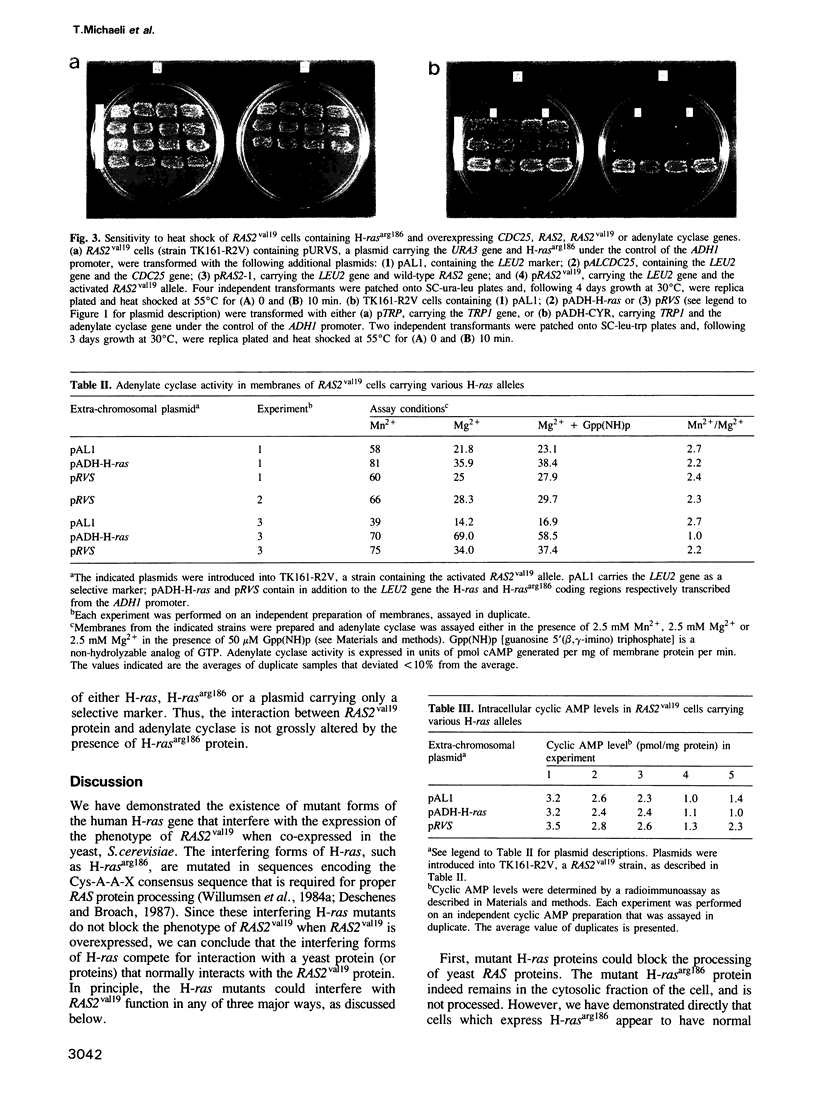
Abstract
We report a class of interfering mutants of the human H-ras gene capable of inhibiting phenotypes arising from the expression of the activated RAS2 gene, RAS2val19, in the yeast Saccharomyces cerevisiae. All these mutants encode unprocessed H-ras proteins that remain in the cytoplasm. One of the mutants, H-rasarg186, was examined in detail. H-rasarg186 protein is a competitive inhibitor of RAS2val19 protein. It does not interfere with processing and membrane localization of RAS2val19, nor does it appear to compete with RAS protein for its proposed regulator, the CDC25 protein. By several criteria the RAS2val19 adenylate cyclase interaction is unaffected by H-rasarg186. We infer from our results that H-rasarg186 protein interferes with an alternative function of RAS2val19.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- Broek D., Samiy N., Fasano O., Fujiyama A., Tamanoi F., Northup J., Wigler M. Differential activation of yeast adenylate cyclase by wild-type and mutant RAS proteins. Cell. 1985 Jul;41(3):763–769. doi: 10.1016/s0092-8674(85)80057-x. [DOI] [PubMed] [Google Scholar]
- Broek D., Toda T., Michaeli T., Levin L., Birchmeier C., Zoller M., Powers S., Wigler M. The S. cerevisiae CDC25 gene product regulates the RAS/adenylate cyclase pathway. Cell. 1987 Mar 13;48(5):789–799. doi: 10.1016/0092-8674(87)90076-6. [DOI] [PubMed] [Google Scholar]
- Calés C., Hancock J. F., Marshall C. J., Hall A. The cytoplasmic protein GAP is implicated as the target for regulation by the ras gene product. Nature. 1988 Apr 7;332(6164):548–551. doi: 10.1038/332548a0. [DOI] [PubMed] [Google Scholar]
- Camonis J. H., Kalékine M., Gondré B., Garreau H., Boy-Marcotte E., Jacquet M. Characterization, cloning and sequence analysis of the CDC25 gene which controls the cyclic AMP level of Saccharomyces cerevisiae. EMBO J. 1986 Feb;5(2):375–380. doi: 10.1002/j.1460-2075.1986.tb04222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon J. F., Tatchell K. Characterization of Saccharomyces cerevisiae genes encoding subunits of cyclic AMP-dependent protein kinase. Mol Cell Biol. 1987 Aug;7(8):2653–2663. doi: 10.1128/mcb.7.8.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFeo-Jones D., Scolnick E. M., Koller R., Dhar R. ras-Related gene sequences identified and isolated from Saccharomyces cerevisiae. Nature. 1983 Dec 15;306(5944):707–709. doi: 10.1038/306707a0. [DOI] [PubMed] [Google Scholar]
- DeFeo-Jones D., Tatchell K., Robinson L. C., Sigal I. S., Vass W. C., Lowy D. R., Scolnick E. M. Mammalian and yeast ras gene products: biological function in their heterologous systems. Science. 1985 Apr 12;228(4696):179–184. doi: 10.1126/science.3883495. [DOI] [PubMed] [Google Scholar]
- Deschenes R. J., Broach J. R. Fatty acylation is important but not essential for Saccharomyces cerevisiae RAS function. Mol Cell Biol. 1987 Jul;7(7):2344–2351. doi: 10.1128/mcb.7.7.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig L. A., Cooper G. M. Inhibition of NIH 3T3 cell proliferation by a mutant ras protein with preferential affinity for GDP. Mol Cell Biol. 1988 Aug;8(8):3235–3243. doi: 10.1128/mcb.8.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field J., Broek D., Kataoka T., Wigler M. Guanine nucleotide activation of, and competition between, RAS proteins from Saccharomyces cerevisiae. Mol Cell Biol. 1987 Jun;7(6):2128–2133. doi: 10.1128/mcb.7.6.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field J., Nikawa J., Broek D., MacDonald B., Rodgers L., Wilson I. A., Lerner R. A., Wigler M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol Cell Biol. 1988 May;8(5):2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler R. G., Degnen G. E., Cox E. C. Mutational specificity of a conditional Escherichia coli mutator, mutD5. Mol Gen Genet. 1974;133(3):179–191. doi: 10.1007/BF00267667. [DOI] [PubMed] [Google Scholar]
- Harper J. F., Brooker G. Femtomole sensitive radioimmunoassay for cyclic AMP and cyclic GMP after 2'0 acetylation by acetic anhydride in aqueous solution. J Cyclic Nucleotide Res. 1975;1(4):207–218. [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka T., Powers S., Cameron S., Fasano O., Goldfarb M., Broach J., Wigler M. Functional homology of mammalian and yeast RAS genes. Cell. 1985 Jan;40(1):19–26. doi: 10.1016/0092-8674(85)90304-6. [DOI] [PubMed] [Google Scholar]
- Kataoka T., Powers S., McGill C., Fasano O., Strathern J., Broach J., Wigler M. Genetic analysis of yeast RAS1 and RAS2 genes. Cell. 1984 Jun;37(2):437–445. doi: 10.1016/0092-8674(84)90374-x. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall M. S., Gibbs J. B., Scolnick E. M., Sigal I. S. Regulatory function of the Saccharomyces cerevisiae RAS C-terminus. Mol Cell Biol. 1987 Jul;7(7):2309–2315. doi: 10.1128/mcb.7.7.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K., Uno I., Ishikawa T. Genetic analysis of the role of cAMP in yeast. Yeast. 1985 Sep;1(1):15–24. doi: 10.1002/yea.320010103. [DOI] [PubMed] [Google Scholar]
- Nikawa J., Cameron S., Toda T., Ferguson K. M., Wigler M. Rigorous feedback control of cAMP levels in Saccharomyces cerevisiae. Genes Dev. 1987 Nov;1(9):931–937. doi: 10.1101/gad.1.9.931. [DOI] [PubMed] [Google Scholar]
- Olempska-Beer Z., Freese E. B. Optimal extraction conditions for high-performance liquid chromatographic determination of nucleotides in yeast. Anal Biochem. 1984 Jul;140(1):236–245. doi: 10.1016/0003-2697(84)90159-3. [DOI] [PubMed] [Google Scholar]
- Powers S., Kataoka T., Fasano O., Goldfarb M., Strathern J., Broach J., Wigler M. Genes in S. cerevisiae encoding proteins with domains homologous to the mammalian ras proteins. Cell. 1984 Mar;36(3):607–612. doi: 10.1016/0092-8674(84)90340-4. [DOI] [PubMed] [Google Scholar]
- Powers S., Michaelis S., Broek D., Santa Anna S., Field J., Herskowitz I., Wigler M. RAM, a gene of yeast required for a functional modification of RAS proteins and for production of mating pheromone a-factor. Cell. 1986 Nov 7;47(3):413–422. doi: 10.1016/0092-8674(86)90598-2. [DOI] [PubMed] [Google Scholar]
- Robinson L. C., Gibbs J. B., Marshall M. S., Sigal I. S., Tatchell K. CDC25: a component of the RAS-adenylate cyclase pathway in Saccharomyces cerevisiae. Science. 1987 Mar 6;235(4793):1218–1221. doi: 10.1126/science.3547648. [DOI] [PubMed] [Google Scholar]
- Salomon Y., Londos C., Rodbell M. A highly sensitive adenylate cyclase assay. Anal Biochem. 1974 Apr;58(2):541–548. doi: 10.1016/0003-2697(74)90222-x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass P., Field J., Nikawa J., Toda T., Wigler M. Cloning and characterization of the high-affinity cAMP phosphodiesterase of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9303–9307. doi: 10.1073/pnas.83.24.9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sefton B. M., Trowbridge I. S., Cooper J. A., Scolnick E. M. The transforming proteins of Rous sarcoma virus, Harvey sarcoma virus and Abelson virus contain tightly bound lipid. Cell. 1982 Dec;31(2 Pt 1):465–474. doi: 10.1016/0092-8674(82)90139-8. [DOI] [PubMed] [Google Scholar]
- Sigal I. S., Gibbs J. B., D'Alonzo J. S., Temeles G. L., Wolanski B. S., Socher S. H., Scolnick E. M. Mutant ras-encoded proteins with altered nucleotide binding exert dominant biological effects. Proc Natl Acad Sci U S A. 1986 Feb;83(4):952–956. doi: 10.1073/pnas.83.4.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taparowsky E., Shimizu K., Goldfarb M., Wigler M. Structure and activation of the human N-ras gene. Cell. 1983 Sep;34(2):581–586. doi: 10.1016/0092-8674(83)90390-2. [DOI] [PubMed] [Google Scholar]
- Toda T., Cameron S., Sass P., Wigler M. SCH9, a gene of Saccharomyces cerevisiae that encodes a protein distinct from, but functionally and structurally related to, cAMP-dependent protein kinase catalytic subunits. Genes Dev. 1988 May;2(5):517–527. doi: 10.1101/gad.2.5.517. [DOI] [PubMed] [Google Scholar]
- Toda T., Uno I., Ishikawa T., Powers S., Kataoka T., Broek D., Cameron S., Broach J., Matsumoto K., Wigler M. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell. 1985 Jan;40(1):27–36. doi: 10.1016/0092-8674(85)90305-8. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trahey M., McCormick F. A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science. 1987 Oct 23;238(4826):542–545. doi: 10.1126/science.2821624. [DOI] [PubMed] [Google Scholar]
- Trahey M., Wong G., Halenbeck R., Rubinfeld B., Martin G. A., Ladner M., Long C. M., Crosier W. J., Watt K., Koths K. Molecular cloning of two types of GAP complementary DNA from human placenta. Science. 1988 Dec 23;242(4886):1697–1700. doi: 10.1126/science.3201259. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Vogel U. S., Dixon R. A., Schaber M. D., Diehl R. E., Marshall M. S., Scolnick E. M., Sigal I. S., Gibbs J. B. Cloning of bovine GAP and its interaction with oncogenic ras p21. Nature. 1988 Sep 1;335(6185):90–93. doi: 10.1038/335090a0. [DOI] [PubMed] [Google Scholar]
- Wigler M., Field J., Powers S., Broek D., Toda T., Cameron S., Nikawa J., Michaeli T., Colicelli J., Ferguson K. Studies of RAS function in the yeast Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol. 1988;53(Pt 2):649–655. doi: 10.1101/sqb.1988.053.01.074. [DOI] [PubMed] [Google Scholar]
- Willumsen B. M., Christensen A., Hubbert N. L., Papageorge A. G., Lowy D. R. The p21 ras C-terminus is required for transformation and membrane association. Nature. 1984 Aug 16;310(5978):583–586. doi: 10.1038/310583a0. [DOI] [PubMed] [Google Scholar]
- Willumsen B. M., Norris K., Papageorge A. G., Hubbert N. L., Lowy D. R. Harvey murine sarcoma virus p21 ras protein: biological and biochemical significance of the cysteine nearest the carboxy terminus. EMBO J. 1984 Nov;3(11):2581–2585. doi: 10.1002/j.1460-2075.1984.tb02177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis: a simple method using two oligonucleotide primers and a single-stranded DNA template. DNA. 1984 Dec;3(6):479–488. doi: 10.1089/dna.1.1984.3.479. [DOI] [PubMed] [Google Scholar]