Abstract
Through exomic sequencing of 32 intrahepatic cholangiocarcinomas, we discovered frequent inactivating mutations in multiple chromatin-remodeling genes (including BAP1, ARID1A and PBRM1), and mutation in one of these genes occurred in almost half of the carcinomas sequenced. We also identified frequent mutations at previously reported hotspots in the IDH1 and IDH2 genes encoding metabolic enzymes in intrahepatic cholangiocarcinomas. In contrast, TP53 was the most frequently altered gene in a series of nine gallbladder carcinomas. These discoveries highlight the key role of dysregulated chromatin remodeling in intrahepatic cholangiocarcinomas.
Carcinomas of the biliary tract are aggressive malignancies, with 5-year survival of less than 10% (ref. 1). These carcinomas arise throughout the biliary tree and are anatomically classified as either intrahepatic or extrahepatic cholangiocarcinomas2. In addition to cholangiocarcinomas, gallbladder carcinomas also arise from the biliary tree. Although often grouped with cholangiocarcinomas owing to the relative rarity of both diseases, gallbladder carcinomas have distinct natural histories compared to cholangiocarcinomas, suggesting different underlying tumor biology. Although a subset of individuals with biliary tract cancers has identifiable risk factors such as primary sclerosing cholangitis or liver fluke infestation, the majority lack such risk factors2. There is currently no way to screen effectively for early disease, and, other than surgery, there are no effective therapies.
Previous studies of the molecular alterations in biliary tract cancers have focused on small sets of selected genes, usually those known to be altered in pancreatic ductal adenocarcinoma. Somatic alterations in the KRAS, TP53, CDKN2A and SMAD4 (DPC4) genes have been reported in cholangiocarcinoma3–7. The prevalence of these alterations varies widely among studies, perhaps in part owing to an inability to analyze the anatomical subtypes of cholangiocarcinoma separately. Mutations in genes coding for components of the phosphatidylinositide 3-kinase (PI3K) cell signaling pathway, including PIK3CA, PTEN and AKT1, have also been reported in cholangiocarcinoma, as have mutations in previously identified hotspots in IDH1 and IDH2 (encoding isocitrate dehydrogenase 1 and 2, respectively)3–5. Interestingly, mutations in these latter genes encoding metabolic enzymes occur frequently in tumors of the central nervous system and in leukemias but have not been identified in any other gastrointestinal malignancy studied so far8. The molecular alterations in gallbladder carcinoma are also poorly understood. As with cholangiocarcinoma, some studies have identified alterations in KRAS, TP53, CDKN2A and SMAD4 (DPC4), but the prevalence of these alterations varies among studies3,9. Rare mutations in CTNNB1 (the gene encoding β-catenin) and PIK3CA have also been reported in gallbladder carcinomas3,10.
To shed light on the molecular mechanisms underlying carcino-genesis in the biliary tract, we sequenced the exomes of a series of clinically and pathologically well-characterized intrahepatic cholangiocarcinoma and gallbladder carcinoma specimens. We collected data on clinical, pathological and surgical details; no subjects had known risk factors for the development of cholangiocarcinoma or gallbladder carcinoma. Survival was calculated using the Kaplan-Meier method and compared using the log-rank test and Cox proportional hazards regression. All tumor samples were macrodissected to enrich for neoplastic cellularity. In the discovery screen, we sequenced approximately 21,000 protein-coding genes in matched tumor and normal DNA from 32 intrahepatic cholangiocarcinomas and 9 gallbladder carcinomas (Supplementary Table 1a,b). We captured coding sequences from individual paired-end libraries for each sample using the Agilent SureSelect Paired-End Version 2.0 Human Exome kit, and we sequenced captured libraries using the Illumina HiSeq 2000 next-generation sequencing platform. This sequencing effort produced 38 Mb in total of captured sequence per library with an average depth of coverage of 130-fold in the targeted region, and >90% of targeted bases were represented by at least ten reads (Supplementary Table 2).
Using stringent criteria, we identified 1,259 somatic mutations in 1,128 genes in the 32 intrahepatic cholangiocarcinomas (Supplementary Tables 3 and 4). Intrahepatic cholangiocarcinomas had a mean of 39 somatic mutations per sample, with a range of 13 to 300. However, all samples except one contained less than 60 non-synonymous somatic mutations. One of the 9 gallbladder carcinomas contained almost 3,000 nonsynonymous somatic mutations, and this tumor, which clearly possessed an unusual mutator phenotype, was eliminated from further analyses. In the remaining 8 gallbladder carcinomas, we identified 724 somatic mutations in 695 genes: gallbladder carcinomas had an average of 91 somatic mutations per sample, with a range of 42 to 252 (Supplementary Tables 3 and 4).
Several chromatin-remodeling genes, including BAP1, ARID1A and PBRM1, frequently harbored inactivating mutations in the discovery screen intrahepatic cholangiocarcinomas (Table 1). Somatic mutations in BAP1, which encodes a nuclear deubiquitinase involved in chromatin remodeling, occurred in 8 of 32 intrahepatic cholangiocarcinomas (25%). Somatic mutations in BAP1 have previously been reported in renal cell carcinoma, uveal melanoma and malignant mesothelioma but have not been reported in any gastrointestinal cancer11. We also identified somatic ARID1A mutations in 6 of 32 intrahepatic cholangiocarcinomas (19%). ARID1A encodes a subunit of the SWI/SNF chromatin-remodeling complexes, and mutations in ARID1A have been reported in several tumor types, including ovarian, colorectal and gastric carcinomas, although they have not been reported in cholangiocarcinomas12. Somatic PBRM1 mutations were identified in 5 of 32 intrahepatic cholangiocarcinomas (17%). As with ARID1A, PBRM1 encodes a subunit of the ATP-dependent SWI/SNF chromatin-remodeling complexes, and mutations in PBRM1 have previously been reported in approximately 40% of clear-cell renal cell carcinomas but have not yet been described in biliary cancers13. The frequent alterations in BAP1, ARID1A and PBRM1 in cholangiocarcinomas highlight the key role of chromatin remodeling in this tumor type. These genes were dramatically enriched for inactivating mutations, with nonsense, frameshift and splice-site mutations accounting for the majority of mutations in each gene (Fig. 1 and Table 1). At least one chromatin-remodeling gene was altered in 15 of 32 cholangiocarcinomas (47%). However, these mutations were not mutually exclusive: three tumors contained mutations in multiple chromatin-remodeling genes, suggesting a lack of overlap in function and an additive effect in epigenetic changes in the cancer cell. Subjects with mutations in one of these chromatin-remodeling genes tended to have shorter survival times than subjects without mutations in these genes, although differences in survival time were not statistically significant (Table 1). When considered together, subjects with a mutation in any one of the three chromatin-remodeling genes (BAP1, ARID1A or PBRM1) trended toward worse survival compared to subjects in whom all three genes were wild type (3-year survival of 47.1% for subjects with mutations compared to 93.3% for subjects without mutations), but these results were not statistically significant (P = 0.1672 by log-rank test). Notably, these mutations may identify additional therapies for cholangiocarcinomas, as the mutations may cause sensitivity to drugs targeting chromatin remodeling, such as histone deacetylase (HDAC) inhibitors, which are already in use or being developed for individuals with cancer14. Intriguingly, previous whole-exome sequencing analysis of liver fluke–associated cholangiocarcinomas did not identify mutations in these chromatin-remodeling genes, highlighting genetic differences in cholangiocarcinomas based on risk factors15.
Table 1.
Genes with frequent somatic alterations in intrahepatic cholangiocarcinoma
| Gene | Number of mutations (DS) (%) | Types of mutations (DS)a | P value (DS)b | Number of mutations (PS) (%) | Types of mutations (PS)a | Mutant 3-year survival (%)c | Wild-type 3-year survival (%)c | P value for 3-year survivalc |
|---|---|---|---|---|---|---|---|---|
| BAP1 | 8 (25) | 3 FS, 1 IF, 2 SS, 2 MS | 7.4 × 10-12 | 5 (16) | 1 SS, 4 MS | 27 | 81 | 0.102 |
| AR1D1A | 6 (19) | 3 NS, 1 FS, 2 MS | 4.3 × 10-3 | 3 (9) | 1 NS, 1 FS, 1 MS | 40 | 79 | 0.51 |
| PBRM1 | 5 (17) | 1 NS, 4 FS | 1.8 × 10-5 | 3 (9) | 1 NS, 1 FS, 1 MS | 50 | 76 | 0.75 |
| 1DH1 or 1DH2 | 6 (19) | 6 MS | 5.4 × 10-3 | 7 (22) | 7 MS | 33 | 81 | 0.0034 |
DS, discovery screen; PS, prevalence screen.
FS, frameshift insertion-deletion; IF, in-frame insertion-deletion; SS, splice-site mutation; MS, missense mutation; NS, nonsense mutation.
Calculated using Bonferroni correction for multiple comparisons (see Supplementary Table 6a for additional data).
Calculated using clinical data from discovery screen samples.
Figure 1.
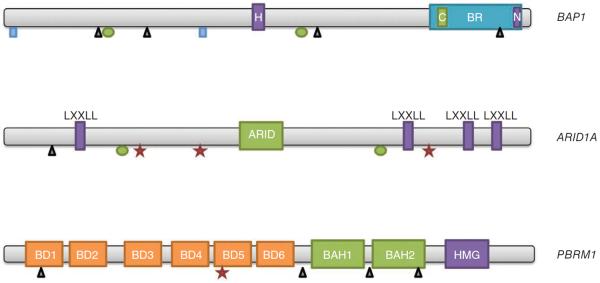
Genes with frequent inactivating mutations in intrahepatic cholangiocarcinoma. Inactivating mutations occurred throughout the coding sequences of BAP1, ARID1A and PBRM1. Rectangles, splice-site mutations; triangles, insertions and deletions; ovals, missense mutations; stars, nonsense mutations. H, HBM-like motif; BR, BRCA1-interacting domain; C, coiled-coil domain; N, nuclear localization signal; LXXLL, LXXLL domain; ARID, ARID domain; BD, bromodomain; BAH, BAH domain; HMG, HMG box.
In addition to identifying inactivating mutations in chromatin-remodeling genes, we also confirmed many of the previous genetic observations in this tumor type3–7. We found somatic mutations in IDH1 in four tumors and in IDH2 in two tumors (19% of the intrahepatic cholangiocarcinomas in total), and these mutations clustered in previously identified hotspots (codons 132 and 172, respectively). The clustering of somatic alterations in mutational hotspots has important implications for early detection: detection of these hotspot IDH gene mutations in plasma could be employed as a screening strategy for populations at high risk. The mutational status of IDH genes was noted to be significantly associated with prognosis—subjects with IDH1 or IDH2 mutations had 3-year survival of 33% compared with 3-year survival of 81% for subjects with wild-type IDH genes (P = 0.0034) (Supplementary Fig. 1). Although subjects with IDH gene mutations were somewhat older and had higher tumor stage (both non-significant; Supplementary Table 5), worse survival persisted, even after adjusting for stage, age and sex using a Cox proportional hazards model (hazards ratio (HR) = 7.37, 95% confidence interval (CI) = 1.13–48.29, P = 0.037). This effect on survival is opposite to the one previously reported for IDH gene mutations in cholangiocarcinoma16. As our sample size was much smaller than that used in the previous study that showed improved survival for cholangiocarcinomas with IDH gene mutation, this discrepancy should be assessed using another independent sample of individuals with intrahepatic cholangiocarcinoma.
In addition, four somatic mutations were identified in FGFR2 in intrahepatic cholangiocarcinoma (13%). Germline activating mutations in this gene have been reported in several craniosynostosis and chondrodysplasia syndromes, and somatic mutations in the same gene regions occur in several tumor types. Two mutations were identified in this study at residues previously reported to be mutated in endometrial carcinoma17. Moreover, FGFR2 gene fusions were recently reported in cholangiocarcinoma, further highlighting the possible importance of this gene in biliary tumorigenesis18. Notably, these FGFR2 alterations represent possible therapeutic targets, with multiple FGFR inhibitors currently in clinical trials.
Mutations were also identified in several other well-known cancer genes. Somatic mutations were identified in several components of the PI3K pathway, including two mutations in PIK3CA, two mutations in PTEN, two mutations in PIK3C2G and one mutation in PIK3C2A (22% of cholangiocarcinomas in total). Two somatic mutations were identified in TP53 (6%), and a single inactivating mutation in CDK2NA as well as a single oncogenic hotspot KRAS mutation were also identified. In addition, NRAS was somatically mutated in a single tumor. No mutations in SMAD4 (DPC4) were identified in intrahepatic cholangiocarcinomas, although a single tumor harbored a mutation in TGFBR2.
To objectively analyze the significance of the mutations identified in the 32 intrahepatic cholangiocarcinomas in the discovery screen, we calculated a combined P value for the mutations in each gene based on three distinct measures of mutation significance: the count of single-base substitutions, the count of insertion-deletion mutations and the relative count of nonsynonymous single-base substitutions. These calculations incorporated gene-specific background mutation rates based on sequence composition and adjusted for multiple comparisons by applying the Bonferroni and Benjamini-Hochberg correction methods (see Online Methods for details). These unbiased analyses (along with the striking enrichment for inactivating mutations) showed that chromatin-remodeling genes BAP1, ARID1A and PBRM1 are clearly drivers in intrahepatic cholangiocarcinoma, as the P values for their observed mutations were 7.4 × 10−12, 4.3 × 10−3 and 1.8 × 10−5, respectively, using the conservative Bonferroni correction for multiple comparisons (Table 1 and Supplementary Table 6a). These analyses also confirmed the key role of IDH1 in intrahepatic cholangiocarcinoma, with a P value of 5.4 × 10−3, and highlighted the potential importance of FGFR2, with a P value of 4.8 × 10−2.
To more accurately determine the mutation frequency of genes identified in the initial sequencing analyses in intrahepatic cholangiocarcinoma (discovery screen), we next sequenced a subset of the potential driver genes (AKT1, ARID1A, BAP1, CDKN2A, CTNNB1, IDH1, IDH2, KRAS, NRAS, PBRM1, PIK3C2A, PIK3C2G, PIK3CA, PTEN, SMAD4, TGFBR2 and TP53) in an independent set of 32 intrahepatic cholangiocarcinoma samples (prevalence screen). As was observed in the discovery screen, multiple mutations were identified in chromatin-remodeling genes: BAP1 was mutated in 5 of 32 cholangiocarcinomas (16%), ARID1A was mutated in 3 of 32 cholangiocarcinomas (9%), and PBRM1 was mutated in 3 of 32 cholangiocarcinomas (9%) (Table 1 and Supplementary Table 7a). In the prevalence screen, mutations in these genes were mutually exclusive, and mutation in one of these genes occurred in 11 of 32 cholangiocarcinomas (34%). Combining the discovery and prevalence screens, BAP1 was mutated in 13 of 64 tumors (20%), ARID1A was mutated in 9 of 64 tumors (14%), and PBRM1 was mutated in 8 of 64 tumors (13%), adding up to 26 cholangiocarcinomas with mutations in chromatin-remodeling genes out of 64 total cholangiocarcinomas (41%). Frequent mutations in IDH1 and IDH2 hotspots were also identified in this prevalence screen, as mutations in these genes were identified in 7 of 32 cholangiocarcinomas (22%; overall prevalence of 20% when both the discovery and prevalence screens were combined). The prevalence screen identified additional mutations in KRAS, NRAS and components of the PI3K pathway (PIK3CA and PI3KC2G), confirming the key role of RAS and PI3K signaling in this tumor type.
Overall, these data on somatic mutations in intrahepatic cholangiocarcinomas highlight the genetic heterogeneity of the disease. No single gene was mutated in >25% of the tumors sequenced. However, even considering this intertumoral genetic heterogeneity, several pathways were frequently targeted–for example, genes involved in chromatin remodeling (including BAP1, ARID1A and PBRM1) were somatically altered in almost half of the intrahepatic cholangiocarcinomas analyzed.
TP53 was the most frequently mutated gene in gallbladder carcinoma, with somatic mutations in five of eight tumors without a mutator phenotype (63%). Somatic mutations in PBRM1 were identified in two of eight gallbladder carcinomas (25%). Another chromatin-remodeling gene, KMT2C (MLL3), was mutated in two tumors (25%); no mutations were identified in this gene in cholangiocarcinomas in this study. One somatic mutation was identified in PIK3CA in a gallbladder carcinoma sample, and one mutation was identified in SMAD4. No mutations in BAP1, ARID1A, IDH1 or IDH2 were identified in gallbladder carcinoma in the discovery screen. We performed similar statistical analysis of the mutations in these eight gallbladder carcinomas: only TP53 had a P value of less than 0.05 with Bonferroni correction (Supplementary Table 6b). To estimate the frequency of somatic mutations in potential driver genes more accurately, we sequenced the same subset of potential driver genes in an independent set of eight additional gallbladder carcinomas (prevalence screen). Intriguingly, we identified not only two additional mutations in PBRM1 (25%) but also one mutation each in BAP1 (13%) and ARID1A (13%) (Supplementary Table 7b). When both the discovery and prevalence screens were combined, the overall mutation prevalence in gallbladder carcinoma for BAP1, ARID1A and PBRM1 was 6%, 6% and 25%, respectively. No mutations were identified in IDH1 or IDH2 in the eight additional gallbladder carcinomas analyzed. In addition, the prevalence screen confirmed the key role of TP53 mutations in gallbladder carcinoma, with mutations identified in two of eight tumors (25%; overall prevalence of 44% when both the discovery and prevalence screens were combined). The number of gallbladder carcinomas analyzed in the current study is too small to make definitive conclusions about the genetic relationships between gallbladder carcinoma and intrahepatic cholangiocarcinoma.
In this study, we report whole-exome sequencing results in two biliary tract carcinomas—intrahepatic cholangiocarcinoma and gall-bladder carcinoma. These data highlight the key role of mutations in chromatin-remodeling genes in cholangiocarcinoma: frequent inactivating mutations occur in three different genes, affecting almost half of the tumors sequenced. These studies point to chromatin remodeling as a crucial area for future investigation, including possible therapeutic targeting. Moreover, comparison of somatic mutation data for cholangiocarcinoma and gallbladder carcinoma suggests that, although both tumor types arise from biliary epithelium, they are genetically distinct.
URL
dbSNP database, http://www.ncbi.nlm.nih.gov/projects/SNP/; 1000 Genomes Project database, http://browser.1000genomes.org/index.html.
ONLINE METHODS
Preparation of clinical samples (discovery screen)
Fresh-frozen tumor and matched normal tissues were obtained from subjects under protocols approved by the institutional review boards at the Johns Hopkins Hospital, Memorial Sloan-Kettering Cancer Center, Mayo Clinic, Fundeni Clinical Institute and University Hospital Trust of Verona. Although direct informed consent was not obtained for this study, the use of deidentified, previously banked samples for this study was approved by all institutional review boards. Tumor tissue was analyzed by frozen section to assess neoplastic cellularity—tumors were macrodissected to remove residual normal tissue and enhance neoplastic cellularity, as confirmed by the examination of multiple frozen sections. Normal samples were analyzed by frozen section to confirm that no tumor tissue was present.
Preparation of Illumina genomic DNA libraries (discovery screen)
Genomic DNA libraries were prepared following the suggested protocol from Illumina with the following modifications.
Genomic DNA (3 μg) from tumor or normal cells in 100 μl of Tris/EDTA buffer was fragmented in a Covaris sonicator to a size of 100–500 bp. To remove fragments of <150 bp, DNA was mixed with 25 μl of 5× Phusion HF buffer, 416 μl of double-distilled water and 84 μl of NT binding buffer and loaded onto a NucleoSpin column (636972, Clontech). The column was centrifuged at 14,000g in a desktop centrifuge for 1 min, washed once with 600 μl of washing buffer (NT3 from Clontech) and centrifuged again for 2 min to dry completely. DNA was eluted in 45 μl of the elution buffer included in the kit.
Purified, fragmented DNA was mixed with 40 μl of water, 10 μl of End-Repair Reaction buffer and 5 μl of End-Repair Enzyme Mix (E6050, NEB). The 100-μl end-repair mixture was incubated at 20 °C for 30 min, and DNA was purified with a PCR purification kit (28104, Qiagen) and eluted with 42 μl of elution buffer.
For A-tailing, 42 μl of end-repaired DNA was mixed with 5 μl of 10× dA Tailing Reaction Buffer and 3 μl of Klenow (exo–, E6053, NEB). The mixture was incubated at 37 °C for 30 min before DNA was purified with a MinElute PCR purification kit (28004, Qiagen). Purified DNA was eluted with 25 μl of elution buffer warmed to 70 °C.
For adaptor ligation, 25 μl of A-tailed DNA was mixed with 10 μl of PE adaptor (Illumina), 10 μl of 5× Ligation buffer and 5 μl of Quick T4 DNA ligase (E6056, NEB). The ligation mixture was incubated at 20 °C for 15 min.
To purify adaptor-ligated DNA, 50 μl of ligation mixture from step 4 was mixed with 200 μl of buffer NT from a NucleoSpin Extract II kit (636972, Clontech) and loaded onto a NucleoSpin column. The column was centrifuged at 14,000g in a desktop centrifuge for 1 min, washed once with 600 μl of wash buffer (NT3 from Clontech) and centrifuged again for 2 min to dry completely. DNA was eluted in 50 μl of the elution buffer included in the kit.
To obtain an amplified library, ten PCR runs of 50 μl each were set up, each including 29 μl of water, 10 μl of 5× Phusion HF buffer, 1 μl of a dNTP mix containing 10 mM of each dNTP, 2.5 μl of DMSO, 1 μl of Illumina PE primer 1, 1 μl of Illumina PE primer 2, 0.5 μl of Hot-Start Phusion polymerase and 5 μl of the DNA from step 5. The PCR program used included an initial incubation at 98 °C for 2 min; 6 cycles of 98 °C for 15 s, 65 °C for 30 s and 72 °C for 30 s; and a final incubation at 72 °C for 5 min. To purify the PCR product, 500 μl of PCR mixture (from the ten PCR runs) was mixed with 1,000 μl of NT buffer from a NucleoSpin Extract II kit and purified as described in step 1. Library DNA was eluted with elution buffer warmed to 70 °C, and DNA concentration was estimated by absorption at 260 nm with a Nanodrop.
Exome DNA capture (discovery screen)
The protocol from the Agilent's SureSelect Paired-End version 2.0 Human Exome kit was used for human exome capture with the following modifications.
A hybridization mixture was prepared containing 25 μl of SureSelect Hyb 1, 1 μl of SureSelect Hyb 2, 10 μl of SureSelect Hyb 3 and 13 μl of SureSelect Hyb 4.
The paired-end DNA library described above (3.4 μl, 0.5 μg), 2.5 μl of SureSelect Block 1, 2.5 μl of SureSelect Block 2 and 0.6 μl of SureSelect Block 3 were loaded into one well of a 384-well Diamond PCR plate (AB-1111, Thermo-Scientific), sealed with microAmp clear adhesive film (4306311, Applied Biosystems) and placed in a GeneAmp PCR system 9700 thermocycler (Life Sciences) for 5 min at 95 °C and then held at 65 °C (with the heated lid on).
Hybridization buffer from step 1 (25–30 μl) was heated for at least 5 min at 65 °C in another sealed plate with the heated lid on.
SureSelect Oligo Capture Library (5 μl), 1 μl of nuclease-free water and 1 μl of diluted RNase block (prepared by diluting RNase block 1:1 with nuclease-free water) were mixed and heated at 65 °C for 2 min in another sealed 384-well plate.
While keeping all reactions at 65 °C, 13 μl of hybridization buffer from step 3 was added to the 7 μl of the SureSelect Capture Library Mix from step 4 and then to the entire contents (9 μl) of the library from step 2. The mixture was slowly pipetted up and down 8–10 times.
The 384-well plate was sealed tightly, and the hybridization mixture was incubated for 24 h at 65 °C with a heated lid.
After hybridization, the following five steps were performed to recover and amplify the captured DNA library.
To prepare magnetic beads for recovering captured DNA, 50 μl of Dynal MyOne Streptavidin C1 magnetic beads (650.02, Invitrogen Dynal) was placed in a 1.5-ml microfuge tube and vigorously resuspended on a vortex mixer. Beads were washed three times by adding 200 μl of SureSelect Binding buffer, mixing on a vortex for 5 s and then removing the supernatant after placing the tubes in a Dynal magnetic separator. After the third wash, beads were resuspended in 200 μl of SureSelect Binding buffer.
To bind captured DNA, the entire hybridization mixture described above (29 μl) was transferred directly from the thermocycler to the bead solution and mixed gently. The hybridization mix–bead solution was incubated in an Eppendorf thermomixer at 850 r.p.m. for 30 min at room temperature.
To wash beads, the supernatant was removed from beads after applying a Dynal magnetic separator, and beads were resuspended in 500 μl of Sure-Select wash buffer 1 through mixing by vortex for 5 s and incubation for 15 min at room temperature. Wash buffer 1 was then removed from the beads after magnetic separation, and beads were further washed three times, with each wash consisting of 500 μl of prewarmed SureSelect wash buffer 2 that was incubated at 65 °C for 10 min. After the final wash, SureSelect wash buffer 2 was completely removed.
To elute captured DNA, beads were suspended in 50 μl of SureSelect elution buffer, mixed by vortex and incubated for 10 min at room temperature. The supernatant was removed after magnetic separation, collected in a new 1.5-ml microfuge tube and mixed with 50 μl of SureSelect neutralization buffer. DNA was purified with a Qiagen MinElute column and eluted in 17 μl of elution buffer warmed to 70 °C to obtain 15 μl of captured DNA library.
To amplify the captured DNA library, 15 PCR runs, each containing 9.5 μl of water, 3 μl of 5× Phusion HF buffer, 0.3 μl of 10 mM dNTP, 0.75 μl of DMSO, 0.15 μl of Illumina PE primer 1, 0.15 μl of Illumina PE primer 2, 0.15 μl of Hot-Start Phusion polymerase and 1 μl of captured exome library, were set up. The PCR program used involved an incubation at 98 °C for 30 s; 14 cycles of 98 °C for 10 s, 65 °C for 30 s and 72 °C for 30 s; and a final incubation at 72 °C for 5 min. To purify PCR products, 225 μl of PCR mixture (from 15 PCR runs) was mixed with 450 μl of NT buffer from the NucleoSpin Extract II kit and purified as described above. The final library DNA was eluted with 30 μl of elution buffer warmed to 70 °C, and DNA concentration was estimated by absorption at 260 nm.
Somatic mutation identification by massively parallel sequencing (discovery screen)
Captured DNA libraries were sequenced with the Illumina HiSeq genome analyzer. Sequencing reads were analyzed and aligned to human genome hg18 with the Eland algorithm in CASAVA 1.6 software (Illumina). A mismatched base was identified as a mutation only when the following occurred: (i) the mismatched base was identified by five or more distinct pairs; (ii) the number of distinct tags containing a particular mismatched base was at least 15% of the total number of distinct tags; and (iii) the mismatched base was not present in greater than 0.2% of the tags in the matched normal sample. All mutations in genes with more than one mutation were confirmed by visual inspection of the sequencing data. Somatic mutation data for each sample are included in Supplementary Table 4. Raw sequencing data, which include germline information, will be made available to qualified investigators upon request provided they can meet the data security requirements of our institutional review board. In addition, 50 mutations were also validated by conventional Sanger sequencing—all 50 mutations were also present in the Sanger sequencing data, confirming that our sequencing assay and mutation identification strategy are reliable (Supplementary Table 8). The SNP search databases included SNPs from dbSNP and the 1000 Genomes Project database.
Survival analyses
Summary statistics were obtained using established methods and are presented as percentages, means or median values. Survival was estimated using the Kaplan-Meier method, and differences were compared using the log-rank test. Cox proportional hazards regression was used to adjust for confounding by subject and tumor characteristics in the analysis of IDH gene mutations.
Statistical analyses of significantly mutated genes (discovery screen)
Using stringent criteria, 1,259 and 725 somatic mutations were identified in the protein-coding sequences of cholangiocarcinoma and gallbladder cancer samples, respectively. We implemented the following statistical framework to identify significantly mutated genes by incorporating background mutation rates, gene length and base composition.
Inspired by previous works19,20, our model defines gene-specific background mutation rates, which capture exome-wide as well as gene-specific sequence-based parameters. We defined eight exhaustive and disjoint sequence-based dinucleotide contexts: C in CpG, G in CpG, C in TpC, G in GpA and all other A, G, C and T nucleotides. We represented the occurrences of each context in the entire protein-coding sequence by Ni and in each gene of interest by gi. Subsequently, we identified the dinucleotide context for all single-base substitution (SBS) somatic mutations identified in each sample cohort and derived the counts of mutations in each context over all coding sequence (protein-coding sequence) (ni). We derived the expected probability of observing a mutation in a base occurring in the coding sequence of a gene of interest as follows
| (1) |
| (2) |
where fi denotes the fraction of bases in dinucleotide context i in the entire coding sequence, where a mutation has been observed in the cohort. It is noteworthy to mention that the context parameters ni and gi are defined as the total number of occurrences of each context sequenced across the cohort; therefore, following the simplifying assumption of full coverage of the entire protein-coding sequence and assuming K samples in the cohort, these parameters will be K times those of a single haploid exome.
Following the definition of fi, we derived the background probability of observing at least mg,obs mutations in a gene of interest, using the binomial tail probability of Lg trials with mg,obs successes and Pmut probability of success in each trial. Here Lg represents the length of the coding sequence of the gene across the cohort.
| (3) |
We used an equivalent formulation to model the statistical significance of observing qg,obs insertions-deletions (indels) in a gene of interest. The background indel frequency (Pindel) was defined as the number of indels recovered in the entire coding sequence of the cohort divided by the length of the entire coding sequence available in the cohort.
| (4) |
The two statistical tests described above (equations 3 and 4) reflect the significance of mutation counts in a gene but are blind to the protein-level consequences of mutations. To capture the impact of mutations on protein, we applied an extension of the tests above that examines enrichment for nonsynonymous mutations in the set of SBS mutations identified in a gene of interest. We defined a background gene-specific ratio of nonsynonymous to synonymous (NS/S) mutations, given the exome-wide NS/S ratio in each dinucleotide context (ri) and the sequence composition of each gene as follows. Note that gi has the same definition as in equation 1.
| (5) |
Given the NS/S ratio for a gene of interest, the probability of an observed mutation in the gene being nonsynonymous is
| (6) |
Following this step, the binomial tail probability of observing from the total of mg,obs mutations in a gene of interest is
| (7) |
The three test statistics (equations 3, 4 and 7) rely on three distinct measures for calling a gene significantly mutated: the counts of SBSs, the counts of indels and the relative counts of nonsynonymous SBSs. Assuming the independence of these measures, given gene-specific parameters of gi and Lg, we combine them using Fisher's combined probability test to derive a measure of overall significance for each gene of interest (combined P value). We acknowledge the fact that Fisher's combined probability test is best suited to P values derived from continuous probability distribution functions; however, it has been shown that its application to P values derived from discrete probability distributions results in conservative estimates of P value21.
Finally, we applied Bonferroni and Benjamini-Hochberg correction methods to the combined P values to control for multiple testing.
Prevalence screen
Exome sequencing results were independently validated in a series of 32 intrahepatic cholangiocarcinomas and 8 gallbladder carcinomas from Verona University Hospital in Italy by investigating the mutational status of the following 17 cancer-associated genes, listed in alphabetical order: AKT1, ARID1A, BAP1, CDKN2A, CTNNB1, IDH1, IDH2, KRAS, NRAS, PBRM1, PIK3C2A, PIK3C2G, PIK3CA, PTEN, SMAD4, TGFBR2 and TP53.
DNA (40 ng) was used for multiplex PCR amplification with a custom Ion AmpliSeq Panel (Life Technologies) that explores the coding regions of the aforementioned genes (covered regions: 90% of total). Mean read depth was 1,276×, with 88.9% of target bases being covered by above 100×.
Emulsion PCR was performed with the OneTouch OT2 system (Life Technologies). The quality of the obtained library was evaluated by Agilent 2100 Bioanalyzer on-chip electrophoresis (Agilent Technologies). Sequencing was run on the Ion Torrent Personal Genome Machine (PGM, Life Technologies) loaded with a 318 chip. Data analysis, including alignment to the hg19 human reference genome and variant calling, was performed using Torrent Suite Software v.3.6 (Life Technologies). Filtered variants were annotated using SnpEff software v.3.1 (alignments visually verified with the Integrative Genomics Viewer; IGV v.2.1, Broad Institute).
Supplementary Material
ACKNOWLEDGMENTS
The authors wish to acknowledge the following funding sources: the Virginia and D.K. Ludwig Fund for Cancer Research; the Lustgarten Foundation for Pancreatic Cancer Research; US National Institutes of Health (NIH) grants P50 CA62924, K08DK090154 and EDRN U01CA086402; Associazione Italiana Ricerca Cancro (AIRC grants 12182, 11930, 6421 and IG 12214); and Italian Cancer Genome Project FIRB RBAP10AHJB.
Footnotes
Accession codes. Raw sequencing data, which include germline information, will be made available to qualified investigators upon request, provided they can meet the data security requirements of the institutional review boards of the institutions contributing samples to this study.
AUTHOR CONTRIBUTIONS T.M.P., R.A.A., A.S., R.K., V.E.V., R.H.H., B.V., K.W.K., N.P. and L.D.W. jointly supervised research. Y.J., F.M.S., A. Maitra, M.F., M.S., A. Mafficini, P.C., R.T.L., A.R., A.G., G.T., F.d.B., A.S., R.K., V.E.V., R.H.H., B.V., K.W.K., N.P. and L.D.W. conceived and designed experiments. Y.J., M.M.S., A. Maitra, G.J.A.O., J.C.R., L.R.R., G.J.G., I.P., S.T.A., S.D., M.F., M.S., A. Mafficini, P.C., R.T.L., A.R., A.G., G.T., F.d.B., A.S., D.K., B.V., K.W.K., N.P. and L.D.W. performed experiments. T.M.P., R.A.A., D.J.L., N.N., V.B.G., R.K., K.W.K., N.P. and L.D.W. performed statistical analysis. Y.J., T.M.P., R.A.A., D.J.L., N.N., V.B.G., M.F., M.S., A. Mafficini, P.C., R.T.L., A.R., A.G., G.T., F.d.B., A.S., W.J., D.K., R.K., R.H.H., B.V., K.W.K., N.P. and L.D.W. analyzed data. F.M.S., M.M.S., A. Maitra, P.A., G.J.A.O., J.C.R., L.R.R., G.J.G., I.P., S.T.A., S.D., M.F., M.S., A. Mafficini, P.C., R.T.L., A.R., A.G., G.T., F.d.B., A.S., W.J., D.K., R.K., V.E.V., R.H.H., B.V., K.W.K., N.P. and L.D.W. contributed reagents, materials or analysis tools. Y.J., T.M.P., R.A.A., F.M.S., A. Maitra, D.K., R.H.H., B.V., K.W.K., N.P. and L.D.W. wrote the manuscript.
COMPETING FINANCIAL INTERESTS The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Everhart JE, Ruhl CE. Burden of digestive diseases in the United States Part III: Liver, biliary tract, and pancreas. Gastroenterology. 2009;136:1134–1144. doi: 10.1053/j.gastro.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 2.Blechacz B, et al. Clinical diagnosis and staging of cholangiocarcinoma. Nat. Rev. Gastroenterol. Hepatol. 2011;8:512–522. doi: 10.1038/nrgastro.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borger DR, et al. Frequent mutation of isocitrate dehydrogenase IDH1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist. 2012;17:72–79. doi: 10.1634/theoncologist.2011-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voss JS, et al. Molecular profiling of cholangiocarcinoma shows potential for targeted therapy treatment decisions. Hum. Pathol. 2013;44:1216–1222. doi: 10.1016/j.humpath.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 5.Xu RF, et al. KRAS and PIK3CA but not BRAF genes are frequently mutated in Chinese cholangiocarcinoma patients. Biomed. Pharmacother. 2011;65:22–26. doi: 10.1016/j.biopha.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 6.Chuang SC, et al. Immunohistochemical study of DPC4 and p53 proteins in gallbladder and bile duct cancers. World J. Surg. 2004;28:995–1000. doi: 10.1007/s00268-004-7447-8. [DOI] [PubMed] [Google Scholar]
- 7.Tannapfel A, et al. Genetic and epigenetic alterations of the INK4a-ARF pathway in cholangiocarcinoma. J. Pathol. 2002;197:624–631. doi: 10.1002/path.1139. [DOI] [PubMed] [Google Scholar]
- 8.Yan H, et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parwani AV, et al. Immunohistochemical and genetic analysis of non-small cell and small cell gallbladder carcinoma and their precursor lesions. Mod. Pathol. 2003;16:299–308. doi: 10.1097/01.MP.0000062656.60581.AA. [DOI] [PubMed] [Google Scholar]
- 10.Yanagisawa N, et al. More frequent β-catenin exon 3 mutations in gallbladder adenomas than in carcinomas indicate different lineages. Cancer Res. 2001;61:19–22. [PubMed] [Google Scholar]
- 11.Murali R, et al. Tumours associated with BAP1 mutations. Pathology. 2013;45:116–126. doi: 10.1097/PAT.0b013e32835d0efb. [DOI] [PubMed] [Google Scholar]
- 12.Jones S, et al. Somatic mutations in the chromatin remodeling gene ARID1A occur in several tumor types. Hum. Mutat. 2012;33:100–103. doi: 10.1002/humu.21633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Varela I, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma X, et al. Histone deacetylase inhibitors: current status and overview of recent clinical trials. Drugs. 2009;69:1911–1934. doi: 10.2165/11315680-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 15.Ong CK, et al. Exome sequencing of liver fluke–associated cholangiocarcinoma. Nat. Genet. 2012;44:690–693. doi: 10.1038/ng.2273. [DOI] [PubMed] [Google Scholar]
- 16.Wang P, et al. Mutations in isocitrate dehydrogenase 1 and 2 occur frequently in intrahepatic cholangiocarcinomas and share hypermethylation targets with glioblastomas. Oncogene. 2013;32:3091–3100. doi: 10.1038/onc.2012.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pollock PM, et al. Frequent activating FGFR2 mutations in endometrial carcinomas parallel germline mutations associated with craniosynostosis and skeletal dysplasia syndromes. Oncogene. 2007;26:7158–7162. doi: 10.1038/sj.onc.1210529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu YM, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3:636–647. doi: 10.1158/2159-8290.CD-13-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sjöblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 20.Kan Z, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 21.Mielke PW, et al. Combining probability values from independent permutation tests: a discrete analog of Fisher's classical method. Psychol. Rep. 2004;95:449–458. doi: 10.2466/pr0.95.2.449-458. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


