Abstract
Background
Chronic persistent oxidative stress has been proposed as a mechanism for late radiation injury to normal tissue. Using biochemical, histological, and pharmacological techniques, we have not been able to confirm this hypothesis for late renal radiation injury. Gene expression may be more revealing, especially since the initial effects of radiation are to damage DNA.
Methods
Gene array studies were done using kidney tissue from irradiated rats, with particular attention to genes pertinent to oxidative stress. The time points were from 1 to 49 days after irradiation. Cellular RNA and mitochondrial DNA were isolated, for gene expression analysis and common deletion testing, respectively.
Results
For the gene expression studies, and from over 30,000 transcripts, only nine related to oxidative stress had 1.4 fold or greater changes in expression. Mitochondrial DNA showed no changes in the common deletion.
Conclusion
These studies do not support the hypothesis of chronic oxidative stress as a mechanism for radiation nephropathy.
Keywords: Radiation injury, Oxidative stress, Gene array testing, Mitochondrial DNA
Introduction
Normal tissue radiation injury occurs in clinical radiation therapy, and after accidental radiation exposures. It is widely accepted that ionizing radiation causes tissue damage by prompt radiolysis of water and oxidative injury to DNA. It has been proposed that persistent oxidative stress is a significant feature and mechanism of chronic normal tissue radiation injuries [1]. Using an experimental radiation nephropathy model, we have not been able to confirm this hypothesis. Biochemical and histological markers of oxidative stress were absent after irradiation and anti-oxidants were ineffective in the mitigation of radiation nephropathy [2–4]. But it remained possible that the assays were not sensitive enough, and that the anti-oxidants chosen were not appropriate for the model. We therefore sought genomic evidence for chronic oxidative stress in experimental radiation nephropathy.
Materials and Methods
Animals
WAG/RijCmcr male rats were bred and housed in a moderate security barrier in the Biomedical Resource Center (BRC) of the Medical College of Wisconsin. The BRC is fully accredited by the American Association for Accreditation of Laboratory Animal Care. Rats were provided with food and water ad libitum and kept in controlled light and temperature conditions. All experimental procedures complied with the animal protocols that are approved by the Institutional Animal Care and Use Committee of the Medical College of Wisconsin.
Experimental design and animal treatment
A total of 48 newly-weaned male rats were used. Litters of four or more rats were distributed evenly to the two arms of the protocol. At age 7 to 8 weeks, 24 rats underwent 10 Gy total body irradiation (TBI), and 24 had no irradiation. Rats were sacrificed at days 1, 7, 21, and 49 after TBI. Irradiated rats had a syngeneic bone marrow transplant immediately after TBI. Irradiation was done with a 320 kVp orthovoltage source with a half value layer of 1.4 mm Cu [5]. Non-irradiated rats were sham-treated. In this model, there is proteinuria by 6 weeks after irradiation, then progressive azotemia and hypertension, with uremic morbidity by 26 weeks after irradiation. The time points of 1, 7, 21, and 49 days were chosen to identify mechanistically relevant gene expression changes that occurred before evidence of tissue injury.
RNA extraction
Both kidneys were snap-frozen in liquid nitrogen and stored at −80°C until RNA preparation. Extraction of RNA was from kidney cortex from control and experimental animals, and always from the upper part of the right kidney, then the RNA was pooled for each group and time point. Total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s method. The quantity and optical density (OD) ratio 260/280 of total RNA and cRNA was assessed by the NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). RNA samples with a ratio of 1.9 or more were considered to be of acceptable purity.
Hybridization to Affymetrix gene expression array and microarray analysis
The Affymetrix GeneChip rat genome RG230 plus 2.0 array, which interrogates ~31,000 transcripts, was selected for these studies for its overall comprehensive coverage. Purified RNA (~100 ng) was amplified using an Affymetrix Express Kit; cRNA was synthesized, labeled, fragmented, and hybridized to the array in accordance to the Affymetrix GeneChip expression analysis technical manual. After hybridization, arrays were washed, stained with phycoerythrin-conjugated streptavidin (Life Technologies, Grand Island, NY, USA), and scanned. Image data were quantified with Affymetrix Expression Console Software and normalized with Robust Multichip Analysis (RMA; www.bioconductor.org/) to determine signal log ratios. The microarray analyses were repeated in triplicate. The statistical significance of differential gene expression was derived through a Rank Product algorithm [6].
DNA extraction
Total DNA (nuclear and mitochondrial) extraction was performed from kidneys of the same rats that were used for the gene expression studies. Total DNA was isolated using the PureLink Genomic DNA kit (Invitrogen, Carlsbad, CA, USA). Following extraction, DNA samples were quantified and characterized by spectrophotometry. Quantity and the optical density (OD) 260/280 of total DNA were assessed by BioPhotometer Plus (Eppendorf AG, Hamburg, Germany). The absorbance ratio of the DNA at 260 and 280 nm was used to determine DNA purity.
mtDNA deletion analysis
Assessment of radiation-induced mtDNA damage focused on measuring the common deletion (CD). The CD is used as a marker of oxidative damage to mtDNA, and it is reported to accumulate in irradiated cells [7]. CDs were detected and quantified by real time PCR (qPCR). All primers were purchased from Integrated DNA Technologies, Inc., Coralville, IA.
The deletion primers amplified an 81 bp product only in the presence of a 4834 bp mitochondrial common deletion (CD). The D-loop primers amplified an 83 bp product from the region outside of the CD, which is an internal standard for total mtDNA. Beta-actin (BA) primers amplified a 70 bp product from the nuclear DNA, and were used to determine the level of nuclear DNA relative to the mtDNA.
Real time quantitative PCR (qPCR) – mtDNA deletion studies
PCR amplification was performed on Mx3005P™ QPCR System (Startagene, Santa Clara, CA, USA). Each PCR reaction sample consisted of 12.5 microliters of iQ SYBR Green Supermix (Bio-Rad Laboratories, Inc., Hercules, CA, USA), 200 nM for each forward and reverse primer and 50ng of total DNA sample. PCR cycling conditions included an initial phase of 3 min at 95°C to activate hot-start iTag DNA polymerase followed by 40 cycles of 15 sec at 95°C and 1 min at 60°C. Each sample of individual rat DNA was assayed in duplicate and fluorescence spectra were continuously monitored.
Results
In the gene array comparison of irradiated to unirradiated rats, only 376 genes had two-fold or greater changes in expression. Of 84 genes linked to oxidative stress (according to the SA Bio, Oxidative Stress and Antioxidant Defense RT2 Profiler™ PCR Array) one had a two-fold or greater change in expression; this was glutathione peroxidase 2 (Gpx2) at 49 days after irradiation. 2526 genes had 1.4 fold or greater changes in expression, of which 9 were genes linked to oxidative stress (Table 1).
Table 1.
Genes with significant expression changes in kidneys after total body irradiation.
| Gene name | Day of study | function | Fold change | p value |
|---|---|---|---|---|
| Scd1- Stearoyl-Coenzyme A desaturase 1 | 1, 7 | fatty acid metabolism | −1.9, −1.7 | 3*10−7, 1.1*10−4 |
| Apoe- Apolipoprotein E | 7, 49 | lipoprotein metabolism | 1.6, 1.5 | 4.5*10−6, 1.6*10−4 |
| Dhcr24- 24-dehydrocholesterol reductase | 7 | cholesterol biosynthesis | −1.7 | 3.8*10−5 |
| Nqo1- NAD(P)H dehydrogenase, quinone 1 | 7 | reduces quinones to hydroquinones | 1.5 | 3.1*10−5 |
| Gpx2- Glutathione peroxidase 2 | 49 | hydrogen peroxide-reducing activity | 2.1 | <10−8 |
| Ptgs2- Prostaglandin-endoperoxide synthase 2 (Cox2) | 49 | biosynthesis of the prostanoids | 1.6 | 3.8*10−5 |
| Sod3- Superoxide dismutase 3 | 49 | dismutation of superoxide radicals | −1.9 | 1.3*10−6 |
| Cygb- Cytoglobin | 49 | reactive oxygen species scavenging | 1.6 | 4*10−5 |
| Vim-Vimentin | 49 | transport of low-density lipoproteins | 1.4 | 5*10−4 |
The incidence of the common deletion in mtDNA did not differ in irradiated kidneys compared to unirradiated kidneys (Figure 1).
Figure 1.
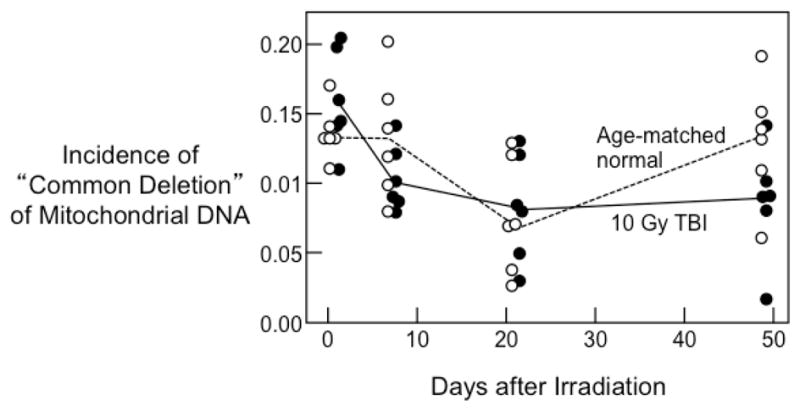
Incidence of the “Common Deletion” in renal mitochondrial DNA as a function of time after irradiation; incidence is a ratio to the number of copies of total mtDNA. Data shown for individual irradiated animals (●) and their age-matched controls (○). The ratio of incidence in TBI versus age-matched normal is 0.88 (95% C.L 0.77–1.02). There is no significant trend with time (both p>0.20).
Discussion
The present studies do not show any evidence for chronic oxidative stress in experimental radiation nephropathy. Of oxidative stress related genes interrogated by the Affymetrix chip, only 9 showed 1.4 fold or greater change after 10 Gy TBI, in comparison to rats that were not irradiated. Seven showed an increase of expression, while four showed a decrease. We have no confirmation of corresponding protein expression changes, but the lack of a consistent upward or downward trend is clear. The lack of change in the CD abundance is also not in favor of persistent, chronic oxidative stress. The complete lack of changes in gene expression at 21 days is striking. This is at a time after irradiation when there is still no evidence of renal injury (i.e., no azotemia, no hypertension, and no proteinuria). Gene expression changes at this time would thus be mechanistically informative for the pathogenesis of radiation nephropathy, because changes in expression would not be affected by renal injury itself. Only three of the genes relevant to oxidative stress had more than 1.4 fold expression changes before the 49 day time point, i.e. before the expression of radiation injury. By 49 days in this model, there is proteinuria and there is evidence of histological injury. The elevation of Gpx2 at that time could well be secondary to injury, and not be causing the injury. The present studies do not show evidence that chronic oxidative stress plays an important role in the pathogenesis of experimental radiation nephropathy.
Gene expression studies of murine radiation nephropathy have also not shown evidence for chronic oxidative stress [2,8]. Kruse et al. [2] showed a broad array of gene expression changes after 16 Gy single fraction bilateral kidney irradiation, including genes relevant to inflammation, proliferation, apoptosis, and other cellular functions. None of the genes cited in that report were genes related to oxidative stress. Zhao et al. [8] tested gene expression at 8 and 24 hours after 10 Gy TBI of mice, and also found no evidence of gene expression changes that indicated oxidative stress. At very low dose rates of 0.65 to 13 microGy/minute, Taki et al. report mitochondrial oxidative phosphorylation gene expression changes [9], but these dose rates are six orders of magnitude less than those of usual exposures and of uncertain significance. Gene expression changes that mechanistically explain radiation nephropathy remain obscure.
We have tested for biochemical and histopathological evidence of chronic oxidative stress in kidneys and urine at time points up to 49 days after 10 Gy TBI, and found no definitive evidence for it [3]. We also tested three anti-oxidant drugs as mitigators of radiation nephropathy in this model and found them ineffective [4]. Together with the present gene expression data, we conclude that there is no evidence for a role for chronic oxidative stress in radiation nephropathy. Treatments and mitigators for this and other normal tissue radiation injuries cannot be solely directed at the attenuation of oxidative stress.
Acknowledgments
These studies were supported by a grant from the National Institutes of Health (NIH/NIAID 1U19AI067734, P.I. John E Moulder).
References
- 1.Robbins ME, Zhao W. Chronic oxidative stress and radiation-induced late normal tissue injury: A review. Int J Radiat Biol. 2004;80:251–259. doi: 10.1080/09553000410001692726. [DOI] [PubMed] [Google Scholar]
- 2.Kruse JJ, te Poele JA, Velds A, Kerkhoven RM, Boersma LJ, et al. Identification of differentially expressed genes in mouse kidney after irradiation using microarray analysis. Radiat Res. 2004;161:28–38. doi: 10.1667/rr3097. [DOI] [PubMed] [Google Scholar]
- 3.Lenarczyk M, Cohen EP, Fish BL, Irving AA, Sharma M, et al. Chronic oxidative stress as a mechanism for radiation nephropathy. Radiat Res. 2009;171:164–172. doi: 10.1667/RR1454.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen EP, Fish BL, Irving AA, Rajapurkar MM, Shah SV, et al. Radiation nephropathy is not mitigated by antagonists of oxidative stress. Radiat Res. 2009;172:260–264. doi: 10.1667/RR1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen EP, Fish BL, Sharma M, Li XA, Moulder JE. Role of the angiotensin II type-2 receptor in radiation nephropathy. Transl Res. 2007;150:106–115. doi: 10.1016/j.trsl.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hong F, Breitling R, McEntee CW, Wittner BS, Nemhauser JL, et al. RankProd: A bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics. 2006;22:2825–2827. doi: 10.1093/bioinformatics/btl476. [DOI] [PubMed] [Google Scholar]
- 7.Prithivirajsingh S, Story MD, Bergh SA, Geara FB, Ang KK, et al. Accumulation of the common mitochondrial DNA deletion induced by ionizing radiation. FEBS Lett. 2004;571:227–232. doi: 10.1016/j.febslet.2004.06.078. [DOI] [PubMed] [Google Scholar]
- 8.Zhao W, Chuang EY, Mishra M, Awwad R, Bisht K, et al. Distinct effects of ionizing radiation on in vivo murine kidney and brain normal tissue gene expression. Clin Cancer Res. 2006;12:3823–3830. doi: 10.1158/1078-0432.CCR-05-2418. [DOI] [PubMed] [Google Scholar]
- 9.Taki K, Wang B, Nakajima T, Wu J, Ono T, et al. Microarray analysis of differentially expressed genes in the kidneys and testes of mice after long-term irradiation with low-dose-rate gamma-rays. J Radiat Res. 2009;50:241–252. doi: 10.1269/jrr.09011. [DOI] [PubMed] [Google Scholar]


