Abstract
Neurodevelopmental disorders can be caused by many different genetic abnormalities that are individually rare but collectively common. Specific genetic causes, including certain copy number variants and single-gene mutations, are shared among disorders that are thought to be clinically distinct. This evidence of variability in the clinical manifestations of individual genetic variants and sharing of genetic causes among clinically distinct brain disorders is consistent with the concept of developmental brain dysfunction, a term we use to describe the abnormal brain function underlying a group of neurodevelopmental and neuropsychiatric disorders and to encompass a subset of various clinical diagnoses. Although many pathogenic genetic variants are currently thought to be variably penetrant, we hypothesise that when disorders encompassed by developmental brain dysfunction are considered as a group, the penetrance will approach 100%. The penetrance is also predicted to approach 100% when the phenotype being considered is a specific trait, such as intelligence or autistic-like social impairment, and the trait could be assessed using a continuous, quantitative measure to compare probands with non-carrier family members rather than a qualitative, dichotomous trait and comparing probands with the healthy population.
Introduction
Neurodevelopmental disorders encompass a highly heterogeneous group of diseases characterised by impairments in cognition, communication, behaviour, and motor functioning as a result of atypical brain development. In the proposed framework for the fifth edition of the Diagnostic and Statistical Manual of Mental Disorders (DSM-5), the category of neurodevelopmental disorders includes intellectual developmental disorders, communication disorders, autism spectrum disorder, attention deficit hyperactivity disorder (ADHD), specific learning disorder, and motor disorders.1 Neurodevelopmental disorders, also extends to include neuropsychiatric disorders, such as schizophrenia and bipolar disorder, and to conditions outside of the realm of the DSM-5, such as cerebral palsy and epilepsy.2–4 Categorical clinical diagnoses are determined by the pattern and severity of impairments resulting from the underlying brain dysfunction and modifying influences, such as beneficial and deleterious experiences.2,5 However, there is substantial clinical heterogeneity, co-occurrence of symptoms and syndromes, and diagnostic overlap among neurodevelopmental disorders that are empirically defined and categorically classified as independent disorders without respect to the validity of the biological construct.2
Epidemiological studies show that co-occurrence of neurodevelopmental disorders is the rule rather than the exception; essentially, all neurodevelopmental disorders coexist with other neurodevelopmental disorders much more commonly than would be expected by chance.6 For example, up to 50% of individuals with ADHD present with movement difficulties consistent with developmental coordination disorder,7 10–40% have affective disorders, and 20–30% have tic disorders.8 Twin studies found that reading disorder, mathematics disorder, and ADHD are familial and heritable, and that the cause of co-occurrence of reading and mathematics disorders (28–64%), reading disorder and ADHD (10–40%), and mathematics disorder and ADHD (12–36%) is primarily explained by common genetic factors.9 There is substantial overlap at the diagnostic, neuropsychological, and aetiological levels among speech-sound disorders, language impairment, and reading disorders.10 Cerebral palsy, although defined by impairment in movement and posture, is associated with high rates of intellectual disability, learning disabilities, speech and language disorders, ADHD, autism spectrum disorders, epilepsy, visual impairment, and hearing impairment.11,12 Epilepsy is strongly associated not only with developmental disabilities such as cerebral palsy, intellectual disability, and autism spectrum disorders, but also with psychiatric disorders including major depression, bipolar disorder, and schizophrenia.3 Although the definition of psychiatric disorders varies across epidemiological studies, most include ADHD and other disruptive behaviour disorders in addition to mood disorders, psychoses, and other DSM diagnoses. Based on this approach, psychiatric disorders are present in 30–50% of children and adolescents with intellectual disability, with a relative risk of 2·8–4·5.13 However, coexisting psychiatric disorders are common even among individuals with neurodevelopmental disorders who do not have intellectual disability. For example, approximately 50% of individuals with high-functioning autism spectrum disorder meet criteria for at least one psychiatric disorder.14 Epidemiological data suggest that rather than being considered as causally and pathophysiologically distinct, neurodevelopmental disorders should be thought of as different patterns of symptoms or impairments of a common underlying neurodevelopmental continuum. The aim of our article is twofold: to propose the concept of developmental brain dysfunction as the common denominator underlying neurodevelopmental and neuropsychiatric disorders based on epidemiological and genetic data; and to highlight the importance of quantitative trait analyses to study families with individuals affected by neurodevelopmental disorders.
Historical perspective
The recognition of co-occurrence of multiple neurodevelopmental disorders in individuals and within families is not new. As early as the mid-19th century, epilepsy was reported to often be inherited with a range of other disorders, including neuropsychiatric diseases, intellectual impairment, and movement disorders. This inherited tendency, or diathesis, which could manifest differently in members or generations of the same family, became known as the neurological taint.3 Although the recognition that a variety of distinct clinical disorders probably shared a common, heritable aetiologic factor was remarkable, synthesis of this concept with the notions of degeneration and atavism contributed to the idea that this inherited tendency resulted in progressive mental, physical, and moral deterioration over generations, which unfortunately was adopted by the eugenics movement in the early 20th century and used as a rationale for segregation, compulsory sterilisation, and genocide.
A century later, in the early 1960s, the terms minimal brain dysfunction in the USA, and minimal cerebral dysfunction in the UK, were introduced to describe formes frustes of well known chronic disabilities of childhood, as a way to acknowledge the common overlap among these disorders seen in clinical practice and to emphasise the role of the brain (organicity) in their aetiology, as opposed to the prevailing psychogenicity models.15 Minimal brain dysfunction and minimal cerebral dysfunction encompassed disturbances of perception; conceptualisation; language; memory; and control of attention, impulse, and motor function. These disturbances are now encompassed within the following diagnostic categories: specific learning disabilities, language disorders, ADHD, and developmental coordination disorder. Minimal brain or cerebral dysfunction did not include the more severe disorders such as intellectual disability, cerebral palsy, epilepsy, autism spectrum disorders, and the brain damage behaviour syndrome described by Strauss and colleagues in the 1940s.16 The word minimal did not refer to a necessarily minor impairment, but rather to brain involvement that was not readily and grossly demonstrable.17
In the 1960s and 1970s—building on the influences of Heinz Werner and others—Zigler and colleagues18 found that in children whose intellectual disability could not be definitively traced to organic causes (referred to as the cultural-familial form of intellectual disability), development followed sequences that were qualitatively similar to typically developing individuals, differing only in the rate and developmental endpoint achieved.18 In the late 1970s and 1980s, this model of developmental continuity was relevant in understanding intellectual disability with known causes (organic intellectual disability)19–21 and recognised the importance of genetic background, particularly parental cognitive abilities, and the effect of specific genetic abnormalities on patterns of cognition and behaviour.
In the 1970s to 1980s, influenced by the work of Alfred Strauss on brain damage in childhood and by other advances in developmental psychology, Capute and colleagues22,23 set out a range of clinical neurodevelopmental disability syndromes of varying severity. This spectrum included the disorders subsumed under minimal brain dysfunction or minimal cerebral dysfunction and the more severe disorders that affect primarily the cognitive, motor, or neurobehavioural streams of development (eg, intellectual disability, cerebral palsy, and autism spectrum disorders, respectively), all representing characteristics of under lying brain dysfunction. Capute and colleagues22,23 also emphasised the continuum across streams of development, such that careful examination of an individual with one disability typically showed additional impairments in the other functional domains. According to this model, the developmental profile of an individual can be thought of as an iceberg, with one or more visible tips representing the defining features of the primary diagnoses, and the submerged portion representing the larger range of characteristics of underlying brain dysfunction.22,23 In 2001, Gilger and Kaplan24 proposed the term atypical brain development to describe developmental variation of brain function and to account for the high comorbidity and causative and neuroanatomic variability among the disorders previously encompassed by minimal brain dysfunction or minimal cerebral dysfunction, and to account for phenomena such as exceptional skills; however, this term has not been widely used.24,25 In 2010, Gillberg25 proposed the acronym ESSENCE to refer to Early Symptomatic Syndromes Eliciting Neurodevelopmental Clinical Examinations, an umbrella term describing the coexistence in preschoolers of significant impairment in multiple areas of functioning, including general development, speech and language, reciprocal social interaction, and motor coordination as well as problems with attention, activity and impulse regulation, mood, sleep, and feeding that encompassed many clinical diagnoses.25 He also emphasised the importance of identifying and treating the various specific impairments rather than compartmentalising syndrome diagnoses and systems of care.
Shared genetic causes for neurodevelopmental disorders
Recent studies of large cohorts of individuals (around 30 000) with a broad range of neurodevelopmental disorders provide strong evidence for several rare genetic causes for this group of disorders; they also show a positive association between the burden of genetic insult, quantified by the number and size of copy number variants, and the severity of the phenotypes.26–28 Furthermore, studies using exome sequencing for gene discovery suggest that hundreds of genes may be associated with neurodevelopmental disorders such as autism spectrum disorders,29,30 intellectual disability,31 and schizophrenia.32 In addition, it has also been recognised that multiple genetic causes are shared among a number of apparently different neurodevelopmental disorders. Genome-wide copy number variant analyses have found variable phenotype expressivity for several recurrent microdeletions and microduplications, arising through nonallelic homologous recombination between highly homologous segmental duplications—ie, the same copy number variant can be associated with several apparently different neurodevelopmental disorders (table 1). Variable expressivity is not limited to copy number variants, which typically contain multiple genes, but also occurs for single gene variants. Like copy number variants, functional mutations in the same gene have been reported in different individuals with seemingly distinct clinical presentations (table 2). Future research will be necessary to clarify whether variable expressivity to the degree that a copy number variant or single gene mutation can result in widely disparate neurodevelopmental disorders is the rule or the exception.
Table 1.
Variable expressivity in selected microdeletion syndromes
| Frequency in clinical cohorts* | Intellectual disability or developmental delay | Autism spectrum disorder | Schizophrenia | Epilepsy | |
|---|---|---|---|---|---|
| 22q11.2 | 1 in 167 | ✓ | ✓ | ✓ | ✓ |
| 16p11.2 | 1 in 241 | ✓ | ✓ | ·· | ✓ |
| 1q21.1 | 1 in 309 | ✓ | ✓ | ✓ | ✓ |
| 15q13.2-q13.3 | 1 in 358 | ✓ | ✓ | ✓ | ✓ |
| 7q11.23 | 1 in 415 | ✓ | ✓ | ·· | ✓ |
| 15q11.2-q13 | 1 in 553 | ✓ | ✓ | ✓ | ✓ |
| 17q21.31 | 1 in 700 | ✓ | ✓ | ·· | ✓ |
| 16p13.11 | 1 in 788 | ✓ | ✓ | ✓ | ✓ |
| 17q12 | 1 in 985 | ✓ | ✓ | ✓ | ✓ |
| 17p11.2 | 1 in 985 | ✓ | ✓ | ·· | ✓ |
| 8p23.1 | 1 in 1854 | ✓ | ✓ | ·· | ✓ |
| 5q35 | 1 in 1970 | ✓ | ✓ | ·· | ✓ |
| 3q29 | 1 in 2101 | ✓ | ✓ | ✓ | ·· |
Frequency in individuals referred for chromosomal microarray testing. Common indications for testing include neurodevelopmental disorders and multiple congenital anomalies.33
Table 2.
Variable expressivity in selected single gene mutations
| Intellectual disability or developmental delay | Autism spectrum disorder | Schizophrenia | Epilepsy | |
|---|---|---|---|---|
| A2BP1 | ✓ | ✓ | ✓ | ✓ |
| AUTS2 | ✓ | ✓ | ·· | ✓ |
| CACNA1C | ✓ | ✓ | ✓ | ✓ |
| CASK | ✓ | ·· | ✓ | ✓ |
| CDKL5 | ✓ | ✓ | ·· | ✓ |
| CNTNAP2 | ✓ | ✓ | ✓ | ✓ |
| DISC1 | ✓ | ✓ | ✓ | ·· |
| EHMT1 | ✓ | ✓ | ✓ | ✓ |
| FMR1 | ✓ | ✓ | ✓ | ✓ |
| FOXP1 | ✓ | ✓ | ·· | ✓ |
| FOXP2 | ✓ | ✓ | ✓ | ·· |
| GRIN2B | ✓ | ✓ | ✓ | ·· |
| MBD5 | ✓ | ✓ | ·· | ✓ |
| MECP2 | ✓ | ✓ | ✓ | ✓ |
| NRXN1 | ✓ | ✓ | ✓ | ✓ |
| PCDH19 | ✓ | ✓ | ✓ | ✓ |
| PTEN | ✓ | ✓ | ·· | ✓ |
| SCN1A | ✓ | ✓ | ·· | ✓ |
| SCN2A | ✓ | ✓ | ·· | ✓ |
| SHANK3 | ✓ | ✓ | ✓ | ·· |
| TCF4 | ✓ | ·· | ✓ | ✓ |
| TSC1 | ✓ | ✓ | ·· | ✓ |
| TSC2 | ✓ | ✓ | ·· | ✓ |
Variable expressivity of copy number and single-gene mutations
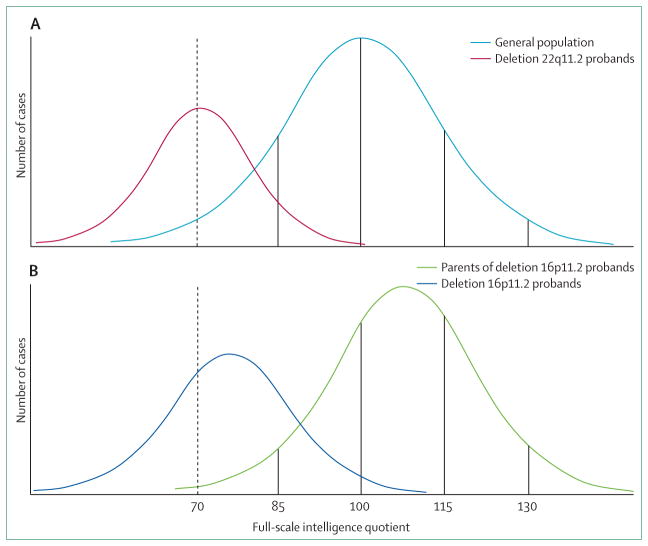
Among all deletion or duplication copy number variants detected during clinical genetic testing of children with unexplained developmental disabilities, the most common is deletion 22q11.2 (DiGeorge and velocardiofacial syndromes), with an incidence of one in 4000 livebirths. This deletion is also one of the best characterised copy number variants in terms of phenotypic heterogeneity. Characteristics of individuals with this deletion include variable degrees of intellectual disability and developmental delay, mild facial dysmorphism, conotruncal cardiac anomalies, cleft palate or velopharyngeal incompetence, hypo calcaemia, immunodeficiency, and thymic aplasia or hypoplasia. Neuropsychiatric disorders such as autism spectrum disorders, schizophrenia, bipolar disorder, ADHD, anxiety, and depression are commonly reported in people with this deletion and, notably, they are variable in severity.34 The mean full-scale intelligence quotient score in individuals with deletion 22q11.2 is 71·2 (SD 12·8), which results in a Gaussian or bell-shaped curve shifted 2 SD to the left of the healthy population distribution mean of 100 (SD 15; figure 1A).35,36 When an individual is diagnosed with deletion 22q11.2 and parental studies are performed, the same deletion might be identified in one of the parents and interpreted as incomplete penetrance if the parent has not independently come to medical attention.34 However, detailed examination commonly shows subtle subclinical features of the syndrome in the carrier parent, which is consistent with variable expressivity.
Figure 1. Cognitive effects of copy number variant syndromes in full-scale intelligence quotient scores.
(A) The intelligence quotient (IQ) distribution curve in individuals with deletion 22q11.2 (red line) is shifted 2 SD to the left of the IQ distribution in the general population (mean 100; SD: 15; light blue line).35 (B) Individuals with deletion 16p11.2 have a mean IQ of 76·1 (dark blue line), which is significantly lower than the mean IQ of their non-carrier first-degree relatives (108·3, green line).36 The higher IQ of first-degree relatives compared to the general population was previously discussed by Zufferey and colleagues36 as likely due to ascertainment bias. For both copy number variant syndromes, many deletion carriers have IQ scores within the normal range (>70); this is often referred to as incomplete penetrance when cognitive function is viewed as a qualitative, dichotomous trait (normal intelligence vs intellectual disability) based on a cutoff of 70 points of IQ (dotted line), but may be better interpreted as variable expressivity of a continuous, quantitative trait.
Deletion 16p11.2 is the second most common pathogenic copy number variant among individuals with neurodevelopmental disorders and can show substantial clinical variability.26,27 This deletion was first identified in individuals with autism spectrum disorders or intellectual disability and was subsequently associated with macrocephaly, obesity, seizures, speech and motor delays, congenital anomalies, and paroxysmal kinesigenic dyskinesia.37–41 Deletion carriers show substantial clinical heterogeneity, including apparently healthy individuals, an observation that is usually interpreted as evidence of incomplete penetrance. A collaboration between two research groups (the 16p11.2 European Consortium and the Simons Variation in Individuals Project consortium) recently assessed 285 individuals with the recurrent approximately 600 kb 16p11.2 deletion and obtained detailed cognitive, psychiatric, and behavioural evaluations in 71 patients and 68 non-carrier controls from the same families.36 Cognitive assessment found a mean full-scale intelligence quotient of 76·1 (SD 16·4) in deletion carriers, and a mean full-scale intelligence quotient of 108·3 in their first-degree relatives who did not have the deletion. Mean verbal intelligence quotient was lower (74, SD 17·5) than mean non-verbal intelligence quotient (83·3, SD 18·0) in deletion carriers. Although only 20% of individuals met criteria for intellectual disability (full-scale intelligence quotient ≤70 and concurrent deficits in adaptive functioning), when compared with unaffected intrafamilial controls, full-scale intelligence quotient was 2 SD lower in deletion carriers (figure 1B). Moreover, most individuals (59 [84·3%] of 70) had neurodevelopmental diagnoses, including autism spectrum disorders, ADHD, anxiety disorders, mood disorders, gross motor delay, and epilepsy.
Additional evidence for recurrent copy number variants with variable expressivity comes from deletion 1q21.1, the third most frequently observed pathogenic copy number variant in paediatric clinical populations.26,27 This deletion was initially described in patients with congenital heart defects,42 and then identified in individuals with schizophrenia43,44 and in children with disorders with variable clinical presentations including intellectual disability, autism spectrum disorders, ADHD, seizures, microcephaly, and mild dysmorphic features.45–47 Deletion 15q13.3 is associated with idiopathic generalised epilepsy, intellectual disability, autism spectrum disorders, schizophrenia, ADHD, bipolar disorder, anxiety and mood disorders, and dysmorphic features.43,44,48–50 Deletion 17q12 has been reported in individuals with autism spectrum disorders, intellectual disability, schizophrenia, bipolar disorder, structural brain malformations, macrocephaly, renal cysts and diabetes syndrome (caused by haploinsufficiency of HNF1B, one of the 15 genes included in the deleted region) and Müllerian aplasia.51–53
Like variably expressed copy number variants, there is increasing evidence from sequencing studies showing that pathogenic sequence variants in the same gene might be associated with various neurodevelopmental phenotypes (table 2). For example, mutations in CNTNAP2, the gene encoding contactin-associated protein-like 2, were initially described in a family with three individuals with Tourette’s syndrome and obsessive-compulsive disorder.54 Subsequently, a homozygous mutation in this gene was reported in Old Order Amish children with cortical dysplasia, focal epilepsy, relative macrocephaly, language regression, hyperactivity, impulsive or aggressive behaviour, and intellectual disability.55 Furthermore, mutations in CNTNAP2 are also associated with schizophrenia,56 autism spectrum disorders,57 and Pitt-Hopkins-like syndrome (intellectual disability, seizures, hyperbreathing pattern).58
Structural and sequence mutations in NRXN1 (the gene that encodes neurexin 1, a presynaptic protein required for effective neurotransmission) are associated with many neurodevelopmental, psychiatric, and neurological phenotypes, including developmental delay or intellectual disability, autism spectrum disorders, schizophrenia, seizures, speech and language disorders, and hypotonia.59–62 Likewise, mutations in PCDH19, which encodes protocadherin 19, a cell-adhesion molecule primarily expressed in the brain, were first described in seven families with X-linked epilepsy and intellectual disability limited to women,63 then reported in 11 unrelated female patients with sporadic infantile epileptic encephalopathy resembling Dravet syndrome,64 and subsequently associated with a range of neuropsychiatric disorders including autism spectrum disorder, depression, panic attacks, aggressive or impulsive behaviour, hyperactivity, and anxiety.65 Similarly, functional sequence variants in several other genes, including DISC1, GRIN2A, and GRIN2B, can result in variable neurodevelopmental phenotypes (table 2).66,67
Developmental brain dysfunction: a conceptual framework
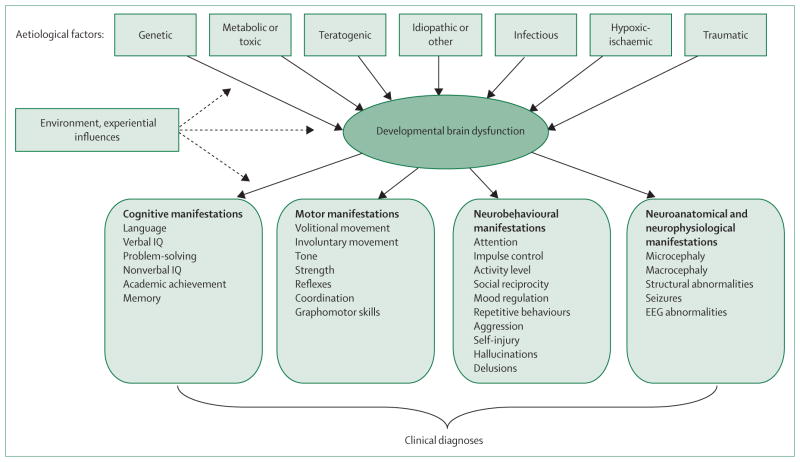
We have previously used the term developmental brain dysfunction to describe the conceptual abnormality of brain function underlying Capute’s spectrum and continuum of developmental disabilities.5,22,23 Expanding on this, we propose that developmental brain dysfunction results in clinical manifestations that include the less severe disorders once encompassed by minimal brain dysfunction or minimal cerebral dysfunction (eg, learning disabilities, language disorders, developmental coordination disorder, and ADHD), the more severe classic neurodevelopmental disabilities (eg, intellectual disability, cerebral palsy, and autism spectrum disorders), and also at least a subset of neuropsychiatric disorders that were regarded as part of the neurological taint more than 150 years ago (eg, schizophrenia and possibly major affective disorders). Developmental brain dysfunction, whether genetic or caused by an insult to the developing central nervous system—such as exposure to a teratogen, trauma, infection, severe nutritional deficiency, or hypoxia-ischaemia—is typically manifested as impairments in cognitive, neuromotor, or neurobehavioural functioning and, in some cases, observable anatomic or neurophysiological findings (figure 2). An individual’s clinical diagnosis is determined by the specific pattern of impairments, but the common denominator is developmental brain dysfunction. This is a broad conceptual categorisation, and not a final diagnosis, since categorical diagnoses and specific impairments must be identified to guide treatment.
Figure 2. Model of developmental brain dysfunction.
Adapted from Myers SM.5
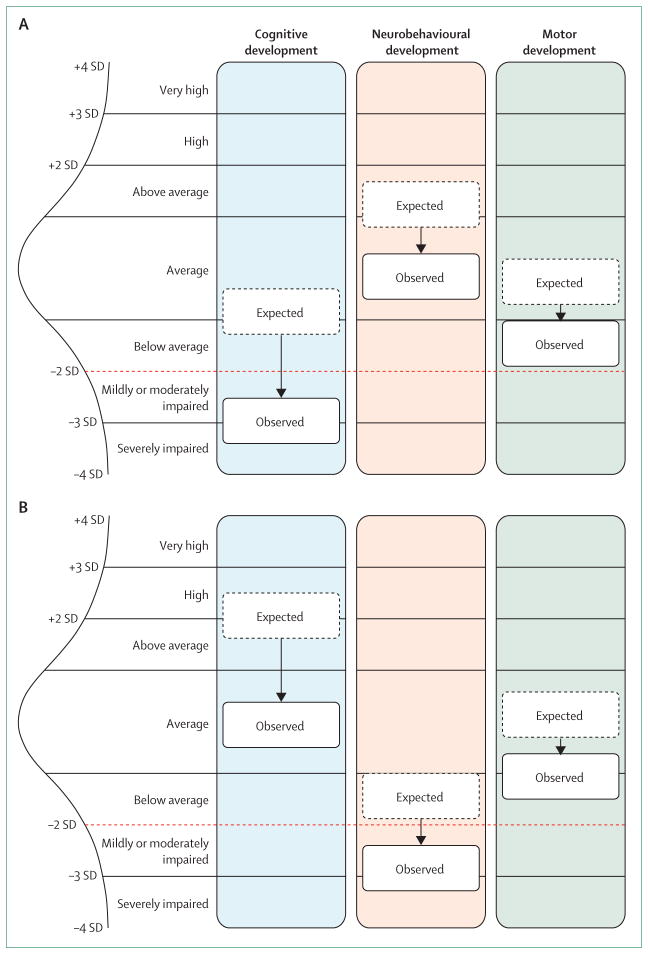
The developmental brain dysfunction model predicts that a substantial subset of various categorical neurodevelopmental and neuropsychiatric diagnoses share similar risk factors and commonly coexist, which is supported by the epidemiology of these disorders. This model also predicts that each particular cause can manifest as a spectrum of impairments of varying severity. The notable variation in the phenotypic expression of recurrent copy number variants and single gene mutations supports the notion of combining a number of neurodevelopmental and neuropsychiatric phenotypes together under the term developmental brain dysfunction. In cases with deletion 16p11.2 or deletion 22q11.2, evidence suggests the impact of the deleterious genetic variant is applied to the affected individual’s expected performance level based on genetic background, resulting in standardised cognitive (intelligence quotient) scores that are substantially lower than expected. Other copy number variants, single gene mutations, and non-genetic insults to the developing brain can also be associated with cognitive, neurobehavioural, and neuromotor deleterious impacts. However, the degree of deleterious effects can vary among the three streams of development—cognitive, neurobehavioural, and motor—and among the components of an individual stream. The same deleterious impact might result in different specific impairments reaching clinical diagnostic threshold in different individuals, depending on the profile of genetic background from which the deleterious impact is extracted (figure 3). It might be possible to estimate the genetic potential, or expectation, profile using parental and sibling profiles as a proxy.
Figure 3. The effect, or deleterious impact, of genetic or other insults on an individual’s neurodevelopmental profile across cognitive, neurobehavioural, and motor streams of development.
In these examples, the actual observed profile of abilities in probands (black solid lines) shows the deleterious effect of a copy number variant on the expected profile based on genetic background (dotted lines). The effect of a particular copy number variant might be different in each stream of development and is arbitrarily represented in these examples as a 2, 1·5, and 1 SD deleterious impact in the cognitive, neurobehavioural, and motor streams, respectively. The red dotted line represents the diagnostic threshold (depicted here as 2 SD below the mean). (A) In this example, the deleterious effect of the copy number variant on quantitative cognitive traits (eg, intelligence quotient) results in reaching the threshold for a diagnosis of intellectual disability, whereas neurobehavioural and motor features do not fall within the clinically impaired range. (B) In this example, because of a different starting potential (based on genetic background), the deleterious effect of the copy number variant on quantitative neurobehavioural traits (eg, social responsiveness scale score) results in reaching the threshold for a diagnosis of a neurobehavioural disorder (eg, autism spectrum disorder) without intellectual disability or motor impairment, since cognitive and motor abilities are not within the impaired range.
Conclusions and future perspectives
For many years, neurodevelopmental disorders have been recognised as clinically and aetiologically heterogeneous, to have overlapping symptoms, and to frequently co-occur. Despite these observations and extensive epidemiological data supporting the notion of a neurodevelopmental continuum, current diagnostic and classification systems are based on descriptive criteria, which were developed to improve the reliability of diagnosis and are largely atheoretical in terms of cause and pathophysiology. However, recent genetic evidence from whole genome copy number variant analyses and sequencing studies shows that identical genetic causes are common among apparently different disorders, as predicted by the developmental brain dysfunction model. These findings provide strong evidence in support of not only reviving the old concept of minimal brain dysfunction or minimal cerebral dysfunction but also expanding the scope to include a broad spectrum of neurodevelopmental and neuropsychiatric disorders, which could be encompassed by the term developmental brain dysfunction.
Since the clinical manifestations of most copy number variant syndromes are diverse, ranging from severe neurodevelopmental disorders to apparently unaffected individuals (even among individuals with identical mutations from the same family), it has been challenging to determine whether incomplete penetrance (some carriers are unaffected) or variable expressivity (all carriers are affected to different degrees) is responsible for the seemingly discordant phenotypes. Moreover, as neurodevelopmental disorders are complex disorders typically affecting more than one aspect of development, individuals with the same mutation can present with different neurodevelopmental profiles (a clinical feature can be present with variable degrees of severity, or absent). Therefore, if phenotypic traits are analysed independently, a genetic abnormality can be considered to be variably expressed for some functional domains and incompletely penetrant for others. However, when discrete deficits are viewed as manifestations of a common, underlying developmental impairment that we refer to as developmental brain dysfunction, we hypothesise that the penetrance of most copy number variant syndromes is likely to approach 100% across the lifespan.
To adequately discriminate incomplete penetrance from variable expressivity, it is important to determine the nature of the variables of interest, as they might be qualitative, dichotomous traits (ie, present or absent, normal or abnormal) or they might be continuous, quantitative variables. For example, cognitive function can be evaluated with standard intelligence quotient tests, which result in scores that are normally distributed in the general population. If intelligence quotient is viewed as a dichotomous trait with a cutoff of 70, representing 2 SD below the mean, individuals with scores above this threshold are judged to have normal intelligence whereas those below are deemed intellectually disabled (figure 1). However, the cutoff is arbitrary and the functional level of an individual with an intelligence quotient of 71 (normal) might not be notably different from someone with an intelligence quotient of 69 (intellectual disability), so it makes sense to approach this aspect of cognitive function as a continuous trait rather than setting cutoffs to assign categorical diagnoses.
Analogous to measures of cognition, methods allowing empirically based assessment of various core neurobehavioural competencies have been developed. Many neurobehavioural traits, including the core social deficiency of autism spectrum disorders, as measured by the Social Responsiveness Scale, are quantitative traits that are continuously distributed across the population.68,69 Various other quantitative scales allow non-categorical evaluation of different aspects of neurobehavioural functioning. For example, the Achenbach system of empirically based assessment includes scales for the assessment of competencies, adaptive functioning, and behavioural, emotional, and social problems across the lifespan; and the childhood routines inventory measures repetitive behaviours.70,71 Motor abilities can also be assessed using quantitative clinical assessment methods such as the physical and neurological examination for subtle signs and the Bruininks-Oseretsky Test of Motor Proficiency.72,73 By using these and similar methods in the evaluation of individuals with neurodevelopmental disorders we could delineate a quantitative neurodevelopmental profile of skill strengths and weaknesses, which can be used to guide treatment and interventions.
Furthermore, quantitative measures can be used in a research setting to compare the neurodevelopmental profile of individuals with a defined genetic mutation to their carrier and non-carrier relatives (genetic background) and quantify the effect, or deleterious impact, of the mutation across different functional domains, allowing measurable genotype-phenotype correlations. The importance of comparing affected individuals to their unaffected parents and siblings to quantify the deleterious effect of a copy number variant is illustrated by the intelligence quotient analyses conducted by Zufferey and colleagues36 in individuals with deletion 16p11.2 and their parents and siblings. If the mean full-scale intelligence quotient identified in probands (76·1) is compared with that in the general population (100), rather than in first-degree relatives of the probands (108·3), the effect of this copy number variant on cognitive function would have been underestimated (23·9 vs 32·2 points on average, respectively). The predictive value of using parental scores on appropriate quantitative measures to estimate the child’s expected performance profile, or starting point from which the deleterious impact of a copy number variant is subtracted, is an important area for future research. For example, if the profile of deleterious impacts of a copy number variant or single gene mutation is known (eg, deletion 22q11.2 has a deleterious impact of 2 SD of full-scale intelligence quotient) and the parents are properly evaluated, it may be possible to predict which developmental domains are likely to be more severely affected and even anticipate which categorical diagnoses might be present, or develop later in life, in an individual with a well studied mutation (figure 3).
The current guidelines of the American Academy of Pediatrics recommend routine, standardised developmental screening in all children at their 9-month, 18-month, and 30-month well-child preventive care visit.74 Other guidelines, from the American Academy of Neurology, Child Neurology Society, and American College of Medical Genetics, recommend chromosomal microarray analysis for children with unexplained global developmental delay.75,76 Therefore, the likelihood is that more children with developmental brain dysfunction of genetic origin will be identified early in life. Based on current research, it is tempting to propose that once a child is shown to have a specific genetic abnormality in clinical practice, parental core functional domains should be evaluated to more accurately predict the phenotypic consequences expected in the child and plan early intervention and treatment accordingly, for a truly personalised medicine approach.
Search strategy and selection criteria.
References for this Personal View were identified through searches of PubMed with the search terms “neurodevelopmental disorders”, “neuropsychiatric disorders”, “minimal brain dysfunction”, “minimal cerebral dysfunction”, “neurological taint”, “genetics”, “genomics”, “genes”, “copy number variants”, “sequence mutations”, “incomplete penetrance”, and “variable expressivity” from inception until December, 2012. Articles were also identified through searches of the reference lists of the articles found with the above cited search terms and of the authors’ own files. Only manuscripts published in English were reviewed. The final reference list was generated on the basis of originality and relevance to the scope of this Personal View.
Acknowledgments
This work was funded in part by grants MH074090 and HD064525 (to DHL) from the National Institutes of Health.
Footnotes
Contributors
AM-D-L and SMM prepared the first draft of this Personal View. TDC, DM-D-L, DWE, and DHL were involved in the writing and revision of the manuscript. All authors approved the final version.
Conflicts of interest
We declare that we have no conflicts of interest.
References
- 1.American Psychiatric Association. [accessed Sept 17, 2012];DSM-5 Development. 2012 http://www.dsm5.org.
- 2.Reiss AL. Childhood developmental disorders: an academic and clinical convergence point for psychiatry, neurology, psychology and pediatrics. J Child Psychol Psychiatry. 2009;50:87–98. doi: 10.1111/j.1469-7610.2008.02046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson MR, Shorvon SD. Heredity in epilepsy: neurodevelopment, comorbidity, and the neurological trait. Epilepsy Behav. 2011;22:421–27. doi: 10.1016/j.yebeh.2011.07.031. [DOI] [PubMed] [Google Scholar]
- 4.Rapoport JL, Giedd JN, Gogtay N. Neurodevelopmental model of schizophrenia: update 2012. Mol Psychiatry. 2012;17:1228–38. doi: 10.1038/mp.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Myers SM. Diagnosis of developmental disabilities. In: Batshaw ML, Lotrecchiano GR, Roizen NJ, editors. Children with disabilities. 7. Baltimore: Paul H. Brookes Publishing Co; 2013. pp. 243–66. [Google Scholar]
- 6.Yeargin-Allsopp M, Boyle C, van Naarden-Braun K, Trevathan E. The epidemiology of developmental disabilities. In: Accardo PJ, editor. Capute and Accardo’s neurodevelopmental disabilities in infancy and childhood. 3. Baltimore: Paul H. Brookes Publishing Co; 2008. pp. 61–104. [Google Scholar]
- 7.Piek JP, Pitcher TM, Hay DA. Motor coordination and kinaesthesis in boys with attention deficit-hyperactivity disorder. Dev Med Child Neurol. 1999;41:159–65. doi: 10.1017/s0012162299000341. [DOI] [PubMed] [Google Scholar]
- 8.Taurines R, Schmitt J, Renner T, Conner AC, Warnke A, Romanos M. Developmental comorbidity in attention-deficit/hyperactivity disorder. Attent Defic Hyperact Disord. 2010;2:267–89. doi: 10.1007/s12402-010-0040-0. [DOI] [PubMed] [Google Scholar]
- 9.Willcutt EG, Pennington BF, Duncan L, et al. Understanding the complex etiologies of developmental disorders: behavioral and molecular genetic approaches. J Dev Behav Pediatr. 2010;31:533–44. doi: 10.1097/DBP.0b013e3181ef42a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pennington BF, Bishop DV. Relations among speech, language, and reading disorders. Annu Rev Psychol. 2009;60:283–306. doi: 10.1146/annurev.psych.60.110707.163548. [DOI] [PubMed] [Google Scholar]
- 11.Odding E, Roebroeck ME, Stam HJ. The epidemiology of cerebral palsy: incidence, impairments and risk factors. Disabil Rehabil. 2006;28:183–91. doi: 10.1080/09638280500158422. [DOI] [PubMed] [Google Scholar]
- 12.Moreno-De-Luca A, Ledbetter DH, Martin CL. Genetic insights into the causes and classification of the cerebral palsies. Lancet Neurol. 2012;11:283–92. doi: 10.1016/S1474-4422(11)70287-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Einfeld SL, Ellis LA, Emerson E. Comorbidity of intellectual disability and mental disorder in children and adolescents: a systematic review. J Intellect Dev Disabil. 2011;36:137–43. doi: 10.1080/13668250.2011.572548. [DOI] [PubMed] [Google Scholar]
- 14.Mazefsky CA, Oswald DP, Day TN, et al. ASD, a psychiatric disorder, or both? Psychiatric diagnoses in adolescents with high-functioning ASD. J Clin Child Adolesc Psycho. 2012;41:516–23. doi: 10.1080/15374416.2012.686102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denckla MB. Minimal brain dysfunction. In: Chall JS, Mirsky AF, editors. Education and the brain. Chicago, IL: University of Chicago Press; 1978. pp. 223–68. [Google Scholar]
- 16.Strauss AS, Lehtinen MA. Psychopathology and education of the brain-injured child. New York: Grune and Stratton; 1947. [Google Scholar]
- 17.Clemmens RL. Syndromes of minimal brain dysfunction in children. Md State Med J. 1966;15:139–40. [PubMed] [Google Scholar]
- 18.Zigler E. Familial mental retardation: a continuing dilemma. Science. 1967;155:292–98. doi: 10.1126/science.155.3760.292. [DOI] [PubMed] [Google Scholar]
- 19.Weisz JR, Zigler E. Cognitive development in retarded and nonretarded persons: Piagetian tests of the similar sequence hypothesis. Psychol Bull. 1979;86:831–51. [PubMed] [Google Scholar]
- 20.Cicchetti D, Pogge-Hesse P. Possible contributions of the study of organically retarded persons to developmental theory. In: Zigler E, Balla D, editors. Mental retardation: the developmental-difference controversy. Hillsdale, NJ: Erlbaum; 1982. pp. 277–318. [Google Scholar]
- 21.Hodapp RM, Zigler E. Applying the developmental perspective to Down syndrome. In: Cicchetti D, Beeghly M, editors. Children with Down syndrome: A developmental perspective. New York: Cambridge University Press; 1990. pp. 1–28. [Google Scholar]
- 22.Capute AJ, Palmer FB. A pediatric overview of the spectrum of developmental disabilities. J Dev Behav Pediatr. 1980;1:66–69. [PubMed] [Google Scholar]
- 23.Capute AJ, Accardo PJ. A Neurodevelopmental perspective on the continuum of developmental disabilities. In: Capute AJ, Accardo PJ, editors. Developmental disabilities in infancy and childhood. Baltimore: Paul H Brookes Publishing Co; 1991. [Google Scholar]
- 24.Gilger JW, Kaplan BJ. Atypical brain development: a conceptual framework for understanding developmental learning disabilities. Dev Neuropsychol. 2001;20:465–81. doi: 10.1207/S15326942DN2002_2. [DOI] [PubMed] [Google Scholar]
- 25.Gillberg C. The ESSENCE in child psychiatry: early symptomatic syndromes eliciting neurodevelopmental clinical examinations. Res Dev Disabil. 2010;31:1543–51. doi: 10.1016/j.ridd.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 26.Kaminsky EB, Kaul V, Paschall J, et al. An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genet Med. 2011;13:777–84. doi: 10.1097/GIM.0b013e31822c79f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooper GM, Coe BP, Girirajan S, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–46. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Girirajan S, Brkanac Z, Coe BP, et al. Relative burden of large CNVs on a range of neurodevelopmental phenotypes. PLoS Genet. 2011;7:e1002334. doi: 10.1371/journal.pgen.1002334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanders SJ, Murtha MT, Gupta AR, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–41. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Roak BJ, Vives L, Girirajan S, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–50. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Najmabadi H, Hu H, Garshasbi M, et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature. 2011;478:57–63. doi: 10.1038/nature10423. [DOI] [PubMed] [Google Scholar]
- 32.Xu B, Roos JL, Dexheimer P, et al. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat Genet. 2011;43:864–68. doi: 10.1038/ng.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moreno-De-Luca D, Sanders S, Willsey A, et al. Using large clinical datasets to infer pathogenicity for rare copy number variants in autism cohorts. Mol Psychiatry. 2012 doi: 10.1038/mp.2012.138. published online Oct 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bassett AS, McDonald-McGinn DM, Devriendt K, et al. Practical guidelines for managing patients with 22q11. 2 deletion syndrome. J Pediatr. 2011;159:332–39. doi: 10.1016/j.jpeds.2011.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moss EM, Batshaw ML, Solot CB, et al. Psychoeducational profile of the 22q11. 2 microdeletion: a complex pattern. J Pediatr. 1999;134:193–98. doi: 10.1016/s0022-3476(99)70415-4. [DOI] [PubMed] [Google Scholar]
- 36.Zufferey F, Sherr E, Beckmann N, et al. A 600 kb deletion syndrome at 16p11. 2 leads to energy imbalance and neuropsychiatric disorders. J Med Genet. 2012;49:660–68. doi: 10.1136/jmedgenet-2012-101203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar RA, KaraMohamed S, Sudi J, et al. Recurrent 16p11. 2 microdeletions in autism. Hum Mol Genet. 2008;17:628–38. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 38.Weiss LA, Shen Y, Korn JM, et al. Association between microdeletion and microduplication at 16p11. 2 and autism. N Engl J Med. 2008;358:667–75. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 39.Shinawi M, Liu P, Kang SH, et al. Recurrent reciprocal 16p11. 2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J Med Genet. 2010;47:332–41. doi: 10.1136/jmg.2009.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dale RC, Grattan-Smith P, Fung VS, Peters GB. Infantile convulsions and paroxysmal kinesigenic dyskinesia with 16p11. 2 microdeletion. Neurology. 2011;77:1401–02. doi: 10.1212/WNL.0b013e31823152d7. [DOI] [PubMed] [Google Scholar]
- 41.Jacquemont S, Reymond A, Zufferey F, et al. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11. 2 locus. Nature. 2011;478:97–102. doi: 10.1038/nature10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Christiansen J, Dyck JD, Elyas BG, et al. Chromosome 1q21, 1 contiguous gene deletion is associated with congenital heart disease. Circ Res. 2004;94:1429–35. doi: 10.1161/01.RES.0000130528.72330.5c. [DOI] [PubMed] [Google Scholar]
- 43.Stefansson H, Rujescu D, Cichon S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–36. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–41. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mefford HC, Sharp AJ, Baker C, et al. Recurrent rearrangements of chromosome 1q21. 1 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–99. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szatmari P, Paterson AD, Zwaigenbaum L, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–28. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harvard C, Strong E, Mercier E, et al. Understanding the impact of 1q21. 1 copy number variant. Orphanet J Rare Dis. 2011;6:54. doi: 10.1186/1750-1172-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharp AJ, Mefford HC, Li K, et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet. 2008;40:322–28. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pagnamenta AT, Wing K, Sadighi Akha E, et al. A 15q13.3 microdeletion segregating with autism. Eur J Hum Genet. 2009;17:687–92. doi: 10.1038/ejhg.2008.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ben-Shachar S, Lanpher B, German JR, et al. Microdeletion 15q13.3: a locus with incomplete penetrance for autism, mental retardation, and psychiatric disorders. J Med Genet. 2009;46:382–88. doi: 10.1136/jmg.2008.064378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moreno-De-Luca D, Mulle JG, Kaminsky EB, et al. Deletion 17q12 is a recurrent copy number variant that confers high risk of autism and schizophrenia. Am J Hum Genet. 2010;87:618–30. doi: 10.1016/j.ajhg.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagamani SC, Erez A, Shen J, et al. Clinical spectrum associated with recurrent genomic rearrangements in chromosome 17q12. Eur J Hum Genet. 2010;18:278–84. doi: 10.1038/ejhg.2009.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loirat C, Bellanne-Chantelot C, Husson I, et al. Autism in three patients with cystic or hyperechogenic kidneys and chromosome 17q12 deletion. Nephrol Dial Transplant. 2010;25:3430–33. doi: 10.1093/ndt/gfq380. [DOI] [PubMed] [Google Scholar]
- 54.Verkerk AJ, Mathews CA, Joosse M, Eussen BH, Heutink P, Oostra BA. CNTNAP2 is disrupted in a family with Gilles de la Tourette syndrome and obsessive compulsive disorder. Genomics. 2003;82:1–9. doi: 10.1016/s0888-7543(03)00097-1. [DOI] [PubMed] [Google Scholar]
- 55.Strauss KA, Puffenberger EG, Huentelman MJ, et al. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N Engl J Med. 2006;354:1370–77. doi: 10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- 56.Friedman JI, Vrijenhoek T, Markx S, et al. CNTNAP2 gene dosage variation is associated with schizophrenia and epilepsy. Mol Psychiatry. 2008;13:261–66. doi: 10.1038/sj.mp.4002049. [DOI] [PubMed] [Google Scholar]
- 57.O’Roak BJ, Deriziotis P, Lee C, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–89. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zweier C, de Jong EK, Zweier M, et al. CNTNAP2 and NRXN1 are mutated in autosomal-recessive Pitt-Hopkins-like mental retardation and determine the level of a common synaptic protein in drosophila. Am J Hum Genet. 2009;85:655–66. doi: 10.1016/j.ajhg.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schaaf CP, Boone PM, Sampath S, et al. Phenotypic spectrum and genotype-phenotype correlations of NRXN1 exon deletions. Eur J Hum Genet. 2012;20:1240–47. doi: 10.1038/ejhg.2012.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ching MS, Shen Y, Tan WH, et al. Deletions of NRXN1 (neurexin-1) predispose to a wide spectrum of developmental disorders. Am J Med Genet. 2010;153:937–47. doi: 10.1002/ajmg.b.31063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Duong L, Klitten LL, Moller RS, et al. Mutations in NRXN1 in a family multiply affected with brain disorders: NRXN1 mutations and brain disorders. Am J Med Genet. 2012;159:354–58. doi: 10.1002/ajmg.b.32036. [DOI] [PubMed] [Google Scholar]
- 62.Gauthier J, Siddiqui TJ, Huashan P, et al. Truncating mutations in NRXN2 and NRXN1 in autism spectrum disorders and schizophrenia. Hum Genet. 2011;130:563–73. doi: 10.1007/s00439-011-0975-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dibbens LM, Tarpey PS, Hynes K, et al. X-linked protocadherin 19 mutations cause female-limited epilepsy and cognitive impairment. Nat Genet. 2008;40:776–81. doi: 10.1038/ng.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Depienne C, Bouteiller D, Keren B, et al. Sporadic infantile epileptic encephalopathy caused by mutations in PCDH19 resembles Dravet syndrome but mainly affects females. PLoS Genet. 2009;5:e1000381. doi: 10.1371/journal.pgen.1000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Camacho A, Simon R, Sanz R, Vinuela A, Martinez-Salio A, Mateos F. Cognitive and behavioral profile in females with epilepsy with PDCH19 mutation: two novel mutations and review of the literature. Epilepsy Behav. 2012;24:134–37. doi: 10.1016/j.yebeh.2012.02.023. [DOI] [PubMed] [Google Scholar]
- 66.Brandon NJ, Sawa A. Linking neurodevelopmental and synaptic theories of mental illness through DISC1. Nat Rev Neurosci. 2011;12:707–22. doi: 10.1038/nrn3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Endele S, Rosenberger G, Geider K, et al. Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nat Genet. 2010;42:1021–26. doi: 10.1038/ng.677. [DOI] [PubMed] [Google Scholar]
- 68.Constantino JN, Davis SA, Todd RD, et al. Validation of a brief quantitative measure of autistic traits: comparison of the social responsiveness scale with the autism diagnostic interview-revised. J Autism Dev Disord. 2003;33:427–33. doi: 10.1023/a:1025014929212. [DOI] [PubMed] [Google Scholar]
- 69.Constantino JN. The quantitative nature of autistic social impairment. Pediatr Res. 2011;69:55–62. doi: 10.1203/PDR.0b013e318212ec6e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Achenbach TM. The Achenbach system of empirically based assessemnt (ASEBA): development, findings, theory, and applications. Burlington, VT: University of Vermont Research Center for Children, Youth and Families; 2009. [Google Scholar]
- 71.Evans DW, Leckman JF, Carter A, et al. Ritual, habit, and perfectionism: the prevalence and development of compulsive-like behavior in normal young children. Child Dev. 1997;68:58–68. [PubMed] [Google Scholar]
- 72.Denckla MB. Revised neurological examination for subtle signs (1985) Psychopharmacol Bull. 1985;21:773–800. [PubMed] [Google Scholar]
- 73.Bruininks RH, Bruininks BD. Test of motor proficiency. 2. Circle Pines, MN: AGS Publishing; 2005. [Google Scholar]
- 74.American Academy of Pediatrics. Identifying infants and young children with developmental disorders in the medical home: an algorithm for developmental surveillance and screening. Pediatrics. 2006;118:405–20. doi: 10.1542/peds.2006-1231. [DOI] [PubMed] [Google Scholar]
- 75.Manning M, Hudgins L. Array-based technology and recommendations for utilization in medical genetics practice for detection of chromosomal abnormalities. Genet Med. 2010;12:742–45. doi: 10.1097/GIM.0b013e3181f8baad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Michelson DJ, Shevell MI, Sherr EH, Moeschler JB, Gropman AL, Ashwal S. Evidence report: genetic and metabolic testing on children with global developmental delay: report of the Quality Standards Subcommittee of the American Academy of Neurology and the Practice Committee of the Child Neurology Society. Neurology. 2011;77:1629–35. doi: 10.1212/WNL.0b013e3182345896. [DOI] [PubMed] [Google Scholar]