number of DEGs |
|---|---|---|---|---|---|
| (i) Different CFs and the same SFs and TFs | 1 | KYP/WNsKY | 610 | 722 | 1332 |
| 2 | KYP/TZsKY | 971 | 1029 | 2000 | |
| 3 | KYP/WNsKY_TZsKY | 858 | 679 | 1537 | |
| 4 | KYsWN/WNP | 533 | 1100 | 1633 | |
| 5 | KYsTZ/TZP | 857 | 1104 | 1961 | |
| 6 | KYsWN_KYsTZ/WNP_TZP | 648 | 1155 | 1803 | |
| 7 | TZsKY/WNsKY | 631 | 739 | 1370 | |
|
| |||||
| (ii) The same CFs and different SFs and TFs | 8 | KYsWN/KYC | 505 | 330 | 835 |
| 9 | KYsTZ/KYC | 654 | 585 | 1239 | |
| 10 | KYsWN_KYsTZ/KYC | 558 | 604 | 1162 | |
| 11 | WNsKY/WNC | 164 | 242 | 406 | |
| 12 | TZsKY/TZC | 331 | 513 | 844 | |
| 13 | WNsKY_TZsKY/WNC_TZC | 8 | 18 | 26 | |
|
| |||||
| (iii) Different SFs and the same CFs and TFs | 14 | KYsWN/KYP | 462 | 576 | 1038 |
| 15 | KYsTZ/KYP | 514 | 893 | 1407 | |
| 16 | KYsWN_KYsTZ/KYP | 518 | 1041 | 1559 | |
| 17 | WNsKY/WNP | 255 | 395 | 650 | |
| 18 | TZsKY/TZP | 416 | 315 | 731 | |
| 19 | WNsKY_TZsKY/WNP_TZP | 44 | 30 | 74 | |
|
| |||||
| (iv) The same SFs and different CFs and TFs | 20 | WNsKY/KYC | 960 | 506 | 1466 |
| 21 | TZsKY/KYC | 1168 | 737 | 1905 | |
| 22 | WNsKY_TZsKY/KYC | 662 | 543 | 1205 | |
| 23 | KYsWN/WNC | 362 | 850 | 1212 | |
| 24 | KYsTZ/TZC | 668 | 1143 | 1811 | |
| 25 | KYsWN_KYsTZ/WNC_TZC | 317 | 709 | 1026 | |
|
| |||||
| (v) Different TFs and the same CFs and SFs; | 26 | KYP/KYC | 452 | 277 | 729 |
| 27 | WNP/WNC | 289 | 307 | 596 | |
| 28 | TZP/TZC | 151 | 259 | 410 | |
| 29 | KYP_WNP_TZP/KYC_WNC_TZC | 0 | 0 | 0 | |
|
| |||||
| (vi) The same TFs and different CFs and SFs | 30 | KYC/WNC | 523 | 1212 | 1735 |
| 31 | KYC/TZC | 710 | 1412 | 2122 | |
| 32 | KYC/WNC_TZC | 531 | 794 | 1325 | |
| 33 | TZC/WNC | 625 | 821 | 1446 | |
| 34 | KYP/WNP | 750 | 1082 | 1832 | |
| 35 | KYP/TZP | 1034 | 1109 | 2143 | |
| 36 | KYP/WNP_TZP | 1144 | 1065 | 2209 | |
| 37 | TZP/WNP | 529 | 956 | 1485 | |
UR genes: up-regulated genes; DR genes: down-regulated genes. In each combination, DEGs are identified from expression level comparison between samples before and after the slash, using the latter as reference. Underscore between samples indicate that these samples are regarded as an integral whole sample for transcriptomic comparison. For example, the FPKM value of each gene in WNsKY_TZsKY in combination 3 is calculated from samples WNsKY and TZsKY.
2.2. General Trend of DEGs in Tobacco Leaves
To determine the general trend of differentially expressed genes (DEGs) in tobacco leaves exposed to different ecological factors, we identified and analyzed DEGs between the 10 samples in our study (Figure S3 and Table 2). Both up- and down-regulated DEGs were represented with red dots. The number of DEGs between RNA-seq samples retrieved from the same cultivated region was far smaller than those from different cultivated regions, which is in accordance with our PCA and MDC results.
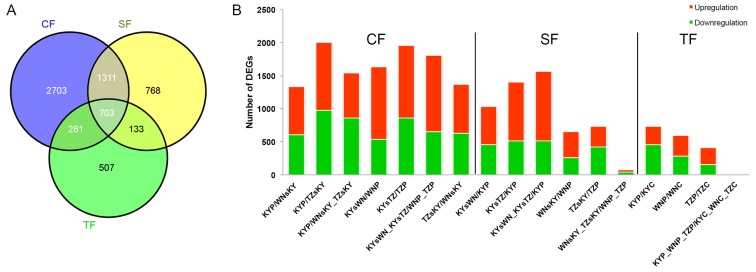
To compare the effect of CFs, SFs, and TFs on the transcriptomes of tobacco leaves, RNA-seq samples were grouped into six different treatments containing 37 pairs of combinations (Tables 2 and S3), including (i) different CFs and the same SFs and TFs; (ii) the same CFs and different SFs and TFs; (iii) different SFs and the same CFs and TFs; (iv) the same SFs and different CFs and TFs; (v) different TFs and the same CFs and SFs; (vi) different TFs and the same CFs and SFs. Based on the screening standard described in Materials and Methods, we found that 6386 genes with FDR <0.5 were differentially expressed in the treatment groups (i), (iii), and (v) (Figure 2), which differ in terms of a single factor. A Venn diagram was created to identify ecological factor-specific DEGs and common DEGs readily affected by changes in ecological factors. The number of CF-, SF-, and TF-specific DEGs was 2703, 768, and 507, respectively, while there were 703 common DEGs in the three treatment groups. In addition, many DEGs were observed between samples that were treated with the same two groups of ecological factors. For instance, CFs and SFs share 1311 common DEGs, whereas TFs have 261 and 133 common DEGs with CFs and SFs, respectively. For samples in group (i) above, KY and TZ (No. 1 and 5 in Table 2) had the most DEGs, while TZ and WN (No. 7 in Table 2) had the fewest DEGs. In group (iii), the number of DEGs in KY soil (No. 14 to 16 in Table 2) was much higher than that in either WN (No. 17 in Table 2) or TZ (No. 18 in Table 2). In group (v), the number of DEGs in KY, WN, and TZ (No. 26 to 28 in Table 2) sequentially decreased, suggesting that changes in TF had diverse effects on the transcriptomes of tobacco leaves sampled from plants cultivated in different regions, and had the greatest effect on plants grown in KY (Table 2 and Figure 2B).
Figure 2.
DEGs identified in three treatment combinations. (A) A Venn diagram was generated to identify CF-, SF-, and TF-specific DEGs and common DEGs within treatment groups (i), (iii), and (v); (B) Number of DEGs in 17 comparison combinations, including seven CF, six SF, and four TF combinations. In each combination, samples were taken from the treatment group with only one different group of ecological factors (CFs, SFs, or TFs). For example, samples of the third comparison combination “KYP/WNsKY_TZsKY” were harvested from the same SFs (KY) and TFs (tillage) and different CFs (KY and sum of WN and TZ). DEGs were determined by comparing the FPKM values of samples before and after slash using the latter as control. An underscore in the sample name indicates an integrated sample. For instance, “WNsKY_TZsKY” represents the average of the sum of WNsKY and TZsKY.
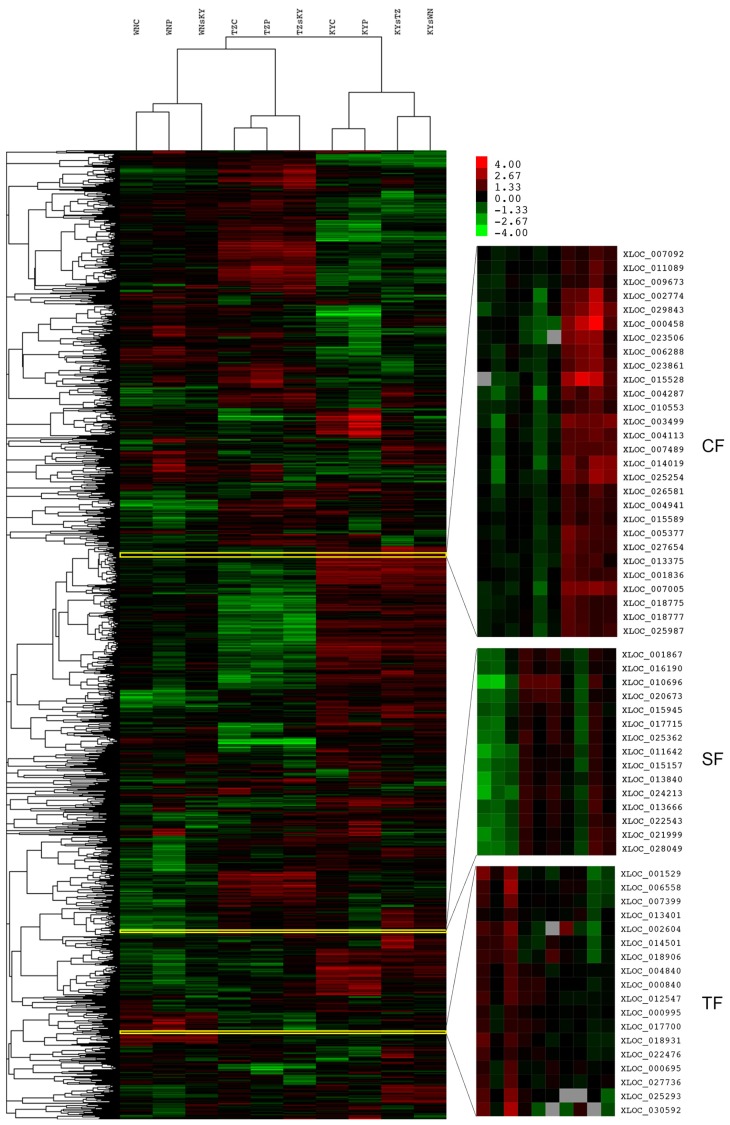
Based on our volcano plot analysis and comparison of number of DEGs affected by different ecological factors (Figure S3), the CFs generally had the greatest effect on transcriptomic variation in tobacco leaves, followed by SFs, and TFs. Cluster analysis of all 6386 DEGs also strongly supported the above conclusion, since expression data of ten samples could be divided into three groups firstly based on three different cultivated regions (CFs), while in the same regions, those between no-tillage and conventional tillage (TFs) were primarily clustered, then grouped with samples derived from soil exchange treatment (Figure 3).
Figure 3.
Hierarchical clustering and Treeview visualization of all DEGs. Ten samples from three different cultivated regions with soil exchange and tillage treatments were collected were subjected to RNA-seq, revealing a total of 6,386 DEGs among treatment groups (i), (iii), and (v). Log2 values were used to cluster all the DEGs in Cluster 3.0 using uncentered correlation and the complete linkage method. Results were visualized using Treeview. Left heatmap represents global visualization of all the DEGs, right gene cluster are representative CF-, SF- and TF-specific DEGs. Red indicates genes that are up-regulated, green indicates genes that are down-regulated.
2.3. Functional Analysis of Common DEGs Affected by CFs, SFs, and TFs
Genes that are induced by ecological factors play crucial roles in plant growth, development, and adaptation to different ecological stresses. In our study, we identified 703 common DEGs that were simultaneously induced by CFs, SFs, and TFs. These inducible genes might be important for maintaining normal growth and development of tobacco plants and for improving resistance to environmental changes. Based on Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) annotations, 21,410 and 9214 unique genes in our study could be assigned GO and KO terms. Enrichment analysis of common DEGs identified a total of 146 GO and 18 KO terms that were significantly over-represented.
In our GO enrichment analysis (Table S4), 153 genes were found to be involved in response to stimulus (GO:0050896), and 125 of these were annotated as response to stress (GO:0006950). These results suggest that several genes related to the plant’s response to environmental stresses are regulated primarily for adaptation to environmental variation in tobacco plants, which is agreement with our previous study [18]. These stress-responsive genes could be further divided into three classes, i.e., genes that are responsive to a temperature stimulus (GO:0009266, 42 genes), light stimulus (GO:0009416, 29 genes), and oxidative stress (GO:0006979, 32 genes). It is noteworthy that 20 of the 29 genes that respond to a light stimulus are responsive to high intensity light. The first two sets of inducible genes are mainly involved in adaptation to climate change through transcriptional regulation of temperature- and high light-responsive genes in tobacco leaves, whereas the oxidative stress-responsive genes might be important for adaptation to different soils, especially those with different water content. Furthermore, we identified 75 and 30 genes that respond to chemical stimulus (GO:0042221) and hormone stimulus (GO:0009725), suggesting that auxin, ethylene, and abscisic acid (ABA) signaling pathway genes have pivotal roles in the tobacco plant’s response to environmental stresses. Many of the 703 common DEGs had functions related to carbohydrate metabolic processes (GO:0005975, 43 genes) and the cell wall (GO:0005618, 47 genes). These genes have important roles in plant cell wall organization, biosynthesis, and modification, and could increase tobacco plant resistance to abiotic or biotic stresses.
In our KEGG enrichment analysis, KEGG pathway annotations of 203 common DEGs were generated and analyzed for statistical significance (Table S5). Common DEGs were mainly over-represented in ancient, conserved, and secondary metabolic pathways, such as flavonoid biosynthesis (ko00941, 6 genes), phenylpropanoid biosynthesis (ko00940, 12 genes), and starch and sucrose metabolism pathways (ko00500, 12 genes). Although nitrogen metabolism (ko00910, 7 genes) and MAPK signaling pathway (ko04010, 7 genes) genes were also significantly over-enriched in our study, DEGs involved in protein processing in the endoplasmic reticulum (ko04141, 42 genes) were more abundant than those associated with any other pathway. Therefore, based on our GO and KEGG enrichment analyses, we propose that common DEGs affected by CFs, SFs, and TFs are the most important for the survival of tobacco plants under various ecological conditions, particularly stress.
Based on the meteorological data at three cultivated regions (Table S6 [27]), measurement of agronomic traits (Table S7) and activities of antioxidant enzyme superoxide dismutase (SOD, EC 1.15.1.1), peroxidase (POD, EC 1.11.1.7) and catalase (CAT, EC 1.11.1.6) (Table 3), it could be observed that tobacco plant height and number of leaves at 70 days after transplanting (DAT) at cultivation region KY were obviously higher than WN and TZ, and stem perimeter and maximum leaf area at 45 and 70 DAT at cultivation region TZ were smaller than KY and WN. In addition, the influence of soil exchange and tillage treatment on agronomic traits also could be found, for example, four tested agronomic traits between KYC and KYP (TFs), and between KYP and KYsTZ (SFs) were quite different (Table S7). To further investigate tobacco plant growth status the activities of antioxidant enzymes SOD, POD and CAT of tobacco leaves were measured. From Table 3, the impact of environmental variation (CFs) on activities of CAT and POD could be seen. For instance, CAT activities of samples from cultivation region TZ were much higher than KY and WN at 45 DAT, but lower at 70 DAT. Soil exchange (SFs) and tillage treatment (TFs) also showed significant influence on variation of POD activities (Table 3). These results indicated that tobacco plants adapted to variation of CFs, SFs and TFs by adjusting enzyme activity and plant growth, which was in accordance with the above-mentioned GO and KEGG pathway analyses.
Table 3.
CAT, POD and SOD activities of tobacco leaves.
| Location | Treatments | 45 DAT | 70 DAT | ||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| CAT | POD | SOD | CAT | POD | SOD | ||
| KY | KYC | 140.8 | 1551.3 | 611.7 | 101.4 | 6298.3 | 481.3 |
| KYP | 128.0 | 2181.9 | 648.2 | 93.5 | 6914.7 | 483.9 | |
| KYsTZ | 116.4 | 4275.2 | 594.0 | 108.8 | 8468.2 | 545.4 | |
| KYsWN | 124.5 | 3929.5 | 654.1 | 74.7 | 9298.9 | 454.1 | |
|
| |||||||
| TZ | TZC | 378.7 | 3929.3 | 511.5 | 94.3 | 4362.9 | 500.7 |
| TZP | 391.9 | 4697.0 | 476.8 | 134.3 | 4474.2 | 449.5 | |
| TZsKY | 361.2 | 3435.4 | 514.5 | 175.9 | 3724.5 | 499.4 | |
|
| |||||||
| WN | WNC | 175.7 | 1780.9 | 509.4 | 142.4 | 5180.0 | 516.9 |
| WNP | 177.7 | 2255.6 | 469.8 | 144.2 | 6428.5 | 470.8 | |
| WNsKY | 208.3 | 1426.6 | 507.7 | 143.5 | 4348.7 | 492.8 | |
Values were expressed in units of enzyme activity per gram wet weight of tissue.
2.4. Functional Analysis of CF-Specific DEGs
Compared with SFs and TFs, CFs had a much greater influence on the transcriptomic variation of tobacco leaves. In our study, a total of 2703 CF-specific DEGs were identified and found to be over-represented in 64 GO terms and 12 KEGG pathways (Table 4). Among the over-enriched KEGG pathways, plant hormone signal transduction (ko04075), with 35 DEGs, was most enriched (Table S8). Although several genes that respond to hormone stimuli were identified in the above-mentioned common DEGs, these genes differed from hormone-responsive CF-specific DEGs (ko04075). We identified several well-studied hormone-responsive genes amongst the CF-specific DEGs, such as ABA-responsive element binding factor ABF3 (XLOC_016015) [28], ABA-activated protein kinase (XLOC_003676), jasmonate receptor CORONATINE INSENSITIVE 1 (COI1) [29], auxin-inducible SAUR gene (XLOC_018600) [30], and cytokinin receptor histidine kinase AHK3 (XLOC_014140) [31]. The large number of hormone-responsive genes specifically affected by CFs may improve the adaptive ability of tobacco plants, allowing them to flourish in various growth regions.
Table 4.
Over-represented GO terms of CF-specific DEGs.
| GO-ID | p-value | Input number | Background number | Description |
|---|---|---|---|---|
| 0048578 | 4.00 × 109 | 8 | 8 | positive regulation of long-day photoperiodism, flowering |
| 0010378 | 4.00 × 109 | 8 | 8 | temperature compensation of the circadian clock |
| 0009813 | 7.20 × 109 | 20 | 52 | flavonoid biosynthetic process |
| 0042398 | 1.51 × 108 | 44 | 200 | cellular amino acid derivative biosynthetic process |
| 0055114 | 5.23 × 108 | 237 | 1916 | oxidation reduction |
| 0010229 | 1.08 × 107 | 11 | 19 | inflorescence development |
| 0006575 | 2.18 × 107 | 57 | 316 | cellular amino acid derivative metabolic process |
| 0009812 | 2.20 × 107 | 20 | 62 | flavonoid metabolic process |
| 0009699 | 4.81 × 107 | 23 | 82 | phenylpropanoid biosynthetic process |
| 0048586 | 5.16 × 107 | 8 | 11 | regulation of long-day photoperiodism, flowering |
| 0006857 | 7.65 × 107 | 17 | 50 | oligopeptide transport |
| 0015833 | 7.65 × 107 | 17 | 50 | peptide transport |
| 0009698 | 9.35 × 107 | 29 | 123 | phenylpropanoid metabolic process |
| 0019748 | 1.01 × 106 | 44 | 230 | secondary metabolic process |
| 0006355 | 1.57 × 106 | 161 | 1262 | regulation of transcription, DNA-dependent |
| 0045449 | 1.99 × 106 | 161 | 1267 | regulation of transcription |
| 0051252 | 2.52 × 106 | 161 | 1272 | regulation of RNA metabolic process |
| 0008215 | 3.27 × 106 | 6 | 7 | spermine metabolic process |
| 0006597 | 3.27 × 106 | 6 | 7 | spermine biosynthetic process |
| 0010556 | 6.96 × 106 | 162 | 1304 | regulation of macromolecule biosynthetic process |
| 0008295 | 7.34 × 106 | 8 | 14 | spermidine biosynthetic process |
| 0031326 | 8.48 × 106 | 166 | 1347 | regulation of cellular biosynthetic process |
| 0009889 | 1.05 × 105 | 166 | 1352 | regulation of biosynthetic process |
| 0008216 | 1.45 × 105 | 8 | 15 | spermidine metabolic process |
| 0006835 | 2.38 × 105 | 7 | 12 | dicarboxylic acid transport |
| 0051171 | 2.62 × 105 | 167 | 1384 | regulation of nitrogen compound metabolic process |
| 0019219 | 2.89 × 105 | 165 | 1367 | regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process |
| 0015798 | 3.15 × 105 | 5 | 6 | myo-inositol transport |
| 0006596 | 6.03 × 105 | 9 | 22 | polyamine biosynthetic process |
| 0006833 | 6.17 × 105 | 11 | 32 | water transport |
| 0042044 | 6.17 × 105 | 11 | 32 | fluid transport |
| 0006725 | 9.44 × 105 | 52 | 341 | cellular aromatic compound metabolic process |
| 0051258 | 1.06 × 104 | 13 | 45 | protein polymerization |
| 0006595 | 1.79 × 104 | 10 | 30 | polyamine metabolic process |
| 0080090 | 1.82 × 104 | 170 | 1466 | regulation of primary metabolic process |
| 0000160 | 1.91 × 104 | 25 | 130 | two-component signal transduction system (phosphorelay) |
| 0015791 | 2.52 × 104 | 5 | 8 | polyol transport |
| 0015850 | 2.52 × 104 | 5 | 8 | organic alcohol transport |
| 0009610 | 2.95 × 104 | 4 | 5 | response to symbiotic fungus |
| 0009873 | 3.36 × 104 | 13 | 50 | ethylene mediated signaling pathway |
| 0010468 | 3.41 × 104 | 162 | 1405 | regulation of gene expression |
| 0019438 | 3.46 × 104 | 33 | 198 | aromatic compound biosynthetic process |
| 0006629 | 3.87 × 104 | 98 | 784 | lipid metabolic process |
| 0042752 | 5.76 × 104 | 8 | 23 | regulation of circadian rhythm |
| 0008610 | 6.35 × 104 | 58 | 422 | lipid biosynthetic process |
| 0008202 | 6.75 × 104 | 12 | 47 | steroid metabolic process |
| 0051552 | 7.97 × 104 | 8 | 24 | flavone metabolic process |
| 0051553 | 7.97 × 104 | 8 | 24 | flavone biosynthetic process |
| 0051554 | 7.97 × 104 | 8 | 24 | flavonol metabolic process |
| 0051555 | 7.97 × 104 | 8 | 24 | flavonol biosynthetic process |
| 0035235 | 8.07 × 104 | 6 | 14 | ionotropic glutamate receptor signaling pathway |
| 0007215 | 8.07 × 104 | 6 | 14 | glutamate signaling pathway |
| 0060255 | 8.65 × 104 | 163 | 1444 | regulation of macromolecule metabolic process |
| 0071369 | 9.03 × 104 | 13 | 55 | cellular response to ethylene stimulus |
| 0009409 | 9.30 × 104 | 42 | 286 | response to cold |
| 0031323 | 9.94 × 104 | 184 | 1661 | regulation of cellular metabolic process |
| 0009755 | 1.12 × 103 | 45 | 315 | hormone-mediated signaling pathway |
| 0032870 | 1.13 × 103 | 46 | 324 | cellular response to hormone stimulus |
| 0042401 | 1.22 × 103 | 12 | 50 | cellular biogenic amine biosynthetic process |
| 0071495 | 1.22 × 103 | 49 | 352 | cellular response to endogenous stimulus |
| 0046148 | 1.36 × 103 | 21 | 116 | pigment biosynthetic process |
| 0010033 | 1.52 × 103 | 116 | 993 | response to organic substance |
| 0009723 | 1.55 × 103 | 22 | 125 | response to ethylene stimulus |
| 0009608 | 1.65 × 103 | 5 | 11 | response to symbiont |
DEGs related to the circadian rhythm (ko04712) were present only amongst the CF-specific DEGs. Given the large number of DEGs related to the circadian rhythm (19 in total; Table S9), we conclude that the circadian rhythm has a considerable effect on the transcriptome of tobacco plants. This result is consistent with a recent study that found that the transcriptome of rice leaves was widely affected by three factors: the circadian clock, environmental stimuli, and plant age. Similar to our study on tobacco plants, the entrained circadian clock and temperature had particularly large effects on the transcriptome [32]. We thus suggest that changes in the circadian rhythm cause CF-specific transcriptomic variations in tobacco plants. Among the CF-specific DEGs, we identified a nuclear zinc-finger gene GIGANTEA (GI, XLOC_014804, XLOC_018429, XLOC_021883, XLOC_023447, XLOC_020085, XLOC_015139 and XLOC_017755), which governs the diurnal rhythm, promoting plant flowering through the CONSTANS (CO)–FLOWERING LOCUS T (FT) regulatory module under long-day conditions [33]. The MYB transcription factors LATE ELONGATED HYPOCOTYL (LHY, XLOC_012886 and XLOC_012888) and CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) are partially redundant and essential for the maintenance of circadian rhythms in constant light conditions [34], and contribute to plant cold tolerance by regulating the C-REPEAT BINDING FACTOR (CBF) cold-response pathway [35]. An active transcriptional repressor of LHY and CCA1, PSEUDO-RESPONSE REGULATOR5 (PRR5, XLOC_011077), could directly downregulate LHY and CCA1 expression, forming an interlocking transcriptional-translational feedback loop of the circadian clock in plants [36]. GI, LHY, and PRR5 could act as key nodes of the circadian clock regulatory network in the leaves of tobacco plants derived from different cultivated regions, playing pivotal roles in plant growth and development and reflecting the adaptive ability of plants to changing environmental cues.
In addition, genes involved in secondary metabolism pathways (phenylpropanoid biosynthesis, starch and sucrose metabolism) were significantly represented in CF-specific DEGs, although these pathways were also overrepresented in common DEGs. Of the CF-specific DEGs involved in phenylpropanoid biosynthesis, genes encoding 4-coumaroyl-CoA synthase 1 (4CL1, XLOC_024395), 4-coumaroyl-CoA synthase 3 (4CL3, XLOC_020018 and XLOC_027613), cinnamic acid 4-hydroxylase (C4H, XLOC_004844 and XLOC_024688), ferulate 5-hydroxylase (F5H, XLOC_004677), phenylalanine ammonia-lyase 1 (PAL1, XLOC_010156), and phenylalanine ammonia-lyase 2 (PAL2, XLOC_026730 and XLOC_010164) are involved in lignin biosynthesis, providing the strength necessary for vertical growth and resistance to biotic stresses and plant diseases. These lignin pathway genes often exist in multi-copy, e.g., four copies of 4CLs and PALs were found in the Arabidopsis genome. To adapt to climate change and to improve plant disease resistance, various members of these multi-gene families may be activated in tobacco leaves, resulting in increased lignin content and, concomitantly, resistance.
2.5. Functional Analysis of SF-Specific DEGs
Soil and nutrients are necessary for the growth and development of tobacco plants in the field. To examine transcriptomic variation in the leaves of plants grown under the same CFs and TFs, but under different SFs, we exchanged soils from the different regions used in this study. Most of the 2915 DEGs attributed to TFs were also differentially expressed in response to changes in CFs and SFs. Only 768 of these DEGs were TF-specific.
KEGG and GO enrichment analyses of all TF-specific DEGs identified 8 and 20 significantly enriched pathways and GO terms, respectively (Tables 5 and S9). Based on these results, DEGs associated with photosynthesis, e.g., antenna proteins (ko00196 and GO:0009523), protein processing in endoplasmic reticulum (ko04141 and GO:0055035), mineral absorption (ko04978 and GO:0000041), and response to stress (GO:0006950) were of special interest. Although stress resistance-related genes were overrepresented in common and CF-specific DEGs, they were also the largest group of SF-specific DEGs. Many SF-specific DEGs were involved in various biotic and abiotic stress responses. Among these genes were HEAT SHOCK FACTOR 4 (HSF4, XLOC_011884) and HEAT SHOCK PROTEIN 70 (HSP70, XLOC_017046 and XLOC_018164), which were both strongly induced by heat stress, and SALT OVERLY SENSITIVE 3 (SOS3, XLOC_008474), which encodes a calcium sensor that is essential for potassium nutrition and salt tolerance. Proteins encoded by the NB-LRR domain-containing disease resistance gene (RPPL1, XLOC_030092) might function with other TIR-NBS proteins involved in salicylic acid biosynthesis and systemic acquired resistance. Stress-responsive SF-specific DEGs did not show a significant preference for a specific stress condition, implying that tobacco plants might have evolved the ability to automatically regulate the transcriptome to withstand changes in soil environments.
Table 5.
Over-represented GO terms of SF-specific DEGs.
| GO-ID | p-value | Input number | Background number | Description |
|---|---|---|---|---|
| 0016036 | 3.89 × 105 | 8 | 52 | cellular response to phosphate starvation |
| 0000041 | 8.71 × 105 | 9 | 74 | transition metal ion transport |
| 0007154 | 1.10 × 104 | 15 | 195 | cell communication |
| 0009765 | 1.97 × 104 | 8 | 65 | photosynthesis, light harvesting |
| 0009867 | 2.73 × 104 | 6 | 37 | jasmonic acid mediated signaling pathway |
| 0071395 | 2.73 × 104 | 6 | 37 | cellular response to jasmonic acid stimulus |
| 0006464 | 3.34 × 104 | 75 | 2049 | protein modification process |
| 0035303 | 4.11 × 104 | 4 | 15 | regulation of dephosphorylation |
| 0006950 | 4.38 × 104 | 79 | 2205 | response to stress |
| 0050896 | 5.30 × 104 | 119 | 3644 | response to stimulus |
| 0006664 | 5.55 × 104 | 6 | 42 | glycolipid metabolic process |
| 0009875 | 5.57 × 104 | 7 | 58 | pollen-pistil interaction |
| 0006827 | 6.15 × 104 | 2 | 2 | high-affinity iron ion transport |
| 0046506 | 6.15 × 104 | 2 | 2 | sulfolipid biosynthetic process |
| 0046505 | 6.15 × 104 | 2 | 2 | sulfolipid metabolic process |
| 0006825 | 6.68 × 104 | 5 | 29 | copper ion transport |
| 0009247 | 7.85 × 104 | 5 | 30 | glycolipid biosynthetic process |
| 0009267 | 8.20 × 104 | 8 | 80 | cellular response to starvation |
| 0009743 | 8.66 × 104 | 12 | 165 | response to carbohydrate stimulus |
| 0043687 | 9.48 × 104 | 68 | 1883 | post-translational protein modification |
Other noteworthy GO terms were closely related to mineral nutrient absorption and transport in the root, such as cellular response to phosphate starvation (GO:0016036), high-affinity iron ion transport (GO:0006827), and copper ion transport (GO:0006825), reflecting the potential differences of mineral content between soils from the three cultivated regions. Nitrogen is the most important nutrient element for normal plant growth and development. In our study, common, CF- and TF-specific DEGs associated with nitrogen metabolism were simultaneously overrepresented. This finding suggests that nitrogen in soil plays a crucial role in tobacco plants’ nutrition from the transcriptome-scale perspective. As a component of the complex nucleic acid structure, the significance of phosphorus for plants is second only to nitrogen. SF-specific DEGs, but not common, CF-, and TF-specific DEGs, involved in the response to phosphate (Pi) deficiency were enriched. The differential expression of these genes might be caused by differences in phosphorus nutrient status in soils of the three cultivated regions, as the available Pi in soil at KY was about three and ten folds of that at WN and TZ, respectively (Table S10 [27]). There were several notable Pi-starvation responsive genes in the SF-specific group, such as SPX (SYG1/Pho81/XPR1, XLOC_002815 and XLOC_000642), PHOSPHOLIPASE D P2 (PLDP2, XLOC_021274), PLDP1 (XLOC_031030), SULFO QUINOVOSYLDIACYLGLYCEROL 1 (SQD1, XLOC_012522 and XLOC_027534), and SQD2 (XLOC_024053 and XLOC_002768). In Arabidopsis thaliana, repression of AtSPX3 led to a decrease in tolerance to Pi starvation and enhanced expression of a subset of Pi-responsive genes. Six SPX genes in rice were found to have diverse functions in plant tolerance to Pi starvation, five of which were responsive to Pi-deficiency in shoots and/or roots, and involved in the regulation of Pi-signaling network in a complex regulatory system [37]. We propose that differences in Pi concentration and distribution in soils in different cultivated regions might result in adaptive changes of Pi homeostasis during the growth and development of tobacco plants, increasing Pi acquisition and absorption by repressing the expression of SPX genes, or vice versa (Table S10). Other than nitrogen- and phosphorus-related genes, we identified seven genes involved in copper acquisition and transport, including copper transporter 1 (COPT1, XLOC_006434 and XLOC_006435), copper transport protein (CCH, XLOC_006386 and XLOC_006380), copper-transporting ATPase (RAN1, XLOC_000748 and XLOC_000749), and haloacid dehalogenase-like hydrolase family protein (PAA2, XLOC_001489). Among these copper homeostasis-related genes, COPT1, CCH, and RAN1 were mainly involved in copper acquisition and transport in leaves, while PAA2 was mainly responsible for metal ion binding and coupled to transmembrane movement of substances. Since these genes were induced by copper deficiency, ozone, and senescence and are required for copper homeostasis and normal plant growth and development, the available copper concentration in the soils of the cultivated regions WN, TZ and KY were 1.29, 1.53 and 1.87 mg/kg, did not show significant difference. Thus, these DEGs involved in copper homeostasis were highly sensitive to change of copper iron concentration could be assumed.
2.6. Functional Analysis of TF-Specific DEGs
To compare the effect of different tillage methods on transcriptomic variation of tobacco leaves, NT and CT approaches were implemented at the three cultivated regions. Overall, TFs had a much smaller effect than CFs and SFs, with a total of 1604 TF-related DEGs being identified, only 507 of which were TF-specific DEGs. KEGG analysis found that TF-specific DEGs involved in photosynthesis (ko00195) showed the highest enrichment level, suggesting that changes in tillage methods would have a significant impact on tobacco plants by altering the transcriptional regulation of photosynthesis genes, especially those related to photosystem I (Tables 6 and S11). More than half of the 13 photosynthesis DEGs encode subunit proteins of the photosystem I reaction center, such as PsaA (XLOC_030806), PasB (XLOC_015064 and XLOC_004737), PasD (XLOC_026534), PasG (XLOC_003245), and PasK (XLOC_014624). Furthermore, TF-specific DEGs encoding ATP synthase B (ATPase B, XLOC_000443), ATPase F (XLOC_001007), ferredoxin (XLOC_007987), and the cytochrome b(6) subunit of the cytochrome b6f complex (PETB, XLOC_007806) and involved in converting light energy into chemical energy, energy absorption, electron transfer, and ATP synthesis, may also contribute to the differences in photosynthetic efficiency of tobacco leaves of plants cultivated by CT or NT. A previous study showed that the photosynthetic rate of rice plants cultivated under NT was significantly higher than that of plants cultivated under CT [38]. This finding is in accordance with the results of our study, which show that TFs affect tobacco plants by modulating the expression of photosynthetic genes.
Table 6.
Over-represented GO terms of TF-specific DEGs.
| GO-ID | p-value | Input number | Background number | Description |
|---|---|---|---|---|
| 0015833 | 1.28 × 104 | 6 | 50 | peptide transport |
| 0042454 | 1.29 × 104 | 3 | 7 | ribonucleoside catabolic process |
2.7. Validation of RNA-Seq Data by qRT-PCR
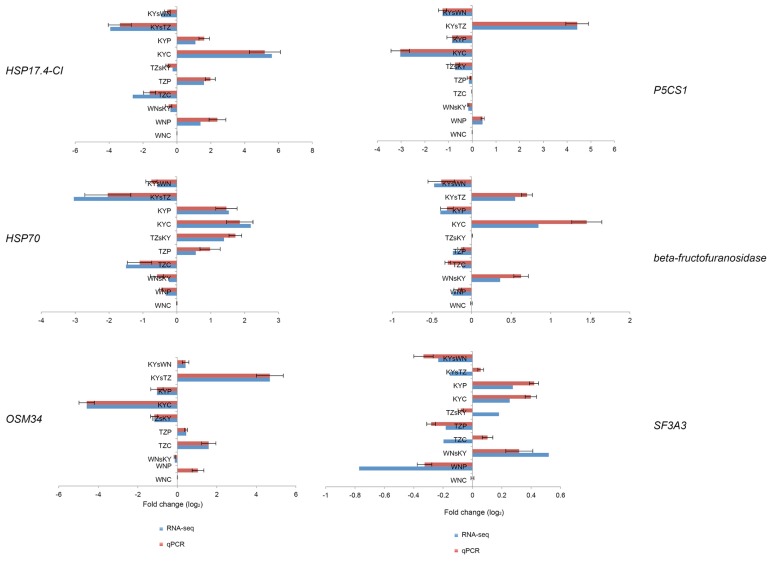
To confirm the transcriptome data generated by RNA-seq, qRT-PCR was carried out on five DEGs and one non-DEG randomly selected for their different expression levels. These genes encoded HSP17.4-CI, HSP70, osmotin-like protein (OSM34), delta 1-pyrroline-5-carboxylate synthetase A (P5CS1), beta-fructofuranosidase, and SF3A3 (splicing factor 3A subunit 3), respectively. All genes except for SF3A3 showed a concordant direction of fold change between RNA-seq and qRT-PCR, using the expression value in WNC to calibrate the data (Figure 4). Although three samples for SF3A3 had different directions of fold change, the remaining seven samples had similar expression patterns. These results confirm the reliability and accuracy of the RNA-seq data in this study.
Figure 4.
Correlation of differential expression between RNA-seq and qRT-PCR. Five DEGs and one non-DEG were chosen as differentially expressed by RNA-seq. The log2-fold change of DEGs obtained from RNA-seq data (blue) versus log2-fold changes of qRT-PCR derived on the basis of expression levels for treatment (pink) averaged from three samples. All the log2-fold changes were calculated using the expression value of WNC as a calibrator. Error bars indicate SD.
3. Experimental Section
3.1. Plant Materials
Tobacco plants (Nicotiana tabacum cv. Yunyan 85) used in this study were kindly provided by the Guizhou Tobacco Research Institute, Guiyang, China. Plants were grown in three different cultivated regions of Guizhou Province, including Longgang Town, Kaiyang County (KY), Niupeng Town, Weining County (WN), and Shexue Town, Tianzhu County (TZ) in 2009, as in our previous study [18]. Main meteorological data at different growth stages and basic physiochemical property of soils at three cultivated regions were listed in Tables S7 and S10. To compare the effect of climate factors (CFs; different cultivated regions), SFs (soils of different regions), and TFs (CT or NT) on the transcriptomes of tobacco leaves, soil from KY was exchanged with soil from WN and TZ, and the soil plow layer (soil depth 20–40 cm) at the three regions was broken (i.e., the plough layer (soil depth 0–20 cm) and plow layer were refilled in turn in their original positions after being dug up). In total, ten RNA-seq samples subjected to various treatments were harvested from the three cultivated regions (Table 7). For each treatment, two to three mature leaves (about 65 cm, taken from the middle parts of tobacco plants) were collected from three plants 70 DAT. To minimize the impact of sampling error, sample collection were carried out at 10:00 to 11:00 a.m. for three consecutive sunny days. All samples were immediately frozen in liquid nitrogen and stored at −80 °C.
Table 7.
RNA-seq samples subjected to various treatments.
| Cultivated regions | CF | SF | TF |
|---|---|---|---|
| KY | KYC | KYsTZ and KYsWN | KYP |
| TZ | TZC | TZsKY | TZP |
| WN | WNC | WNsKY | WNP |
KYC, TZC, and WNC represent samples originating from KY, TZ, and WN without soil exchange and tillage treatment. KYsTZ and KYsWN represent samples harvested from KY grown in soils from TZ and WN, respectively. TZsKY and WNsKY indicate samples collected from TZ and WN and grown on KY soil. KYP, TZP, and WNP represent samples harvested from the corresponding cultivated regions with tillage treatment.
3.2. RNA Preparation
Total RNA was extracted from approximately 100 mg tobacco leaves using a Plant RNA Mini Kit (Watson Biotechnologies, Inc., Shanghai, China), as previously described [39]. RNA was treated with RNase-free DNase I (TaKaRa, Dalian, China) for 30 min at 37 °C to remove all possible DNA contamination. RNA quality was checked by gel electrophoresis (using a 1.2% formaldehyde denaturing agarose gel). RNA concentrations were determined by measuring the absorbance at 260 nm with a NanoDrop ND-2000 Spectrophotometer (Thermo Scientific, Waltham, MA, USA) and an Agilent 2100 BioAnalyzer (Agilent Technologies, Palo Alto, CA, USA). After quantification, RNA samples from each treatment were equivalently pooled for RNA-seq and qRT-PCR analysis.
3.3. RNA-Seq Library Preparation and Sequencing
Library preparation and sequencing reactions were conducted in the Beijing Genome Institute (BGI, Shenzhen, China). Briefly, mRNAs were isolated from purified total RNA using magnetic oligo (dT) beads and fragmented, followed by first-strand cDNA synthesis using random hexamer-primed reverse transcription. The second-strand cDNA was generated using buffer, dNTPs, RNase H, and DNA polymerase I. After purification with a QIAquick Gel Extraction Kit (Qiagen, Frankfurt, Gremany), short fragments were resolved for end reparation and adaptor ligation. Following gel electrophoresis, cDNA fragments of approximately 200 bp were isolated and used for cluster generation. Finally, the samples were sequenced using single end (SE) read sequencing with 50 cycles on an Illumina HiSeq 2000, following the manufacturer’s instructions. Base calling was performed with Illumina software Pipeline 1.4 (Illumina, San Diego, CA, USA). RNA-seq data were deposited in the NCBI Sequence Read Archive (SRA) under accession numbers SRR1040764 to SRR1040773.
3.4. RNA-Seq Analysis
RNA-seq reads were mapped to 84,602 of tobacco SGN Unigene sequences (ftp://ftp.solgenomics.net/unigene_builds/single_species_assemblies/Nicotiana_tabacum/Nicotiana_tabacum_unigene.v2.seq) retrieved from the Solanaceae Genomics Network (SGN) [21] using TopHat, as described by Trapnell et al. [15]. Cufflinks assembled transcripts and quantified transcript abundance in terms of fragments per kilobase of exon per million mapped fragments (FPKM) [22]. Both TopHat and Cufflinks analyses were carried out in default modes. The Cuffdiff program within Cufflinks was used to test for statistically significant differences in transcript expression between 37 comparison pairs (Table 2). Differentially expressed genes (DEGs) were identified using the following two criteria: (i) absolute fold-change >2 and (ii) q-value (false discovery rate (FDR)) < 0.05. To further the analysis of our sequencing data, cluster analysis of expression profiles of all DEGs was performed with Cluster 3.0 software with uncentered correlation and complete linkage hierarchical clustering option [40] and the heatmap was visualized using Java TreeView [41].
To better understand the meaning of differential expression, the function of tobacco SGN UniGenes was annotated using BLASTX against the Arabidopsis thaliana proteome (version TAIR10 database) with an e-value cut-off of 10−5 [42,43]. GO annotations of reference SGN UniGenes were performed to retrieve molecular function, biological process, and cellular component terms using Blast2GO (http://www.blast2go.org/) [44]. Enrichment of GO categories among DEGs was assessed by BinGO v2.4.4, a Cytoscape plugin [45,46]. KEGG-based annotation and pathway enrichment analysis was performed using KOBAS 2.0 program [47], which assigned the enzyme commission (EC) numbers and significantly enriched metabolic pathways in DEGs compared with the whole reference sequences [48]. All GO and KEGG statistical tests were corrected for multiple comparisons (Benjamini Hochberg method) [49]. To cluster the samples based on the similarity of gene expression profiles of tobacco leaves, unsupervised principal component analysis (PCA) and multi-dimensional scaling (MDS) were applied. To visualize patterns in different treatments, SPSS 20 (IBM Corp., Armonk, NY, USA) and the R package CummeRbund v2.0.0 were used [50]. The expression of all detected genes in our RNA-seq data was subjected to PCA and MDS analysis and plotted.
3.5. Quantitative Real-Time PCR (qRT-PCR) Validation
Five DEGs and one non-DEG identified by RNA-seq were assayed by qRT-PCR. Gene-specific primers were designed based on the nucleotide sequence of the chosen unigenes using Primer 3.0 software [51]. Primers used in this study are summarized in Table S12. The same total RNAs were used as those in the RNA-seq experiments. cDNA synthesis, qRT-PCR cycling conditions, amplification efficiency, and specificity assessment were as described in Lei et al. [18]. Briefly, 1 μg of DNaseI-treated total RNA was reverse-transcribed using the PrimeScript RT Master Mix (TaKaRa). qRT-PCR was performed with SYBR Premix Ex Taq II (TaKaRa) using the CFX96 Touch Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). Three independent biological replicates were analyzed per sample. The expression level of each sample was calculated using the 2−ΔΔCt method, with the housekeeping gene NtEF-1α serving as an internal control, as its expression is stable in tobacco plants [52,53].
3.6. Measurement of Agronomic Traits and SOD, CAT and POD Activities of Tobacco Leaves
To assess the impact of different ecological factors on tobacco plants, four agronomic traits, including plant height, stem perimeter, number of leaves and maximum leaf area of tobacco plants were determined at 45 and 70 DAT. A portable leaf area meter (LI-COR, model LI-3000) was used for measuring the area of the maximum leaf of tobacco plant in each treatment. The activities of SOD, POD and CAT were determined spectrophotometrically. For extraction of SOD, POD and CAT, about 1 g of tobacco leaves samples were ground under liquid nitrogen and homogenized in 10 mL of the extraction buffer containing 50 mM phosphate buffer (pH 7.8) and 1% polyvinylpyrrolidone (PVP). The homogenate was centrifuged at 10,000× g at 4 °C for 10 min. The supernatants obtained after centrifugation were used for the enzyme activity analyses. SOD and CAT activities were measured using SOD and CAT Detection Kits (A001 and A007, Nanjing Jiancheng Bioengineering Institute, Nanjing, China) according to the manufacturer’s instructions. Total POD activities were determined by spectrophotometrically monitoring guaiacol oxidation at 470 nm following a previous described method [54]. Values were expressed in units of SOD, POD or CAT activities per gram wet-weight of tissue.
4. Conclusions
In conclusion, we performed RNA-seq analysis on tobacco leaves derived from 10 treatment combinations of three ecological factors, CFs, SFs and TFs. RNA-seq analysis generated 58,466,453 reads, which assembled into 30,688 unique transcripts. We detected 4980, 2916, and 1605 DEGs that were affected by CFs, SFs, and TFs, which included 2703, 768, and 507 specific and 703 common DEGs, respectively. Plots of PCA and MDC, and cluster analysis all revealed that the greatest transcriptomic variation was caused by CFs, followed by SFs, with the ratio of factor-specific impacts of CFs:SFs:TFs being about 5:1.5:1. GO and KEGG enrichment analyses showed that genes involved in abiotic stress responses and secondary metabolic pathways were overrepresented in the common and CF-specific DEGs, implying that these genes mediate adaptation to environmental variation. In addition, we noted enrichment in CF-specific DEGs related to the circadian rhythm, SF-specific DEGs involved in mineral nutrient absorption and transport, and SF- and TF-specific DEGs associated with photosynthesis. Based on these results, we propose a model that explains how plants adapt to various ecological factors at the transcriptomic level. Additionally, these data would be useful in selecting candidate genes for future investigations of stress resistance, the circadian rhythm, nutrient absorption, and photosynthesis in tobacco.
Acknowledgments
This work was supported in part by grants from the Key Special Program of China National Tobacco Corporation (TS-02-20110014), the Program of Guizhou Provincial Tobacco Company (2010-02), the Foundation of Science and Technology of Guizhou Province of China (J[2010]2088 and J[2013]2195), the Fundamental Research Funds for the Central Universities (XDJK2012A009), the Southwest University Research Foundation (SWU110015), Natural Science Foundation of Chongqing (cstc2011jjA80026), National Natural Science Foundation of China (31101192 and 31101175).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Author Contributions
LB, LK, and DF carried out the experiments, analyzed the data, and drafted the manuscript. ZK contributed to data analysis. RZ, CY, ZH, and ZL performed RNA-seq, analyzed sequencing data. QC, GW and WJ performed experiments and reviewed the manuscript. PW designed the study and participated in writing the manuscript.
References
- 1.Baker N.T., Capel P.D. Environmental Factors that Influence the Location of Crop Agriculture in the Conterminous United States. U.S. Geological Survey; Reston, VA, USA: 2011. p. 72. [Google Scholar]
- 2.Criddle R.S., Hopkin M.S., McArthur E.D., Hansen L.D. Plant distribution and the temperature coefficient of metabolism. Plant Cell Environ. 1994;17:233–243. [Google Scholar]
- 3.Hinsinger P., Brauman A., Devau N., Gérard F., Jourdan C., Laclau J.P., le Cadre E., Jaillard B., Plassard C. Acquisition of phosphorus and other poorly mobile nutrients by roots. Where do plant nutrition models fail? Plant Soil. 2011;348:29–61. [Google Scholar]
- 4.Chivenge P., Vanlauwe B., Six J. Does the combined application of organic and mineral nutrient sources influence maize productivity? A meta-analysis. Plant Soil. 2011;342:1–30. [Google Scholar]
- 5.Waters B.M., Sankaran R.P. Moving micronutrients from the soil to the seeds: Genes and physiological processes from a biofortification perspective. Plant Sci. 2011;180:562–574. doi: 10.1016/j.plantsci.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 6.Wang Z.H., Li S.X., Malhi S. Effects of fertilization and other agronomic measures on nutritional quality of crops. J. Sci. Food Agric. 2008;88:7–23. [Google Scholar]
- 7.Nyborg M., Solberg E.D., Izaurralde R.C., Malhi S.S., Molina-Ayala M. Influence of long-term tillage, straw and N fertilizer on barley yield, plant-N uptake and soil-N balance. Soil Till. Res. 1995;36:165–174. [Google Scholar]
- 8.Malhi S.S., Grant C.A., Johnston A.M., Gill K.S. Nitrogen fertilization management for no-till cereal production in the Canadian great plains: A review. Soil Till. Res. 2001;60:101–122. [Google Scholar]
- 9.Vakali C., Zaller J.G., Köpke U. Reduced tillage effects on soil properties and growth of cereals and associated weeds under organic farming. Soil Till Res. 2011;111:133–141. [Google Scholar]
- 10.Rizhsky L., Hongjian L., Mittler R. The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol. 2002;130:1143–1151. doi: 10.1104/pp.006858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rizhsky L., Liang H., Shuman J., Shulaev V., Davletova S., Mittler R. When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol. 2004;134:1683–1696. doi: 10.1104/pp.103.033431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dal Santo S., Tornielli G.B., Zenoni S., Fasoli M., Farina L., Anesi A., Guzzo1 F., Delledonne M., Pezzotti M. The plasticity of the grapevine berry transcriptome. Genome Biol. 2013;14:R54. doi: 10.1186/gb-2013-14-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsirigos A., Haiminen N., Bilal E., Utro F. GenomicTools: A computational platform for developing high-throughput analytics in genomics. Bioinformatics. 2012;28:282–283. doi: 10.1093/bioinformatics/btr646. [DOI] [PubMed] [Google Scholar]
- 14.Kakumanu A., Ambavaram M.M.R., Klumas C., Krishnan A., Batlang U. Effects of drought on gene expression in maize reproductive and leaf meristem tissue revealed by RNA-Seq. Plant Physiol. 2012;160:846–867. doi: 10.1104/pp.112.200444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Rourke J.A., Yang S.S., Miller S.S., Bucciarelli B., Liu J., Rydeen A., Bozsoki Z., Uhde-Stone C., Tu Z.J., Allan D., et al. An RNA-Seq transcriptome analysis of Pi deficient white lupin reveals novel insights into phosphorus acclimation in plants. Plant Physiol. 2013;161:705–724. doi: 10.1104/pp.112.209254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dugas D., Monaco M., Olson A., Klein R., Kumari S., Ware D., Klein P. Functional annotation of the transcriptome of sorghum bicolor in response to osmotic stress and abscisic acid. BMC Genomics. 2011;12:514. doi: 10.1186/1471-2164-12-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaeger P.A., Doherty C., Ideker T. Modeling transcriptome dynamics in a complex world. Cell. 2012;151:1161–1162. doi: 10.1016/j.cell.2012.11.031. [DOI] [PubMed] [Google Scholar]
- 18.Lei B., Zhao X.H., Zhang K., Zhang J., Ren W., Ren Z., Chen Y., Zhao H.N., Pan W.J., Chen W., et al. Comparative transcriptome analysis of tobacco (Nicotiana tabacum) leaves to identify aroma compound-related genes expressed in different cultivated regions. Mol. Biol. Rep. 2013;40:345–357. doi: 10.1007/s11033-012-2067-0. [DOI] [PubMed] [Google Scholar]
- 19.Ali Reza F. The effect of nitrogen and potassium fertilizer on yield, quality and some quantitative characteristics of flue-cured tobacco cv. Coker347. Afr. J. Agric. Res. 2012;7:1827–1833. [Google Scholar]
- 20.Orlando F. Growth and development responses of tobacco (Nicotiana tabacum L.) to changes in physical and hydrological soil properties due to minimum tillage. Am. J. Plant Sci. 2011;2:334–344. [Google Scholar]
- 21.Bombarely A., Menda N., Tecle I.Y., Buels R.M., Strickler S., Fischer-York T., Pujar A., Leto J., Gosselin J., Mueller L.A. The sol genomics network (solgenomics. net): Growing tomatoes using Perl. Nucleic Acids Res. 2011;39:D1149–D1155. doi: 10.1093/nar/gkq866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y., Ghaffari N., Johnson C.D., Braga-Neto U.M., Wang H., Chen R., Zhou H. Evaluation of the coverage and depth of transcriptome by RNA-Seq in chickens. BMC Bioinf. 2011;12:S5. doi: 10.1186/1471-2105-12-S10-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malone J.H., Oliver B. Microarrays, deep sequencing and the true measure of the transcriptome. BMC Biol. 2011;9:34. doi: 10.1186/1741-7007-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edwards K.D., Bombarely A., Story G.W., Allen F., Mueller L.A., Coates S.A., Jones L. TobEA: An atlas of tobacco gene expression from seed to senescence. BMC Genomics. 2010;11:142. doi: 10.1186/1471-2164-11-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen W., Xiong J., Chen Y., Pan W.J., Li Z.Y. Effects of climate and soil on the carotenoid and cuticular extract content of cured tobacco leaves. Acta Ecol. Sin. 2013;33:3865–3877. [Google Scholar]
- 28.Yoshida T., Fujita Y., Sayama H., Kidokoro S., Maruyama K., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010;61:672–685. doi: 10.1111/j.1365-313X.2009.04092.x. [DOI] [PubMed] [Google Scholar]
- 29.Xie D. COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science. 1998;280:1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- 30.Ann T., Kono N., Kosemura S., Yamahura S., Hasegawa K. Isolation and characterization of an auxin-inducible SAUR gene from radish seedlings. Mitochondrial DNA. 1998;9:329–333. [PubMed] [Google Scholar]
- 31.Ha S., Vankova R., Yamaguchi-Shinozaki K., Shinozaki K., Tran L.S. Cytokinins: Metabolism and function in plant adaptation to environmental stresses. Trends Plant Sci. 2012;17:172–179. doi: 10.1016/j.tplants.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 32.Nagano A.J., Sato Y., Mihara M., Antonio B.A., Motoyama R., Itoh H., Nagamura Y., Izawa T. Deciphering and prediction of transcriptome dynamics under fluctuating field conditions. Cell. 2012;151:1358–1369. doi: 10.1016/j.cell.2012.10.048. [DOI] [PubMed] [Google Scholar]
- 33.Sawa M., Kay S.A. GIGANTEA directly activates Flowering Locus T inArabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2011;108:11698–11703. doi: 10.1073/pnas.1106771108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizoguchi T., Wheatley K., Hanzawa Y., Wright L., Mizoguchi M., Song H.-R., Carré I.A., Coupland G. LHY and CCA1 are partially redundant genes required to maintain circadian rhythms inArabidopsis. Dev. Cell. 2002;2:629–641. doi: 10.1016/s1534-5807(02)00170-3. [DOI] [PubMed] [Google Scholar]
- 35.Dong M.A., Farré E.M., Thomashow M.F. Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway inArabidopsis. Proc. Natl. Acad. Sci. USA. 2011;108:7241–7246. doi: 10.1073/pnas.1103741108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakamichi N., Kiba T., Henriques R., Mizuno T., Chua N.H., Sakakibara H. Pseudo-response regulators 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell. 2010;22:594–605. doi: 10.1105/tpc.109.072892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z., Hu H., Huang H., Duan K., Wu Z., Wu P. Regulation of OsSPX1 and OsSPX3 on expression of OsSPX domain genes and Pi-starvation signaling in rice. J. Integr. Plant Biol. 2009;51:663–674. doi: 10.1111/j.1744-7909.2009.00834.x. [DOI] [PubMed] [Google Scholar]
- 38.Chen S., Xia G., Zhao W., Wu F., Zhang G. Characterization of leaf photosynthetic properties for no-tillage rice. Rice Sci. 2007;14:283–288. [Google Scholar]
- 39.Lu K., Chai Y.R., Zhang K., Wang R., Chen L., Lei B., Lu J., Xu X.F., Li J.N. Cloning and characterization of phosphorus starvation inducible Brassica napus PURPLE ACID PHOSPHATASE 12 gene family, and imprinting of a recently evolved MITE-minisatellite twin structure. Theor. Appl. Genet. 2008;117:963–975. doi: 10.1007/s00122-008-0836-x. [DOI] [PubMed] [Google Scholar]
- 40.De Hoon M.J., Imoto S., Nolan J., Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 41.Saldanha A.J. Java Treeview—Extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 42.Lamesch P., Berardini T.Z., Li D., Swarbreck D., Wilks C., Sasidharan R., Muller R., Dreher K., Alexander D.L., Garcia-Hernandez M. The Arabidopsis information resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conesa A., Götz S., García-Gómez J.M., Terol J., Talón M., Robles M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 45.Smoot M.E., Ono K., Ruscheinski J., Wang P.L., Ideker T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maere S., Heymans K., Kuiper M. Bingo: A cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 47.KOBAS 2.0. [(accessed on 4 March 2014)]. Available online: http://kobas.cbi.pku.edu.cn/home.do.
- 48.Xie C., Mao X., Huang J., Ding Y., Wu J., Dong S., Kong L., Gao G., Li C.Y., Wei L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995:289–300. [Google Scholar]
- 50.Goff L., Trapnell C., Kelley D.CummeRbund: Analysis, Exploration, Manipulation, and Visualization of Cufflinks High-Throughput Sequencing DataAvailable online: http://www.bioconductor.org/packages/release/bioc/html/cummeRbund.html 8 May 2013
- 51.Rozen S., Skaletsky H. Primer3 on the www for general users and for biologist programmers. In: Krawetz S., Misener S., editors. Bioinformatics Bethods and Protocols: Methods in Molecular Biology. Vol. 132. Humana Press; Totowa, NJ, USA: 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- 52.Schmidt G.W., Delaney S.K. Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Mol. Genet. Genomics. 2010;283:233–241. doi: 10.1007/s00438-010-0511-1. [DOI] [PubMed] [Google Scholar]
- 53.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 54.Ranieri A., Petacco F., Castagna A., Soldatini G.F. Redox state and peroxidase system in sunflower plants exposed to ozone. Plant Sci. 2000;159:159–167. doi: 10.1016/s0168-9452(00)00352-6. [DOI] [PubMed] [Google Scholar]