Abstract
Drosophila neuroblasts are a model system for studying stem cell self-renewal and the establishment of cortical polarity. Larval neuroblasts generate a large apical self-renewing neuroblast, and a small basal cell that differentiates. We performed a genetic screen to identify regulators of neuroblast self-renewal, and identified a mutation in sgt1(suppressor-of-G2-allele-of-skp1) that had fewer neuroblasts. We found that sgt1 neuroblasts have two polarity phenotypes: failure to establish apical cortical polarity at prophase, and lack of cortical Scribble localization throughout the cell cycle. Apical cortical polarity was partially restored at metaphase by a microtubule-induced cortical polarity pathway. Double mutants lacking Sgt1 and Pins (a microtubule-induced polarity pathway component) resulted in neuroblasts without detectable cortical polarity and formation of “neuroblast tumors.” Mutants in hsp83 (encoding the predicted Sgt1-binding protein Hsp90), LKB1, or AMPKα all show similar prophase apical cortical polarity defects (but no Scribble phenotype), and activated AMPKα rescued the sgt1 mutant phenotype. We propose that an Sgt1/Hsp90-LKB1-AMPK pathway acts redundantly with a microtubule-induced polarity pathway to generate neuroblast cortical polarity, and the absence of neuroblast cortical polarity can produce neuroblast tumors.
INTRODUCTION
The precise regulation of stem cell self-renewal versus differentiation is essential for normal development, required for tissue homeostasis, and may suppress tumorigenesis. Despite its importance, the molecular mechanisms regulating stem self-renewal are only beginning to be uncovered. Recently, Drosophila larval neuroblasts have proven to be an effective model for characterizing the mechanisms regulating stem cell self-renewal (Doe, 2008; Januschke and Gonzalez, 2008).
Drosophila larval neuroblasts undergo repeated asymmetric cell divisions that involve formation of molecularly distinct apical and basal cortical domains, and alignment of the mitotic spindle along the apical/basal polarity axis. The apical cortex contains two protein complexes: the Par complex (Bazooka, Baz; atypical protein kinase C, aPKC; and Partitioning defective-6, Par-6) and the Pins complex (Partner of Inscuteable, Pins; Gαi/o, and Discs large, Dlg). These two complexes are thought to be linked by the protein Inscuteable (Insc) (Doe, 2008; Knoblich, 2008). The differentiation factors Numb, Brain tumor (Brat), and Prospero (Pros) accumulate on the basal surface of neuroblasts; the adaptor protein Miranda (Mira) is required for both Brat and Pros localization (Doe, 2008; Knoblich, 2008). In larval neuroblasts, apical polarity is first established at late G2/early prophase, whereas basal cortical polarity is first detectable at the prophase/metaphase transition (Siller et al., 2006). The apical domain is partitioned into the larger daughter cell which self-renews as a neuroblast, while the basal domain segregates into the smaller daughter cell which has a more restricted developmental potential (Doe, 2008; Knoblich, 2008).
Despite the importance of the apical polarity proteins in regulating neuroblast self-renewal, it remains unknown exactly how the Par and Pins complexes are initially localized to the apical cortex. The conserved polarity protein Cdc42 binds Par-6 and is required for apical localization of Par-6 and aPKC, but cdc42 mutants still localize Baz to the apical cortex, albeit weakly (Atwood et al., 2007). The only mutant reported to abolish Baz apical localization is lkb1 (Bonaccorsi et al., 2007), which encodes a serine/threonine kinase in the Par-4 kinase family; however, the authors only observed weak Baz localization in wild type neuroblasts, so the significance of the mutant phenotype is difficult to interpret.
Here we describe a forward genetic screen that identified a mutant with a reduced number of brain neuroblasts and defects in the earliest steps of generating neuroblast cortical polarity. We mapped the mutant to a region of the third chromosome and used a novel method to identify the mutant lesion, revealing a small deletion in the sgt1 gene. Sgt1 is an evolutionarily conserved protein that directly interacts with Hsp90 using its “CHORD/Sgt1” domain (Lee et al., 2004); it regulates kinetochore assembly in yeast and mammalian cells; cell cycle progression in yeast, mammals, and Drosophila; and pathogen sensing in plant and animal cells (da Silva Correia et al., 2007; Kitagawa et al., 1999; Martins et al., 2009; Mayor et al., 2007; Shirasu, 2009; Steensgaard et al., 2004).
RESULTS
A genetic screen for neuroblast self-renewal mutants identifies sgt1
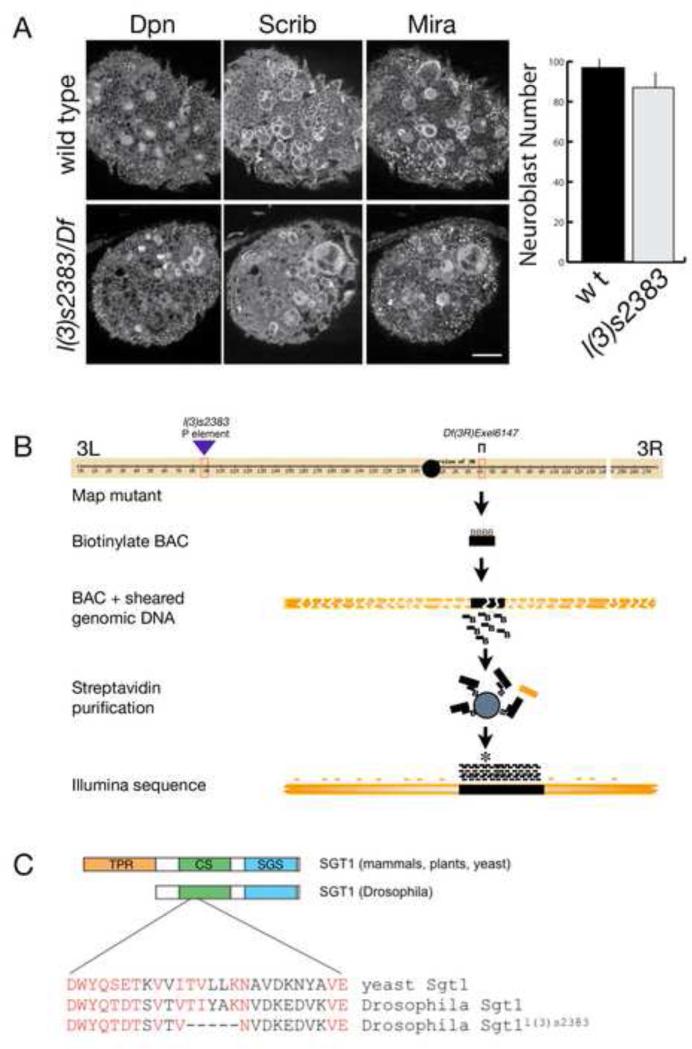
To find potential regulators of neuroblast self-renewal, we screened homozygous P-element induced mutations for altered larval neuroblast numbers, using Miranda (Mira) and Deadpan (Dpn) to identify neuroblasts and Scribble (Scrib) to outline the neuroblast cell cortex and highlight general neuroanatomical landmarks (Lee et al., 2006c) (Figure 1A). One mutant that showed a modest decrease in neuroblast number, and larval/pupal lethality, was l(3)s2383 which contains a P element at 66E1-66E2 on the left arm of the third chromosome (Figure 1A,B). To confirm that the P element insertion caused the phenotype, we assayed the l(3)s2383 chromosome in trans to a deficiency that removed chromosomal region 66E1-66E2. Surprisingly, these larvae were completely normal for neuroblast number and viability, showing that the mutation mapped to elsewhere on the third chromosome. We used deficiency mapping to localize the mutation to the 84F6-84F13 region of the right arm of the third chromosome (Figure 1B). We found that l(3)s2383 / Df(3R)6147 larvae showed the same neuroblast number and larval/pupal lethality phenotypes as l(3)s2383 homozygotes. Phenotypic analysis of overlapping deficiencies in the region allowed us to map the mutation to a 60kb interval, and we confirmed the mutation was in this interval by genomic DNA rescue of the phenotype (data not shown; and see Materials and Methods).
Figure 1. Identification of a mutant in sgt1 that is required for larval brain development.
(A) Wild type and l(3)s2383 / Df(3R)Exel6147 (l(3)s2383 / Df) third instar larval brains stained for the neuroblast markers Miranda (Mira) and Deadpan (Dpn) plus Scribble (Scrib) which decorates the cortex of all cells in the brain. Right: histogram of the average neuroblast number per brain lobe (bar, standard deviation) in each genotype. Note that the mutant brains have fewer neuroblasts and Scrib protein is cytoplasmic. (B) Schematic of the “sequence capture” and deep sequencing strategy used to identify the lesion in the l(3)s2383 mutant chromosome. Although the chromosome was generated in a P element mutagenesis, we found a 15 nucleotide deletion unrelated to the P element insertion that showed the mutant phenotype when transheterozygous to Df(3R)Exel6147. (C) The l(3)s2383 mutation resulted in an in-frame five amino acid deletion in the Drosophila Sgt1 protein, within the N-terminal CS domain (green); the C-terminal Sgt1-specific SGS domain is shown (white).
To identify the DNA lesion responsible for the phenotype, we used a novel “sequence capture” strategy combined with Illumina deep sequencing (Figure 1B) to identify an in frame 15 nucleotide deletion in the first exon of the sgt1 gene, also known as CG9617 (Fig 1C; and see Materials and Methods). We henceforth call this allele sgt1s2383. To confirm that the sgt1s2383 lesion is responsible for the observed neuroblast and viability phenotype, we crossed sgt1s2383 with the known sgt1c01428 mutation (Martins et al., 2009). We found that sgt1s2383 fails to complement sgt1c01428, and that sgt1s2383 / sgt1c01428 larvae have the same phenotype as sgt11s2383 homozygotes. Furthermore, the fact that the phenotype of sgt1s2383 / sgt1c01428 is identical to that of sgt1s2383 / Df(3R)6147 indicated that sgt1s2383 is a strong or null allele for CNS function. The lesion in sgt1s2383 resulted in a five amino acid deletion in the Chord and Sgt1 (CS) domain that mediates Sgt1-Hsp90 protein interactions in other organisms (Zhang et al., 2010). We conclude that Drosophila Sgt1 is required for maintaining normal numbers of larval neuroblasts and for viability.
sgt1 is required for apical protein localization in prophase neuroblasts
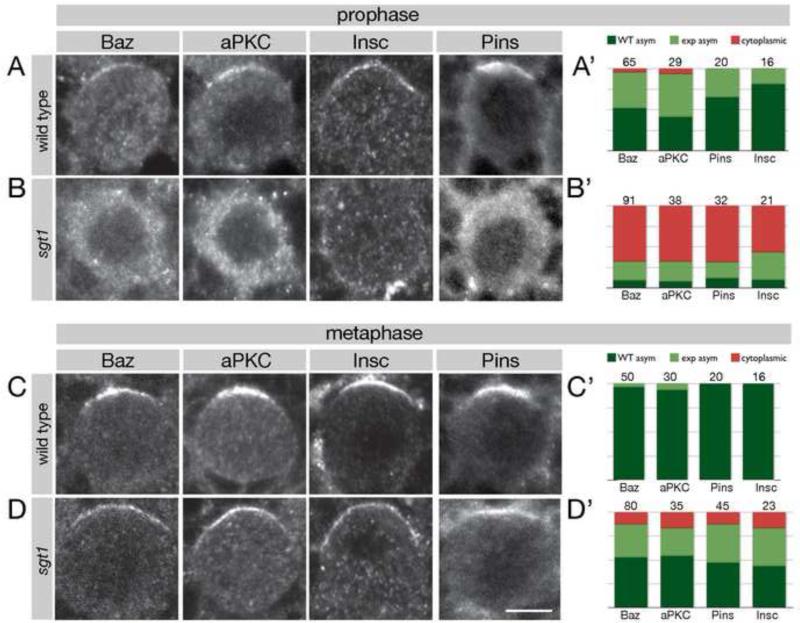
To investigate the cellular origin of the neuroblast depletion phenotype in sgt1 mutants, we assayed neuroblast cell cycle progression and neuroblast cell polarity. Prophase neuroblasts were identified as PH3+ without a bipolar spindle; metaphase neuroblasts were identified as PH3+ with a bipolar spindle. We confirmed a previous report (Martins et al., 2009) that sgt1 mutants have slower cell cycle progression, cytokinesis failure, polyploidy, multiple centrosomes, and malformed mitotic spindles (Supplemental Figure 1 and data not shown). In this paper, we focus on the previously unreported cell polarity aspects of the sgt1 mutant phenotype. First, we assayed neuroblast apical cortical polarity (“apical cortex” is defined as the cortical domain that is partitioned into the sibling neuroblast, whereas “basal cortex” is defined as the domain segregated into the GMC). The first sign of neuroblast asymmetry occurs during late G2 and early prophase when Par complex proteins (e.g. Baz, aPKC), Pins complex proteins (e.g. Pins), and the Par/Pins linker protein Insc are localized to an apical cortical crescent (Figure 2A; quantified in A’). In contrast, sgt1 mutant prophase neuroblasts typically showed cytoplasmic or undetectable localization of these proteins (Figure 2B; quantified in B’). Interestingly, during metaphase we found a substantial rescue of apical cortical polarity (Figure 2C,D; quantified in C’,D’); the basal proteins Miranda and Numb were also fairly normal at metaphase (Supplemental Figure 2). We conclude that Sgt1 is required for establishing apical cortical polarity during prophase, and during metaphase an Sgt1-independent pathway can generate apical/basal cortical protein localization.
Figure 2. sgt1 mutant neuroblasts have defects in establishing apical cortical polarity.
Wild type and sgt1s2383 / sgt1s2383 (sgt1) mutant larval brain neuroblasts stained for the indicated cortical proteins plus α-tubulin (not shown) and phospho-histone H3 (PH3; not shown). Prophase neuroblasts were identified as PH3+ without a bipolar spindle; metaphase neuroblasts were identified as PH3+ with a bipolar spindle. Scale bar, 5 μm.
(A) Wild type prophase neuroblasts show apical enrichment of the Par complex (Baz, aPKC) and Pins complex (Pins) and Insc; quantification in (A’).
(B) sgt1 prophase neuroblasts show a nearly complete loss of apical localized Par complex (Baz, aPKC), Pins complex (Pins) and Insc; quantification in (B’).
(C) Wild type metaphase neuroblasts show apical enrichment of the Par complex (Baz, aPKC) and Pins complex (Pins) and Insc; quantification in (C’).
(D) sgt1 metaphase neuroblasts have recovered substantial apical protein localization compared to their prophase phenotype, although it is not fully back to wild type levels; quantification in (D’).
(A’-D’) Quantification of the phenotypes shown in A-D. Dark green, normal asymmetric; light green, expanded asymmetric; red, cytoplasmic. Number of neuroblasts scored shown in each bar.
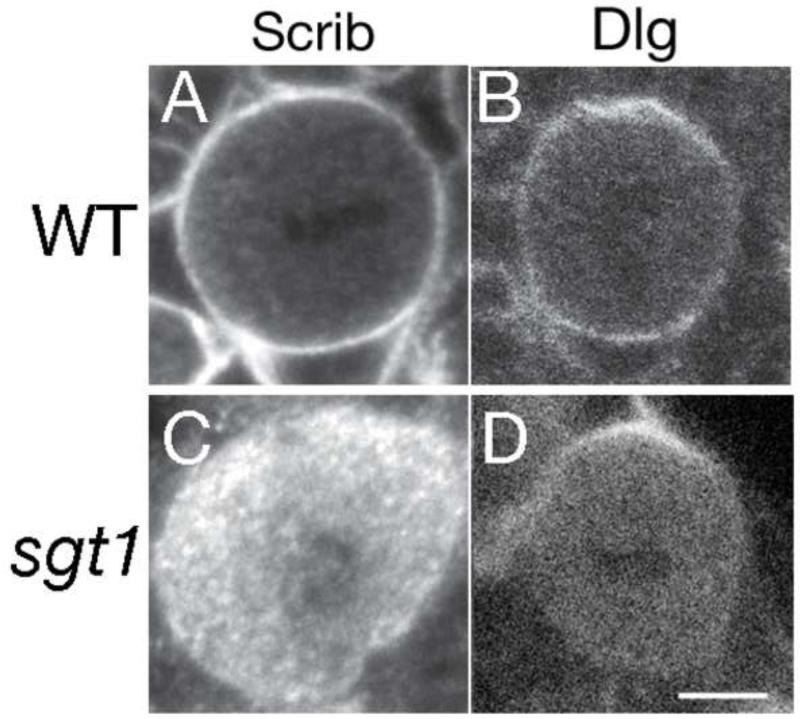
We next analyzed Dlg and Scrib cortical localization in wild type and sgt1 mutant neuroblasts, for two reasons. First, we had observed a complete lack of Scrib cortical localization in our original screen (Figure 1A), and second, Dlg is part of the Pins complex (Siegrist and Doe, 2005) so we wanted to know if it was delocalized in prophase neuroblasts similar to Pins. In wild type interphase and mitotic neuroblasts, Scrib was always uniform cortical (Figure 3A; 100% uniform cortical, n=50), while Dlg was invariably uniform cortical throughout the cell cycle plus enriched with Pins at the apical cortex during mitosis (Figure 3B; 100% uniform cortical n=53, 94% apical enriched, n=34). In contrast, sgt1 mutant neuroblasts showed a strong loss of cortical Scrib localization during metaphase (Figure 3C; 3% cortical, n=75) and interphase (Figure 1A, and data not shown); they also showed a loss of the uniform cortical pool of Dlg protein at interphase and mitosis, with retention of the pool of Dlg asymmetrically co-localized with Pins at the apical cortex at metaphase (Figure 3D; 2% uniform cortical n=45, 83% apical enriched, n=29). We conclude that Sgt1 is essential for targeting Dlg/Scrib to the neuroblast cortex, except where Dlg is co-localized with Pins.
Figure 3. sgt1 mutant neuroblasts have defects in Dlg/Scrib cortical localization.
(A, B) Wild type prophase or metaphase larval neuroblasts; Scrib is uniform cortical, and Dlg is uniform cortical with apical enrichment at metaphase.
(C, D) sgt1s2383 / sgt1s2383 (sgt1) metaphase larval neuroblasts; Scrib is cytoplasmic, and Dlg is mostly cytoplasmic except for persistent apical enrichment at metaphase. Scale bar, 5 μm.
Martins et al. (2009) showed that overexpression of wild type Polo kinase can rescue aspects of the sgt1 neuroblast cell cycle defects; we found that Polo overexpression increases the number of mitotic neuroblasts in both wt and sgt1 mutant brains (Supplemental Figure 1B), but does not rescue neuroblast prophase cortical polarity (7% normal Pins, n=45; 4% normal aPKC, n=27) or Scrib cortical localiation (0% normal Scrib, n=75).
Microtubules induce apical cortical polarity in sgt1 metaphase neuroblasts
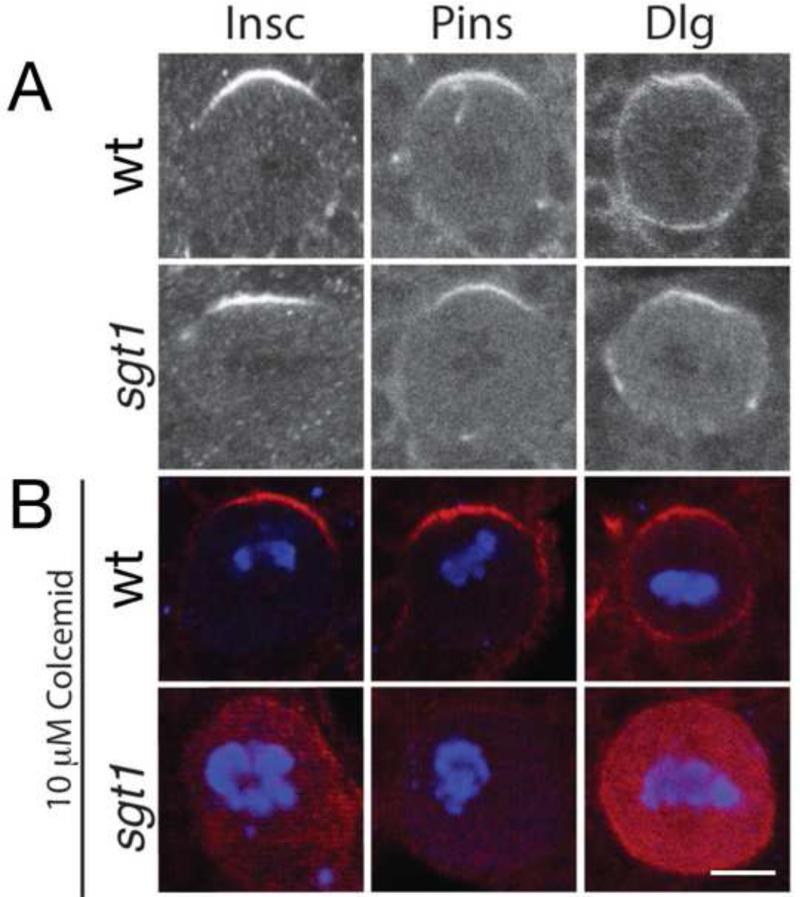
What is the Sgt1-independent pathway that generates Pins and Par complex apical cortical localization in metaphase neuroblasts? We previously defined a microtubule-induced cortical polarity pathway that could induce Pins/Gαi/Dlg cortical polarity (Siegrist and Doe, 2005), so here we test whether this pathway induces Pins and Par complex apical cortical polarity at metaphase in sgt1 mutant neuroblasts. We used the microtubule inhibitor colcemid to abolish spindle microtubules in sgt1 mutant and control neuroblasts. Wild type neuroblasts treated with the colcemid retain apical cortical crescents of Dlg, Pins and Insc (Figure 4B, top row; quantified in figure legend). Strikingly, sgt1 mutant neuroblasts treated with colcemid lack all detectable apical cortical polarity (Figure 4B, bottom row; quantified in figure legend). We conclude that microtubules are required to establish apical cortical polarity in the absence of Sgt1.
Figure 4. Sgt1 and microtubules act in redundant pathways to generate apical protein localization in metaphase neuroblasts.
Wild type and sgt1s2383 / sgt1s2383 (sgt1) metaphase larval neuroblasts stained for the indicated Pins complex proteins and the mitotic marker mitotic marker phospho-histone H3 (not shown in A; blue in B). Scale bar, 5 μm.
(A) Wild type and sgt1 mutant neuroblasts show apical localization of Insc and Pins proteins, and apical enrichment of Dlg protein. Quantification of apical asymmetry: WT (Pins, 100%, n=50; Insc, 100%, n=40; Dlg, 94%, n=34), sgt1 (Pins, 83%, n=35; Insc, 82%, n=23; Dlg, 83%, n=29)
(B) Wild type and sgt1 mutants treated with 10 μM Colcemid to depolymerize microtubules. Wild type neuroblasts show normal apical localization of the Pins complex (Pins, 100%, n=20; Insc, 100%, n=26; Dlg, 100%, n=20), whereas sgt1 mutants lack detectable cortical localization of Pins complex proteins (Pins, 6%, n=31; Insc, 14%, n=22; Dlg, 7%, n=15).
pins sgt1 double mutants neuroblasts lack all detectable cortical polarity and form neuroblast brain “tumors”
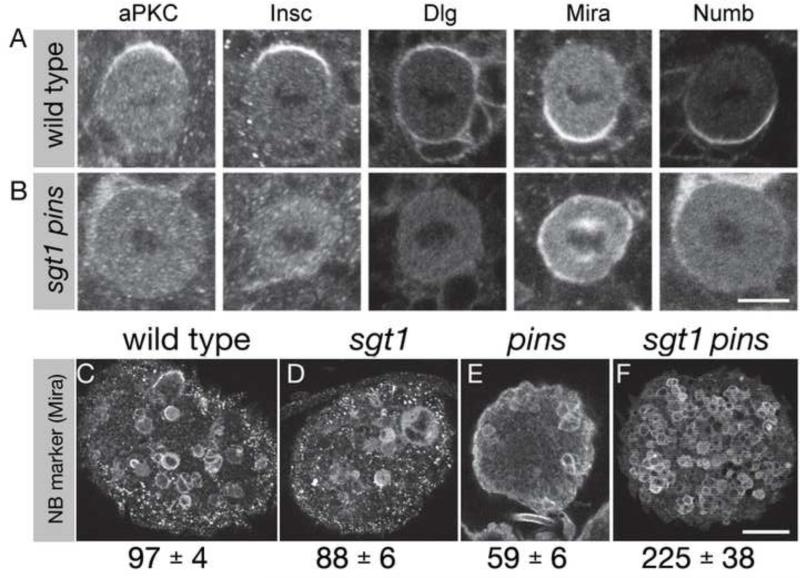
The developmental consequence of a complete loss of neuroblast polarity has never been assayed, and the loss of both Sgt1-dependent and microtubule-dependent cortical polarity pathways is predicted to generate a completely apolar neuroblast (see above). We can not assay sibling cell fate in sgt1- colcemid-treated neuroblasts because they are arrested in mitosis, so we examined a double mutant lacking Sgt1 and the microtubule-induced polarity component Pins. We tested (a) whether sgt1 pins double mutants abolish both Par and Pins complex polarity, and if so (b) what is the fate of such apolar neuroblasts. We found that sgt1 pins double mutant neuroblasts establish little or no apical or basal cortical polarity (Figure 5B and data not shown). The developmental consequences of this loss of polarity are the formation of ectopic neuroblasts throughout the brain (Figure 5F). This is in striking contrast to the sgt1 and pins single mutant brains, which are each smaller with reduced neuroblast numbers (Figure 5D,E). We propose that loss of all cortical polarity in these double mutants results in symmetric neuroblast division to form two neuroblasts and expand the pool of brain neuroblasts (see Discussion).
Figure 5. sgt1 pins double mutants larval brains have ectopic, apolar neuroblasts.
Wild type, sgt1s2383 / sgt1s2383 (sgt1) single mutants, pinsp62 / pinsp62 (pins) single mutants, or sgt1 pins double mutant larval brains stained for the indicated cortical polarity proteins and the mitotic marker phospho-histone H3 (not shown). Scale bar, 5 μm (A-J) and 25 μm (K-N).
(A) Wild type metaphase neuroblasts showing normal cortical polarity; apical up. Quantification of normal asymmetric localization for each protein: aPKC, 100%, n=30; Insc, 100%, n=16; Dlg, 83%, n=29; Mira, 100%, n=20; Numb, 100%, n=20.
(B) sgt1 pins double mutant metaphase neuroblasts lack all cortical polarity. Quantification of asymmetric localization for each protein: aPKC, 6%, n=64; Insc, 5%, n=39; Dlg, 0%, n=45; Mira, 10%, n=79; Numb, 7%, n=41.
(C-F) Third instar larval brains stained for the neuroblast marker Miranda (Mira); the number of central brain neuroblasts in each genotype is given below each panel (average +/- standard deviation). (C) Wild type showing normal neuroblast numbers. (D,E) sgt1 and pins single mutant brains showing reduced neuroblast numbers. (F) sgt1 pins double mutant brain showing a large increase in neuroblast numbers.(Ahmed et al., 2003)
Reducing Hsp90 levels mimics the sgt1 mutant phenotype
To determine the mechanism by which Sgt1 promotes apical cortical polarity and Dlg/Scrib cortical localization, we started by determining if Sgt1 worked together with its evolutionarily-conserved binding partner, Hsp90. Sgt1 directly interacts with Hsp90 via its CS domain in all organisms tested (Catlett and Kaplan, 2006; Lee et al., 2004; Lingelbach and Kaplan, 2004; Nony et al., 2003), and our sgt1s2383 allele generates an Sgt1 protein lacking 5 amino acids within CS domain (Figure 1C). We obtained mutants in hsp83 (which encodes the Hsp90 protein) and examined the larval brain neuroblast phenotype in hsp8313F3/hsp83582 transheterozygotes (Lange et al., 2000). We find that hsp83 mutants have neuroblast phenotypes that are similar to that of sgt1 mutants: loss of apical cortical polarity at prophase (Figure 6B,C; quantified in Figure 6G). However, there is no loss of cortical Scrib in hsp83 mutants (Figure 6C), suggesting that Sgt1 promotes Dlg/Scrib localization by an Hsp90-independent mechanism (see Discussion). The similarity of the sgt1 and hsp83 apical cortical polarity phenotypes, and the fact that the Sgt1s2383 protein has a deletion within the Hsp90-binding CS domain, leads us to hypothesize that Sgt1/Hsp90 act together to promote apical cortical polarity in prophase neuroblasts.
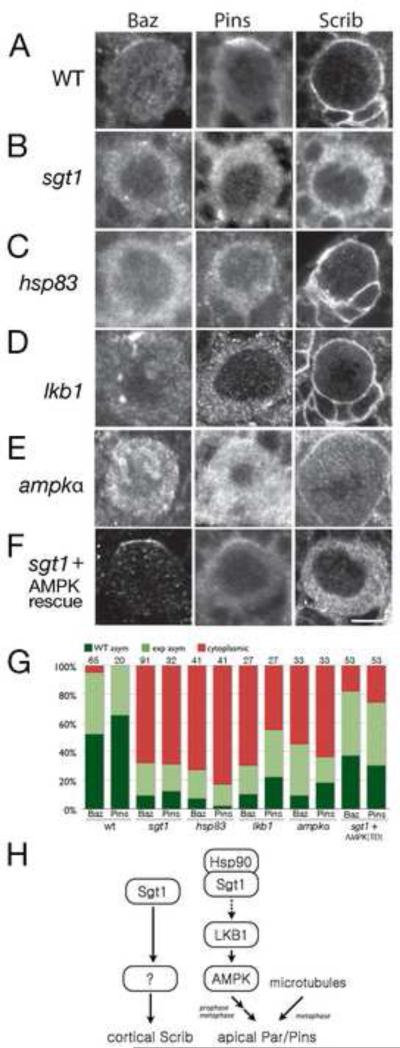
Figure 6. sgt1, hsp83, LKB1, and AMPKαhave similar neuroblast polarity phenotypes.
Prophase larval brain neuroblasts stained for the Par complex protein Bazooka (Baz), the Pins complex protein Pins, and the cortical protein Scribble (Scrib). Quantification in (G). Scale bar, 5 μm.
(A) Wild type. Baz and Pins form apical cortical crescents; Scrib is uniform cortical.
(B) sgt1s2383 / Df(3R)6147 (sgt1). Baz and Pins are delocalized; Scrib is cytoplasmic.
(C) hsp8313F3/hsp83582 (hsp83). Baz and Pins are delocalized; Scrib is uniform cortical.
(D) lkb14A4-2 / lkb14A4-2 (lkb1). Baz and Pins are delocalized; Scrib is uniform cortical.
(E) ampk1 / ampk1 (ampkα). Baz and Pins are delocalized; Scrib is uniform cortical.
(F) sgt1s2383 / Df(3R)6147; tub-gal4 UAS-ampkαTD(sgt1 + AMPK rescue). Baz and Pins form apical cortical crescents; Scrib is cytoplasmic.
(G) Quantification of the Baz and Pins phenotypes for the genotypes listed in A-F. Dark green, normal asymmetric; light green, expanded asymmetric; red, cytoplasmic. Number of neuroblasts scored shown in each bar.
(H) Model for Sgt1 regulation of neuroblast cortical polarity. Sgt1/Hsp90 activate LKB1 which activates AMPK to promote apical Par/Pins complex localization by an unknown mechanism; spindle microtubules provide a redundant pathway generating Par/Pins apical localization at metaphase. Sgt1 acts by an Hsp90/LKB1/AMPK-independent mechanism to promote Scribble (Scrib) uniform cortical localization.
Sgt1 acts via an LKB1/AMPK pathway to generate apical cortical polarity, but not for Scrib cortical localization
Sgt1 has been shown to act through Polo kinase to regulate neuroblast cell cycle progression and cytokinesis (Martins et al., 2009). We confirmed that misexpression of Polo rescues the neuroblast cell cycle phenotypes, but found no rescue of the neuroblast cortical polarity phenotypes (data not shown). Thus, we investigated other downstream effectors of Sgt1 for a role in establishing apical protein localization.
lkb1 mutants are reported to have defects in neuroblast apical polarity that are similar to the sgt1 mutant phenotype (Bonaccorsi et al., 2007). We confirm that lkb1 mutants had a nearly identical neuroblast polarity phenotype as sgt1 mutants, with the striking exception of showing normal Scrib cortical localization (Figure 6D; quantified in Figure 6G). In addition, lkb1 mutants – similar to sgt1 mutants – have slower cell cycle progression, cytokinesis failure, polyploidy, multiple centrosomes, and malformed mitotic spindles (Bonaccorsi et al., 2007)(data not shown). We conclude that Sgt1 and LKB1 are both required to establish apical cortical polarity in neuroblasts.
LKB1 kinase can activate multiple members of the AMP-activated kinase (AMPK) family, such as Par-1, AMPK, Sik, NUAK, and others (Lizcano et al., 2004). Both Par-1 and AMPK activity are required for generating cell polarity in multiple cell types and organisms (Goldstein and Macara, 2007; Williams and Brenman, 2008), so we tested whether Par-1 or AMPK functions downstream of Sgt1/LKB1 in generating apical localization of Par complex proteins in neuroblasts. First we examined the localization and function of Par-1; we observed high levels of the protein in cortex glia that ensheath the neuroblast, but no sign of polarized localization within the neuroblast and no change in neuroblast polarity in par-1 mutant MARCM clones (data not shown). We conclude that loss of Par-1 from larval neuroblasts has no detectable effecton cell polarity.
We next assayed AMPKα mutants, and found that they – like sgt1 and lkb1 mutants – showed a loss of apical protein localization at prophase (Figure 6E; quantified in Figure 6G), slower cell cycle progression, cytokinesis failure, polyploidy, multiple centrosomes, and malformed mitotic spindles (Lee et al., 2007)(data not shown). As with lkb1 mutants, we saw normal cortical localization of Scrib, confirming that Sgt1 acts in an LKB1/AMPK-independent pathway to promote Scrib cortical localization. Importantly, we found that expression of constitutively activated AMPKα (Lee et al., 2007) (tub-gal4 UAS-ampkαTD) substantially rescued the sgt1 mutant phenotype (Figure 6F; quantified in Figure 6G). We conclude that an Sgt1 – Hsp90 – LKB1 – AMPK pathway is used to establish apical cortical polarity in prophase neuroblasts via an unknown link between AMPKα and the apical Par complex, whereas an Sgt1-specific pathway is used to establish cortical Dlg/Scrib localization (Figure 6H).
The AMPK pathway promotes myosin contractility in epithelia, and an activated myosin regulatory light chain, called Sqh in Drosophila (Sqh20E21E), can rescue the ampkα embryonic epithelial polarity phenotype (Lee et al., 2007; Mirouse et al., 2007). Thus, we tested if active Sqh21E (Jordan and Karess, 1997) could rescue the sgt1 neuroblast cortical polarity phenotype, or whether loss of sgt1, lkb1 or ampkα altered cortical myosin activity in larval neuroblasts. First, we found that expression of activated Sqh21E or the non-activatable Sqh20A21A (Jordan and Karess, 1997; Lee et al., 2007) both failed to rescue the sgt1 neuroblast cortical polarity phenotype (Supplementary Figure 3A). Second, we found that all three pathway mutants (sgt1, lkb1, and ampkα) showed an increase in uniform or patchy cortical activated Sqh20E21E (Supplemental Figure 3B-E). In addition, time-lapse imaging of sgt1 mutant larval neuroblasts showed highly abnormal cortical protrusions, misshapen cells, and failure of cytokinesis (Supplemental Figure 3F, Movie 1); these phenotypes are never seen in wild type larval neuroblasts (Cabernard and Doe, 2009; Cabernard et al., 2010). The failure to execute cytokinesis is consistent with a loss of myosin activity, whereas the elevated cortical phospho-Sqh is consistent with an increase in myosin activity; thus all three pathway mutants (sgt1, lkb1, and ampkα) have defects in the regulation of cortical myosin activity. However, the lack of cortical polarity phenotype in neuroblasts homozygous for zipper, which encodes myosin heavy chain (Barros et al., 2003; Peng et al., 2000), and the inability of Sqh21E to rescue the cortical polarity phenotype (this work) suggests that simple loss of myosin activity can not explain the sgt1, lkb1, or ampkα mutant neuroblast polarity defects (Figure 6H).
DISCUSSION
Here we present evidence that the evolutionary-conserved protein Sgt1 acts with Hsp90, LKB1 and AMPK to promote apical localization of the Par and Pins complexes in prophase neuroblasts. We propose that Sgt1/Hsp90 proteins function together based on multiple lines of evidence: (1) they show conserved binding from plants to humans (Catlett and Kaplan, 2006; Lee et al., 2004; Lingelbach and Kaplan, 2004; Nony et al., 2003); (2) our sgt1s2383 mutant results in a five amino acid deletion within the CS domain, which is the Hsp90 binding domain (Catlett and Kaplan, 2006; Zhang et al., 2008); (3) sgt1 and hsp83 have similar cell cycle phenotypes (Supplemental Figure 1)(Lange et al., 2000); and (4) sgt1 and hsp83 have similar neuroblast polarity phenotypes (Figure 6B,C). The Sgt1/Hsp90 complex either stabilizes or activates client proteins (Zuehlke and Johnson, 2010); we suggest that Sgt1 activates LKB1, rather than stabilizing it, because we were unable to rescue the sgt1 mutant phenotype by simply overexpressing wild type LKB1 protein (data not shown). We have not tested for direct interactions between Sgt1 and LKB1 proteins, and thus the mechanism by which Sgt1 activates LKB1 remains unknown.
LKB1 is a “master kinase” that activates at least 13 kinases in the AMPK family (Lizcano et al., 2004). We suggest that LKB1 activates AMPK to promote neuroblast polarity because overexpression of phosphomimetic, activated AMPKα can rescue the lkb1 and sgt1 mutant phenotype (Figure 6F and data not shown). It remains unclear how AMPK activity promotes apical protein localization. An antibody to activated AMPKα (anti-phosphoT385-AMPKα , Mirouse et al., 2007) shows spindle and cytoplasmic staining that is absent in ampkα mutants, and centrosomal staining that persists in AMPKα null mutants, but no sign of asymmetric localization in neuroblasts (data not shown). AMPK activity is thought to directly (Lee et al., 2007) or indirectly (Bultot et al., 2009) activate myosin regulatory light chain to promote epithelial polarity (Lee et al., 2007; Miranda et al., 2010). AMPK is activated by a rise in AMP/ATP levels that occur under energy stress or high metabolism; AMP binds to the γ regulatory subunit of the heterotrimeric complex and results in allosteric activation of the α subunit (Hardie et al., 2007). ampkα mutants grown under energy stress have defects in apical/basal epithelial cell polarity in follicle cells within the ovary (Mirouse et al., 2007). In contrast, AMPKα mutants grown on nutrient rich food still show defects in embryonic epithelial polarity (Lee et al., 2007), neuroblast apical polarity (this work), and visceral muscle contraction (Bland et al., 2010). Larval neuroblasts, embryonic ectoderm, and visceral muscle may have a high metabolic rate, require low basal AMPK activity, or use a different mechanism to activate AMPK than epithelial cells. What are the targets of AMPK signaling for establishing apical cortical polarity in larval neuroblasts? AMPK could directly phosphorylate Baz to destabilize the entire pool of apical proteins, but currently we have no evidence supporting such a direct model. AMPK may act via regulating cortical myosin activity: we have seen clear defects in cortical motility, ectopic patchy activated myosin at the cortex, and failure of cytokinesis in sgt1, lkb1, and ampkα mutants (Movie 1; Supplemental Figure 3). This strongly suggests defects in the regulation of myosin activity, but how or if gain/loss/mispositioning of myosin activity leads to failure to establish apical cortical polarity remains unknown. Lastly, the defects in apical cell polarity seen at prophase could be due to the prometaphase cell cycle delays.
What activates the Sgt1-LKB1-AMPK pathway to promote cell polarity during prophase? In budding yeast, Sgt1 requires phosphorylation on Serine 361 (which is conserved in Drosophila Sgt1) for dimerization and function (Bansal et al., 2009); this residue is conserved in Drosophila Sgt1 but its functional significance is unknown.
Sgt1/Hsp90/LKB1/AMPK are all required for apical Par/Pins complex localization, but Sgt1 must act via a different pathway to promote Dlg/Scrib cortical localization, because only the sgt1 mutant affects Dlg/Scrib localization, and overexpression of activated AMPKα is unable to restore cortical Scrib in sgt1 mutants (Figure 6). The mechanism by which Sgt1 promotes Dlg/Scrib cortical localization is unknown.
We have shown that sgt1 mutants lack Par/Pins apical polarity in prophase neuroblasts, but these proteins are fairly well polarized in metaphase neuroblasts. The rescue of cortical polarity is microtubule dependent, probably occurring via the previously described microtubule-dependent cortical polarity pathway containing Pins, Dlg and Khc-73 (Siegrist and Doe, 2005). The weak polarity defects still observed in sgt1 metaphase neuroblasts may be due to the poor spindle morphology (Supp. Figure 1). The lack of microtubule-induced polarity at prophase, despite a robust microtubule array in prophase neuroblasts, suggests that the microtubule-induced cortical polarity pathway is activated at metaphase. Activation of the pathway could be via expression of the microtubule-binding protein Khc-73; via phosphorylation of Pins, Dlg or Khc-73 by a mitotic kinase like Aurora A (Johnston et al., 2009); or via a yet unknown pathway.
It was somewhat surprising that the sgt1 pins double mutants had increased numbers of brain neuroblasts, because each single mutant had reduced neuroblast numbers (this work)(Lee et al., 2006b). The double mutant phenotype may be due to loss of both Pins and cortical Dlg/Scrib, as the sgt1 pins double mutant phenotype is similar to the dlg pins double mutant phenotype (data not shown). It could also be due to a change in an unknown downstream effector of both Sgt1 and Pins. A not mutually exclusive possibility is that the sgt1 pins double mutant phenotype is due to loss of all Par/Pins cortical polarity. This model is consistent with our observation that sgt1 or pins single mutants retain some neuroblast cortical polarity, whereas the sgt1 pins double mutants lack all known neuroblast cortical polarity. We propose that the apolar double mutant neuroblasts partition cell fate determinants equally to both siblings, and that both siblings frequently assume a neuroblast identity. This is supported by our recent finding that when the neuroblast spindle is aligned orthogonal to a normal apical/basal polarity axis, such that both siblings inherit equal amounts of apical cortical proteins, the siblings always acquire a neuroblast identity (Cabernard and Doe, 2009). Thus, equal partitioning of apical/basal cell fate determinants (in spindle orientation mutants) or failure to establish any cortical polarity (sgt1 pins mutants) may result in neuroblast/neuroblast siblings and an expansion of the neuroblast population.
METHODS
Identification and sequencing of the sgt1s2383 mutant
The sgt1s2383 mutant allele was originally found in a P-element screen of the 3rd chromosome. The P-element mapped to 66E1-2, but deficiency mapping revealed that the lethality and CNS phenotype mapped to Df(3R)6147 at 84F6-F13. To further define the region, rescue constructs were made using gap-repair and ΦC31 mediated transgenesis (Venken et al., 2009). Using the tilling BAC 33N15, we made two overlapping 60kb and 28kb rescue constructs; only the 60kb construct rescued the l(3)s2383 phenotype, narrowing the relevant region to approximately 13 genes. To identify the lesion in this interval, we used a biotin mediated “sequence capture” technique followed by deep sequencing. Genomic DNA from both wild type and mutant larva were prepared for deep sequencing. Using BAC33N15 as a template, biotin incorporated “probes” were created. The biotin labeled probes were individually hybridized with wild type or mutant DNA and purified over streptavidin beads. The genomic DNA was then eluted from the beads, and sequenced using Illumina deep sequencing, revealing a 15 nucleotide in frame deletion within the first exon of sgt1s2383; this allele will be called sgt1 for simplicity. In addition, a P element allele of sgt1 (C01428; from the Harvard Exelexis collection) failed to complement and showed the same phenotype as sgt1s2383.
Fly Stocks
Df(3R)6147 was obtained from the Bloomington stock center. The lkb14A4-2 and lkb14A4-11 mutant stocks (Mirouse et al., 2007) were kindly provided by Daniel St. Johnston (Cambridge, UK); the strong or null ampkα mutants ampk1, ampk2 and the UAS-GFP-lkb1 and UAS-ampkα (T-D) stocks were gifts from Jay Brenman (Chapel Hill, NC); UAS-apkcRNAi was obtained from the Vienna Stock Center. pinsp62 was obtained from Bill Chia (Singapore); hsp8313F3 and hsp83582 were obtained from Howard Lipshitz (Toronto); sqh21E, sqh20A21A, sqh21A were obtained from the Bloomington stock center; UAS-polo was obtained from Claudio Sunkel (Porto, Portugal), and worniu-gal4 UAS-miranda:GFP (Cabernard and Doe, 2009) was used for live imaging. The sgt1s2383 pinsp62 double mutants were generated by recombination. insc-gal4 (aka 1407-gal4), worniu-gal4, and tubulin-gal4 were all used for rescue experiments. Mutant larva were identified by lack of balancer chromosome markers Tubby or GFP.
Antibody staining, drug treatment, and imaging
Third instar larval brains were dissected and fixed as previously described (Lee et al., 2006a). Neuroblast counting and BrdU labeling were done as previously described (Lee et al., 2006a). Dissected brains were washed several times in PBS-T (PBS with 0.1% TritonX-100) and blocked for 1hr in PBS-BT (PBS with 0.1% Triton and 1% BSA). Brain preparations were incubated overnight at 4C with primary antibodies diluted in PBS-BT. All analyses were performed with the mitotic markers rat anti-α-tubulin (1:3000; Serotec, Kidlington, Oxford, UK) or rabbit anti-phospho-histone H3 (1:1000, Upstate, West Grove, PA) and one or more of the following antibodies: guinea pig anti-Bazooka (1:1000, Doe lab), rabbit anti-aPKC (1:1000; Sigma, St. Louis, MO), rabbit anti-Par-6 (1:100; Doe lab), rat anti-Pins (1:500; Doe lab), rabbit anti-Inscuteable (1:500; Bill Chia), rabbit anti-Gαi (1:500; Doe lab), mouse anti-Discs Large (1:500; Developmental Hybridoma Studies Bank [DHSB], Iowa), anti-rabbit Scribble (1:5000; Doe lab), guinea pig anti-Miranda (1:400; Doe lab), rabbit anti-Miranda (1:1000; Doe lab), rabbit anti-phosphoT385AMPKα (1:100; Cell Signaling, Beverly, MA), anti-phospho-Ser19 human MRLC (Cell Signaling, no. 3671 1:100 to detect phospho-Ser22 Sqh (Myosin regulatory light chain), and guinea pig anti-Numb (1:100; Jim Skeath). After 3 washes with PBS-BT the samples were incubated for 2 hours at room temperature with the appropriate secondary antibody (FITC-conjugated IgG, Rhodamine Red-X-conjugated IgG, and Cy5-conjugated IgG; Invitrogen, Eugene, OR). All secondary antibodies were diluted from a 0.5 mg/ml stock solution and used at 1:400. Brains washed in PBT, mounted in VectaShield medium, and examined with a Bio-Rad Radiance 2000 confocal microscope using a 63X 1.4NA oil immersion objective or a Zeiss LSM 700 confocal microscope using a 40X 1.3NA oil immersion objective. Image processing was performed using ImageJ (NIH) and Photoshop (Adobe), and figures were assembled in Illustrator (Adobe).
To inhibit microtubule formation, dissected wild type and sgt1 mutant brains were placed in 10μg/ml Colcemid (Sigma, St. Louis, MO) diluted in Schneider's medium (Sigma, St. Louis, MO) for 1hr. Brains were fixed and antibody stained as described above.
Supplementary Material
Supplemental figure 1. sgt1 mutant neuroblasts have a slow cell cycle, supernumerary centrosomes, and spindle morphology defects.
(A) Wild type or sgt1 mutant brains from larvae exposed to BrdU for two hours (quantified to right). In sgt1 mutants, fewer neuroblasts incorporate BrdU, indicating a slower cell cycle.
(B) Wild type or sgt1 larval brains with and without overexpression of wild type Polo protein (1407-gal4 UAS-polo) stained for the mitotic marker phospho-histoneH3 (PH3); quantified to right. sgt1 mutants show fewer PH3+ neuroblasts; Polo overexpression increases the number of PH3+ neuroblasts.
(C) Larval brain neuroblasts stained for the centrosomal markers Centrosomin (Cnn) and gammatubulin (γtub) plus the neuroblast marker Miranda (Mira). In wild type, virtually all mitotic neuroblasts contain two centrosomes. In sgt1 mutants, many mitotic neuroblasts contain more than two centrosomes.
(D) Larval brain neuroblasts stained for the spindle marker alpha-tubulin (αtub) and the neuroblast marker Miranda (not shown). In wild type, virtually all mitotic neuroblasts contain a bipolar mitotic spindle. In sgt1 mutants, mitotic neuroblasts often contain multi-polar or apolar spindles.
Supplemental figure 2. Basal protein localization in sgt1 mutant metaphase neuroblasts.
Basal localization of Miranda (left) and Numb (right) in wild type and sgt1 mutant metaphase neuroblasts. In sgt1 mutants, Miranda shows normal basal asymmetric localization in the majority of neuroblasts (54%; left) with the remainder showing basal asymmetric localization plus cytoplasmic staining (27%; center), or cytoplasmic staining (20%; right; n=15). Numb typically shows normal basal asymmetric localization (86%, n=22).
Supplemental figure 3. sgt1, LKB1, and AMPKα mutants have abnormal cortical myosin.
(A) Histogram showing that activated myosin regulatory light chain (sqhE21 = sqhE) or non-activatable myosin regulatory light chain (sqhA21 = sqhA; sqhA21A22 = sqhAA) do not rescue the sgt1 neuroblast polarity phenotype. Y axis shows percent mitotic (PH3+) neuroblasts that have normal cortical polarity for the indicated proteins. Number of neuroblasts scored, from left to right in each genotype is 115, 59, 32, 40 (wild type); 171, 73, 44, 77 (sgt1); 24, 24 (sgt1; sqhA); 64, 31, 64, 31 (sgt1; sqhAA); 24, 15, 24, 15 (sgt1; sqhE).
(B-E) Larval brain neuroblasts stained for activated myosin regulatory light chain (antibody recognizes phospho-serine 22 of Spaghetti squash). Representative neuroblast indicated by arrow. (B) Wild type interphase neuroblasts have primarily cytoplasmic staining; (C-E) sgt1 mutant (C), lkb1 mutant (D) and ampkα mutant (E) have primarily cortical staining. Arrow, PH3+ mitotic neuroblasts; arrowhead, PH3- interphase neuroblasts that normally lack cortical activated myosin regulatory light chain. Scale bars, 25 μm.
(F) Still frames from Movie 1 showing cortical blebbing in a larval neuroblast; the cortical marker used is Mira:GFP. Time stamp is in min:sec.
Supplemental movie 1. sgt1 mutants have abnormal cortical contractility.
Wild type neuroblasts expressing Mira:GFP have little or no cortical protrusions during interphase or mitosis (Cabernard and Doe, 2009; Cabernard et al., 2010). In this movie, an sgt1 mutant larval neuroblast expressing Mira:GFP protein exhibits disorganized cortical contractility, with abnormal blebbing and apparent failure of cytokinesis. Time stamp is in min:sec.
Highlights.
We used a novel sequence-capture genomic method to identify sgt1.
sgt1 is required for Bazooka localization in neuroblasts.
sgt1 is required for Scribble localization in neuroblasts.
sgt1 acts at the top of an Hsp90-LKB1-AMPK polarity pathway.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmed A, Chandra S, Magarinos M, Vaessin H. Echinoid mutants exhibit neurogenic phenotypes and show synergistic interactions with the Notch signaling pathway. Development. 2003;130:6295–6304. doi: 10.1242/dev.00796. [DOI] [PubMed] [Google Scholar]
- Atwood Chabu, C., Penkert Doe, Prehoda. Cdc42 acts downstream of Bazooka to regulate neuroblast polarity through Par-6-aPKC. J Cell Sci. 2007 doi: 10.1242/jcs.014902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal P, Mishra A, High A, Abdulle R, Kitagawa K. sgt1 dimerization is negatively regulated by protein kinase CK2-mediated phosphorylation at S361. J Biol Chem. 2009 doi: 10.1074/jbc.M109.012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros CS, Phelps CB, Brand AH. Drosophila nonmuscle myosin II promotes the asymmetric segregation of cell fate determinants by cortical exclusion rather than active transport. Dev Cell. 2003;5:829–840. doi: 10.1016/s1534-5807(03)00359-9. [DOI] [PubMed] [Google Scholar]
- Bland ML, Lee RJ, Magallanes JM, Foskett JK, Birnbaum MJ. AMPK Supports Growth in Drosophila by Regulating Muscle Activity and Nutrient Uptake in the Gut. Dev Biol. 2010 doi: 10.1016/j.ydbio.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaccorsi S, Mottier V, Giansanti MG, Bolkan BJ, Williams B, Goldberg ML, Gatti M. The Drosophila Lkb1 kinase is required for spindle formation and asymmetric neuroblast division. Development. 2007;134:2183–2193. doi: 10.1242/dev.02848. [DOI] [PubMed] [Google Scholar]
- Cabernard C, Doe CQ. Apical/basal spindle orientation is required for neuroblast homeostasis and neuronal differentiation in Drosophila. Dev Cell. 2009;17:134–141. doi: 10.1016/j.devcel.2009.06.009. [DOI] [PubMed] [Google Scholar]
- Cabernard C, Prehoda KE, Doe CQ. A spindle-independent cleavage furrow positioning pathway. Nature. 2010;467:91–94. doi: 10.1038/nature09334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catlett MG, Kaplan KB. sgt1p is a unique co-chaperone that acts as a client adaptor to link Hsp90 to Skp1p. J Biol Chem. 2006;281:33739–33748. doi: 10.1074/jbc.M603847200. [DOI] [PubMed] [Google Scholar]
- da Silva Correia J, Miranda Y, Leonard N, Ulevitch R. sgt1 is essential for Nod1 activation. Proc Natl Acad Sci USA. 2007;104:6764–6769. doi: 10.1073/pnas.0610926104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe C. Neural stem cells: balancing self-renewal with differentiation. Development. 2008;135:1575–1587. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- Goldstein B, Macara IG. The PAR proteins: fundamental players in animal cell polarization. Dev Cell. 2007;13:609–622. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke J, Gonzalez C. Drosophila asymmetric division, polarity and cancer. Oncogene. 2008;27:6994–7002. doi: 10.1038/onc.2008.349. [DOI] [PubMed] [Google Scholar]
- Johnston CA, Hirono K, Prehoda KE, Doe CQ. Identification of an Aurora-A/PinsLINKER/Dlg spindle orientation pathway using induced cell polarity in S2 cells. Cell. 2009;138:1150–1163. doi: 10.1016/j.cell.2009.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P, Karess R. Myosin light chain-activating phosphorylation sites are required for oogenesis in Drosophila. J Cell Biol. 1997;139:1805–1819. doi: 10.1083/jcb.139.7.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa K, Skowyra D, Elledge SJ, Harper JW, Hieter P. sgt1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol Cell. 1999;4:21–33. doi: 10.1016/s1097-2765(00)80184-7. [DOI] [PubMed] [Google Scholar]
- Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Lange BM, Bachi A, Wilm M, González C. Hsp90 is a core centrosomal component and is required at different stages of the centrosome cycle in Drosophila and vertebrates. EMBO J. 2000;19:1252–1262. doi: 10.1093/emboj/19.6.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C-Y, Andersen RO, Cabernard C, Manning L, Tran KD, Lanskey MJ, Bashirullah A, Doe CQ. Drosophila Aurora-A kinase inhibits neuroblast self-renewal by regulating aPKC/Numb cortical polarity and spindle orientation. Genes Dev. 2006a;20:3464–3474. doi: 10.1101/gad.1489406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C-Y, Robinson KJ, Doe CQ. Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature. 2006b;439:594–598. doi: 10.1038/nature04299. [DOI] [PubMed] [Google Scholar]
- Lee C-Y, Wilkinson BD, Siegrist SE, Wharton RP, Doe CQ. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev Cell. 2006c;10:441–449. doi: 10.1016/j.devcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Lee JH, Koh H, Kim M, Kim Y, Lee SY, Karess RE, Lee S-H, Shong M, Kim J-M, Kim J, Chung J. Energy-dependent regulation of cell structure by AMP-activated protein kinase. Nature. 2007;447:1017–1020. doi: 10.1038/nature05828. [DOI] [PubMed] [Google Scholar]
- Lee Y-T, Jacob J, Michowski W, Nowotny M, Kuznicki J, Chazin WJ. Human sgt1 binds HSP90 through the CHORD-sgt1 domain and not the tetratricopeptide repeat domain. J Biol Chem. 2004;279:16511–16517. doi: 10.1074/jbc.M400215200. [DOI] [PubMed] [Google Scholar]
- Lingelbach LB, Kaplan KB. The interaction between sgt1p and Skp1p is regulated by HSP90 chaperones and is required for proper CBF3 assembly. Mol Cell Biol. 2004;24:8938–8950. doi: 10.1128/MCB.24.20.8938-8950.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins T, Maia AF, Steffensen S, Sunkel CE. sgt1, a co-chaperone of Hsp90 stabilizes Polo and is required for centrosome organization. EMBO J. 2009;28:234–247. doi: 10.1038/emboj.2008.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor A, Martinon F, De Smedt T, Petrilli V, Tschopp J. A crucial function of sgt1 and HSP90 in inflammasome activity links mammalian and plant innate immune responses. Nat Immunol. 2007;8:497–503. doi: 10.1038/ni1459. [DOI] [PubMed] [Google Scholar]
- Miranda L, Carpentier S, Platek A, Hussain N, Gueuning M-A, Vertommen D, Ozkan Y, Sid B, Hue L, Courtoy PJ, Rider MH, Horman S. AMP-activated protein kinase induces actin cytoskeleton reorganization in epithelial cells. Biochem Biophys Res Commun. 2010;396:656–661. doi: 10.1016/j.bbrc.2010.04.151. [DOI] [PubMed] [Google Scholar]
- Mirouse V, Swick LL, Kazgan N, St Johnston D, Brenman JE. LKB1 and AMPK maintain epithelial cell polarity under energetic stress. J Cell Biol. 2007;177:387–392. doi: 10.1083/jcb.200702053. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Nony P, Gaude H, Rossel M, Fournier L, Rouault J-P, Billaud M. Stability of the Peutz-Jeghers syndrome kinase LKB1 requires its binding to the molecular chaperones Hsp90/Cdc37. Oncogene. 2003;22:9165–9175. doi: 10.1038/sj.onc.1207179. [DOI] [PubMed] [Google Scholar]
- Peng CY, Manning L, Albertson R, Doe CQ. The tumour-suppressor genes lgl and dlg regulate basal protein targeting in Drosophila neuroblasts. Nature. 2000;408:596–600. doi: 10.1038/35046094. [DOI] [PubMed] [Google Scholar]
- Shirasu K. The HSP90-sgt1 chaperone complex for NLR immune sensors. Annual review of plant biology. 2009;60:139–164. doi: 10.1146/annurev.arplant.59.032607.092906. [DOI] [PubMed] [Google Scholar]
- Siegrist SE, Doe CQ. Microtubule-induced Pins/Galphai cortical polarity in Drosophila neuroblasts. Cell. 2005;123:1323–1335. doi: 10.1016/j.cell.2005.09.043. [DOI] [PubMed] [Google Scholar]
- Steensgaard P, Garrè M, Muradore I, Transidico P, Nigg EA, Kitagawa K, Earnshaw WC, Faretta M, Musacchio A. sgt1 is required for human kinetochore assembly. EMBO Rep. 2004;5:626–631. doi: 10.1038/sj.embor.7400154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K, Carlson J, Schulze K, Pan H, He Y, Spokony R, Wan K, Koriabine M, de Jong P, White K, Bellen H, Hoskins R. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat Methods. 2009 doi: 10.1038/nmeth.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T, Brenman J. LKB1 and AMPK in cell polarity and division. Trends Cell Biol. 2008 doi: 10.1016/j.tcb.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Zhang M, Botër M, Li K, Kadota Y, Panaretou B, Prodromou C, Shirasu K, Pearl LH. Structural and functional coupling of Hsp90- and sgt1-centred multi-protein complexes. EMBO J. 2008;27:2789–2798. doi: 10.1038/emboj.2008.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Kadota Y, Prodromou C, Shirasu K, Pearl LH. Structural basis for assembly of Hsp90-sgt1-CHORD protein complexes: implications for chaperoning of NLR innate immunity receptors. Mol Cell. 2010;39:269–281. doi: 10.1016/j.molcel.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuehlke A, Johnson JL. Hsp90 and co-chaperones twist the functions of diverse client proteins. Biopolymers. 2010;93:211–217. doi: 10.1002/bip.21292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figure 1. sgt1 mutant neuroblasts have a slow cell cycle, supernumerary centrosomes, and spindle morphology defects.
(A) Wild type or sgt1 mutant brains from larvae exposed to BrdU for two hours (quantified to right). In sgt1 mutants, fewer neuroblasts incorporate BrdU, indicating a slower cell cycle.
(B) Wild type or sgt1 larval brains with and without overexpression of wild type Polo protein (1407-gal4 UAS-polo) stained for the mitotic marker phospho-histoneH3 (PH3); quantified to right. sgt1 mutants show fewer PH3+ neuroblasts; Polo overexpression increases the number of PH3+ neuroblasts.
(C) Larval brain neuroblasts stained for the centrosomal markers Centrosomin (Cnn) and gammatubulin (γtub) plus the neuroblast marker Miranda (Mira). In wild type, virtually all mitotic neuroblasts contain two centrosomes. In sgt1 mutants, many mitotic neuroblasts contain more than two centrosomes.
(D) Larval brain neuroblasts stained for the spindle marker alpha-tubulin (αtub) and the neuroblast marker Miranda (not shown). In wild type, virtually all mitotic neuroblasts contain a bipolar mitotic spindle. In sgt1 mutants, mitotic neuroblasts often contain multi-polar or apolar spindles.
Supplemental figure 2. Basal protein localization in sgt1 mutant metaphase neuroblasts.
Basal localization of Miranda (left) and Numb (right) in wild type and sgt1 mutant metaphase neuroblasts. In sgt1 mutants, Miranda shows normal basal asymmetric localization in the majority of neuroblasts (54%; left) with the remainder showing basal asymmetric localization plus cytoplasmic staining (27%; center), or cytoplasmic staining (20%; right; n=15). Numb typically shows normal basal asymmetric localization (86%, n=22).
Supplemental figure 3. sgt1, LKB1, and AMPKα mutants have abnormal cortical myosin.
(A) Histogram showing that activated myosin regulatory light chain (sqhE21 = sqhE) or non-activatable myosin regulatory light chain (sqhA21 = sqhA; sqhA21A22 = sqhAA) do not rescue the sgt1 neuroblast polarity phenotype. Y axis shows percent mitotic (PH3+) neuroblasts that have normal cortical polarity for the indicated proteins. Number of neuroblasts scored, from left to right in each genotype is 115, 59, 32, 40 (wild type); 171, 73, 44, 77 (sgt1); 24, 24 (sgt1; sqhA); 64, 31, 64, 31 (sgt1; sqhAA); 24, 15, 24, 15 (sgt1; sqhE).
(B-E) Larval brain neuroblasts stained for activated myosin regulatory light chain (antibody recognizes phospho-serine 22 of Spaghetti squash). Representative neuroblast indicated by arrow. (B) Wild type interphase neuroblasts have primarily cytoplasmic staining; (C-E) sgt1 mutant (C), lkb1 mutant (D) and ampkα mutant (E) have primarily cortical staining. Arrow, PH3+ mitotic neuroblasts; arrowhead, PH3- interphase neuroblasts that normally lack cortical activated myosin regulatory light chain. Scale bars, 25 μm.
(F) Still frames from Movie 1 showing cortical blebbing in a larval neuroblast; the cortical marker used is Mira:GFP. Time stamp is in min:sec.
Supplemental movie 1. sgt1 mutants have abnormal cortical contractility.
Wild type neuroblasts expressing Mira:GFP have little or no cortical protrusions during interphase or mitosis (Cabernard and Doe, 2009; Cabernard et al., 2010). In this movie, an sgt1 mutant larval neuroblast expressing Mira:GFP protein exhibits disorganized cortical contractility, with abnormal blebbing and apparent failure of cytokinesis. Time stamp is in min:sec.