Abstract
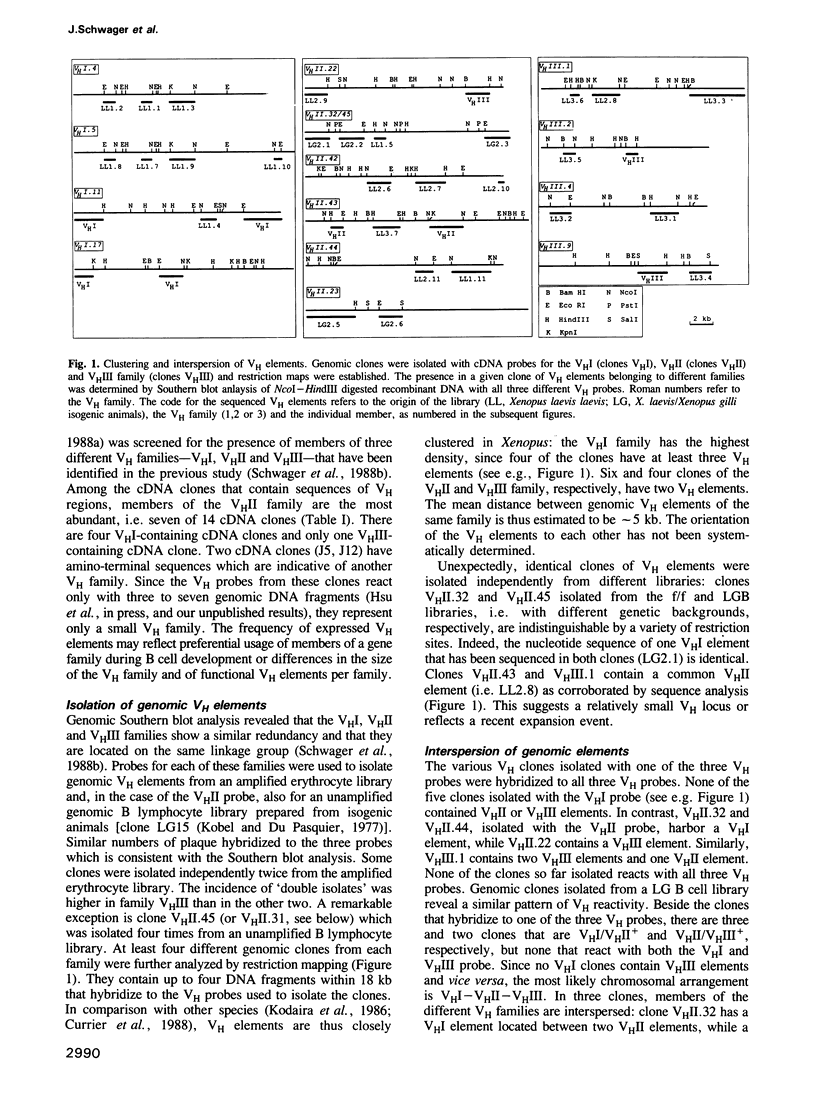
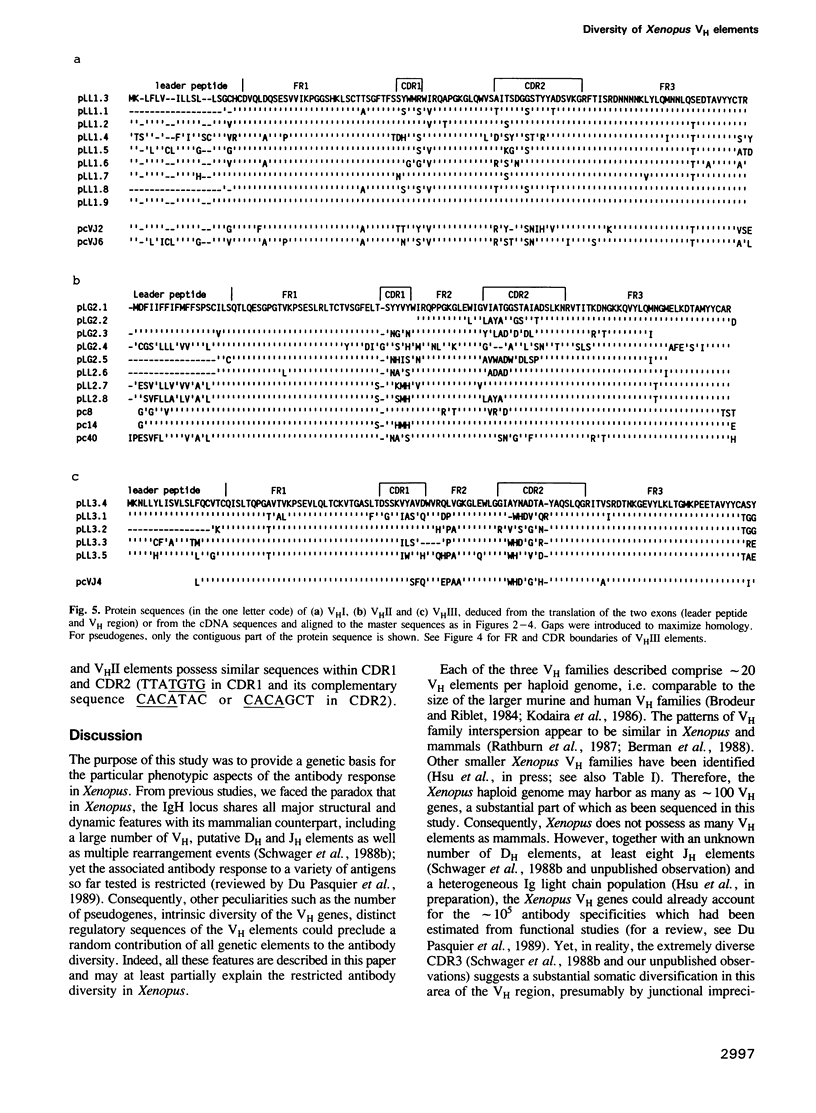
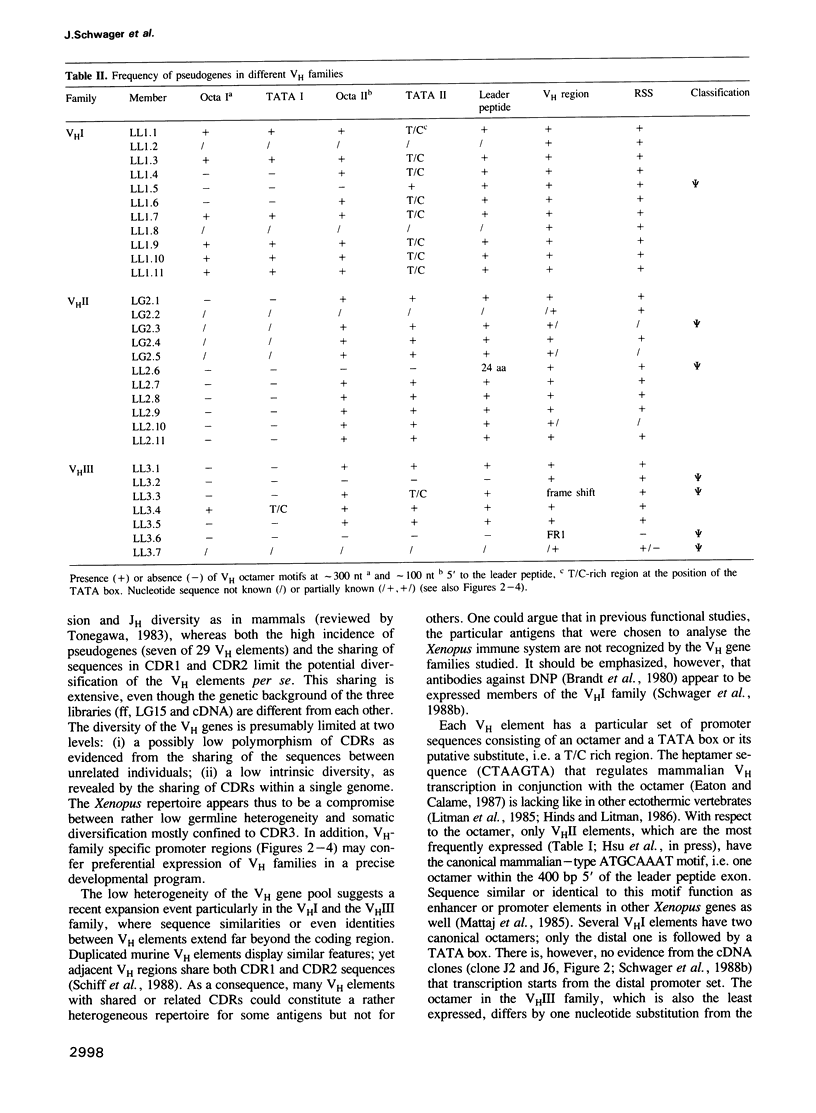
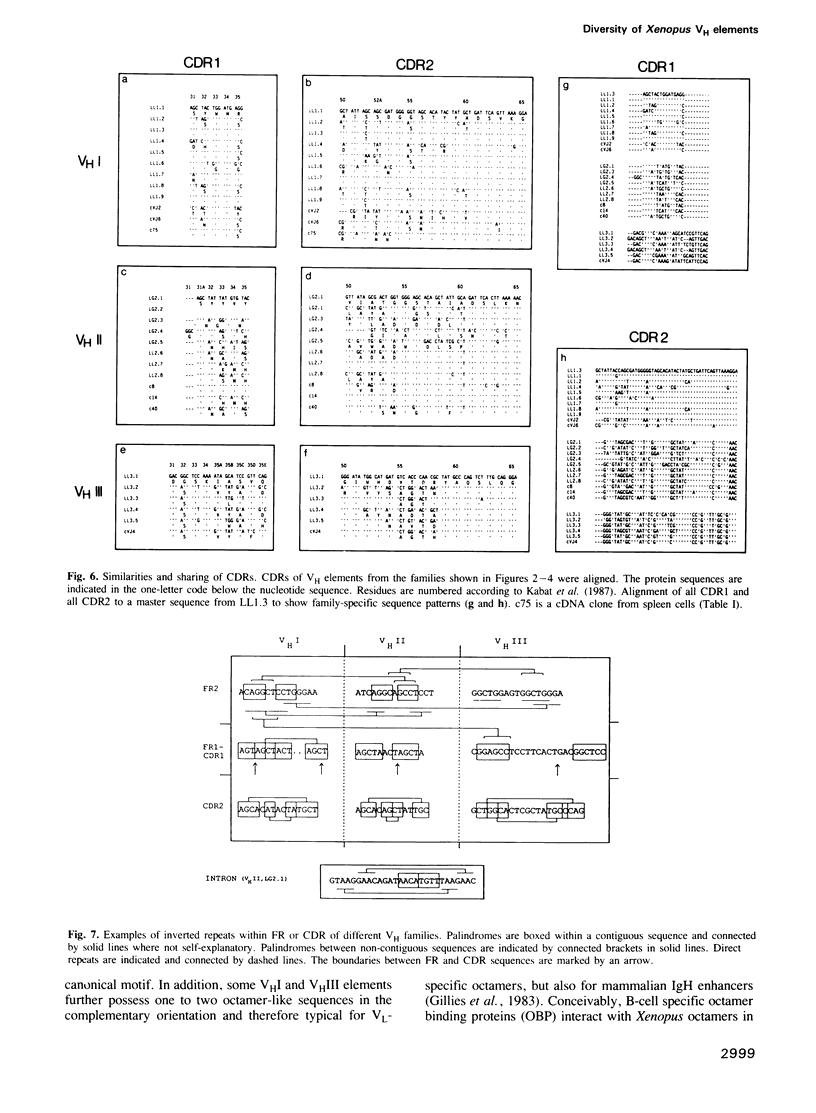
The Xenopus IgH locus includes various variable (VH) families, several putative diversity (DH) and at least seven joining (JH) elements, but--although structurally very similar to the mammalian locus--it contributes to a restricted antibody repertoire. The largest three VH families contain 15-30 VH elements which are interspersed at the VHI-VHII and VHII-VHIII boundaries. Twenty-nine genomic and eight expressed VH regions have been sequenced. Each VH family has distinct promoter elements with up to three octamers (ATGCCTAAAT) in either orientation. The incidence of pseudogenes ranges from less than 15% in VHI and VHII to approximately 50% in VHIII, consistent with their relative expression. CDR1 and CDR2 show low overall diversity with nucleotide divergence limited to parts of the CDRs. Randomly selectedly VH elements share CDR1 and CDR2, in some cases also with expressed VH regions. Thus, the complexity of VH elements is not maximal. Patterns of sequence similarities or identities indicate recombination or gene conversion events; sets of direct and inverted repeats flank the sites of, or lie within FR or CDR sequences where these genetic events may occur. Restricted antibody diversity in Xenopus seems therefore to be at least partially related to low complexity of VH elements, frequence of pseudogenes and expression regulated by specific promoter elements; diversity may potentially be increased by (non)homologous recombination events.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akira S., Okazaki K., Sakano H. Two pairs of recombination signals are sufficient to cause immunoglobulin V-(D)-J joining. Science. 1987 Nov 20;238(4830):1134–1138. doi: 10.1126/science.3120312. [DOI] [PubMed] [Google Scholar]
- Alt F. W., Blackwell T. K., Yancopoulos G. D. Development of the primary antibody repertoire. Science. 1987 Nov 20;238(4830):1079–1087. doi: 10.1126/science.3317825. [DOI] [PubMed] [Google Scholar]
- Ben-Sasson S. A. Immunoglobulin differentiation is dictated by repeated recombination sequences within the V region prototype gene: a hypothesis. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4598–4602. doi: 10.1073/pnas.76.9.4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berek C., Milstein C. Mutation drift and repertoire shift in the maturation of the immune response. Immunol Rev. 1987 Apr;96:23–41. doi: 10.1111/j.1600-065x.1987.tb00507.x. [DOI] [PubMed] [Google Scholar]
- Berman J. E., Mellis S. J., Pollock R., Smith C. L., Suh H., Heinke B., Kowal C., Surti U., Chess L., Cantor C. R. Content and organization of the human Ig VH locus: definition of three new VH families and linkage to the Ig CH locus. EMBO J. 1988 Mar;7(3):727–738. doi: 10.1002/j.1460-2075.1988.tb02869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt D. C., Griessen M., Du Pasquier L., Jaton J. C. Antibody diversity in amphibians: evidence for the inheritance of idiotypic specificities in isogenic Xenopus. Eur J Immunol. 1980 Oct;10(10):731–736. doi: 10.1002/eji.1830101002. [DOI] [PubMed] [Google Scholar]
- Brodeur P. H., Riblet R. The immunoglobulin heavy chain variable region (Igh-V) locus in the mouse. I. One hundred Igh-V genes comprise seven families of homologous genes. Eur J Immunol. 1984 Oct;14(10):922–930. doi: 10.1002/eji.1830141012. [DOI] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Currier S. J., Gallarda J. L., Knight K. L. Partial molecular genetic map of the rabbit VH chromosomal region. J Immunol. 1988 Mar 1;140(5):1651–1659. [PubMed] [Google Scholar]
- Dale R. M., McClure B. A., Houchins J. P. A rapid single-stranded cloning strategy for producing a sequential series of overlapping clones for use in DNA sequencing: application to sequencing the corn mitochondrial 18 S rDNA. Plasmid. 1985 Jan;13(1):31–40. doi: 10.1016/0147-619x(85)90053-8. [DOI] [PubMed] [Google Scholar]
- Du Pasquier L. Antibody diversity in lower vertebrates--why is it so restricted? Nature. 1982 Mar 25;296(5855):311–313. doi: 10.1038/296311a0. [DOI] [PubMed] [Google Scholar]
- Du Pasquier L., Schwager J., Flajnik M. F. The immune system of Xenopus. Annu Rev Immunol. 1989;7:251–275. doi: 10.1146/annurev.iy.07.040189.001343. [DOI] [PubMed] [Google Scholar]
- Du Pasquier L., Wabl M. R. Antibody diversity in amphibians: inheritance of isoelectric focusing antibody patterns in isogenic frogs. Eur J Immunol. 1978 Jun;8(6):428–433. doi: 10.1002/eji.1830080611. [DOI] [PubMed] [Google Scholar]
- Eaton S., Calame K. Multiple DNA sequence elements are necessary for the function of an immunoglobulin heavy chain promoter. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7634–7638. doi: 10.1073/pnas.84.21.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkner F. G., Zachau H. G. Correct transcription of an immunoglobulin kappa gene requires an upstream fragment containing conserved sequence elements. Nature. 1984 Jul 5;310(5972):71–74. doi: 10.1038/310071a0. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Gillies S. D., Morrison S. L., Oi V. T., Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983 Jul;33(3):717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- Golding G. B., Gearhart P. J., Glickman B. W. Patterns of somatic mutations in immunoglobulin variable genes. Genetics. 1987 Jan;115(1):169–176. doi: 10.1093/genetics/115.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadji-Azimi I., Schwager J., Thiebaud C. B-lymphocyte differentiation in Xenopus laevis larvae. Dev Biol. 1982 Apr;90(2):253–258. doi: 10.1016/0012-1606(82)90374-8. [DOI] [PubMed] [Google Scholar]
- Hinds K. R., Litman G. W. Major reorganization of immunoglobulin VH segmental elements during vertebrate evolution. Nature. 1986 Apr 10;320(6062):546–549. doi: 10.1038/320546a0. [DOI] [PubMed] [Google Scholar]
- Kleinfield R., Hardy R. R., Tarlinton D., Dangl J., Herzenberg L. A., Weigert M. Recombination between an expressed immunoglobulin heavy-chain gene and a germline variable gene segment in a Ly 1+ B-cell lymphoma. 1986 Aug 28-Sep 3Nature. 322(6082):843–846. doi: 10.1038/322843a0. [DOI] [PubMed] [Google Scholar]
- Kodaira M., Kinashi T., Umemura I., Matsuda F., Noma T., Ono Y., Honjo T. Organization and evolution of variable region genes of the human immunoglobulin heavy chain. J Mol Biol. 1986 Aug 20;190(4):529–541. doi: 10.1016/0022-2836(86)90239-1. [DOI] [PubMed] [Google Scholar]
- Kolchanov N. A., Solovyov V. V., Rogozin I. B. Peculiarities of immunoglobulin gene structures as a basis for somatic mutation emergence. FEBS Lett. 1987 Apr 6;214(1):87–91. doi: 10.1016/0014-5793(87)80018-2. [DOI] [PubMed] [Google Scholar]
- Krawinkel U., Zoebelein G., Bothwell A. L. Palindromic sequences are associated with sites of DNA breakage during gene conversion. Nucleic Acids Res. 1986 May 12;14(9):3871–3882. doi: 10.1093/nar/14.9.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landy A., Ross W. Viral integration and excision: structure of the lambda att sites. Science. 1977 Sep 16;197(4309):1147–1160. doi: 10.1126/science.331474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litman G. W., Murphy K., Berger L., Litman R., Hinds K., Erickson B. W. Complete nucleotide sequences of three VH genes in Caiman, a phylogenetically ancient reptile: evolutionary diversification in coding segments and variation in the structure and organization of recombination elements. Proc Natl Acad Sci U S A. 1985 Feb;82(3):844–848. doi: 10.1073/pnas.82.3.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I. W., Lienhard S., Jiricny J., De Robertis E. M. An enhancer-like sequence within the Xenopus U2 gene promoter facilitates the formation of stable transcription complexes. Nature. 1985 Jul 11;316(6024):163–167. doi: 10.1038/316163a0. [DOI] [PubMed] [Google Scholar]
- Parslow T. G., Blair D. L., Murphy W. J., Granner D. K. Structure of the 5' ends of immunoglobulin genes: a novel conserved sequence. Proc Natl Acad Sci U S A. 1984 May;81(9):2650–2654. doi: 10.1073/pnas.81.9.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathbun G. A., Capra J. D., Tucker P. W. Organization of the murine immunoglobulin VH complex in the inbred strains. EMBO J. 1987 Oct;6(10):2931–2937. doi: 10.1002/j.1460-2075.1987.tb02597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reth M., Gehrmann P., Petrac E., Wiese P. A novel VH to VHDJH joining mechanism in heavy-chain-negative (null) pre-B cells results in heavy-chain production. 1986 Aug 28-Sep 3Nature. 322(6082):840–842. doi: 10.1038/322840a0. [DOI] [PubMed] [Google Scholar]
- Reynaud C. A., Anquez V., Grimal H., Weill J. C. A hyperconversion mechanism generates the chicken light chain preimmune repertoire. Cell. 1987 Feb 13;48(3):379–388. doi: 10.1016/0092-8674(87)90189-9. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner W. How do different transcription factors binding the same DNA sequence sort out their jobs? Trends Genet. 1989 Feb;5(2):37–39. doi: 10.1016/0168-9525(89)90017-6. [DOI] [PubMed] [Google Scholar]
- Schiff C., Corbet S., Fougereau M. The Ig germline gene repertoire: economy or wastage? Immunol Today. 1988 Jan;9(1):10–14. doi: 10.1016/0167-5699(88)91348-5. [DOI] [PubMed] [Google Scholar]
- Schwager J., Grossberger D., Du Pasquier L. Organization and rearrangement of immunoglobulin M genes in the amphibian Xenopus. EMBO J. 1988 Aug;7(8):2409–2415. doi: 10.1002/j.1460-2075.1988.tb03086.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwager J., Mikoryak C. A., Steiner L. A. Amino acid sequence of heavy chain from Xenopus laevis IgM deduced from cDNA sequence: implications for evolution of immunoglobulin domains. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2245–2249. doi: 10.1073/pnas.85.7.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983 Apr 14;302(5909):575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Turpen J. B., Smith P. B. Precursor immigration and thymocyte succession during larval development and metamorphosis in Xenopus. J Immunol. 1989 Jan 1;142(1):41–47. [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth T., Staudt L., Baltimore D. An octamer oligonucleotide upstream of a TATA motif is sufficient for lymphoid-specific promoter activity. Nature. 1987 Sep 10;329(6135):174–178. doi: 10.1038/329174a0. [DOI] [PubMed] [Google Scholar]






