Abstract
Traditional peptide-mediated siRNA transfection via peptide transduction domains exhibits limited cytoplasmic delivery of siRNA due to endosomal entrapment. This work overcomes these limitations with the use of membrane-destabilizing peptides derived from melittin for the knockdown of NFkB signaling in a model of adult T-Cell leukemia/lymphoma. While the mechanism of siRNA delivery into the cytoplasmic compartment by peptide transduction domains has not been well studied, our analysis of melittin derivatives indicates that concurrent nanocomplex disassembly and peptide-mediated endosomolysis are crucial to siRNA transfection. Importantly, in the case of the most active derivative, p5RHH, this process is initiated by acidic pH, indicating that endosomal acidification after macropinocytosis can trigger siRNA release into the cytoplasm. These data provide general principles regarding nanocomplex response to endocytosis which may guide the development of peptide/siRNA nanocomplex-based transfection.

Keywords: siRNA, endosomolysis, melittin, drug delivery, nanoparticle
Post-transcriptional degradation of mRNA via RNA interference (RNAi) provides a targeted approach for silencing gene expression that may prove beneficial in the treatment of many clinically relevant diseases.1–2 RNAi can be induced by delivery of small-interfering RNA (siRNA) into the cytoplasm of a mammalian cell, after which incorporation of the siRNA into RNA-induced silencing complexes (RISC) leads to sequence-specific cleavage of complementary mRNA.3–4 Given siRNA’s activity in the cytoplasm, siRNA must bypass impermeable cellular membranes to reach the cytoplasmic compartment. Unfortunately, due to siRNA’s large molecular weight (~21kDa) and negative charge, naked siRNA cannot diffuse freely through cell membranes, necessitating an effective delivery system to aid cellular uptake and subsequent endosomal escape.5–8
Common siRNA delivery systems include cationic lipids and polymers, which are efficient, yet hampered by potential toxicity.9–18 Recent work has focused on poly-basic peptides or peptide transduction domains (PTD) for siRNA transfection owing to their lack of toxic side effects.19–27 Unfortunately, many studies have reported only modest success at achieving highly efficient siRNA delivery when complexed with peptides as a consequence of excessive endosomal entrapment.28–32 Acknowledging endosomal entrapment as the primary barrier hindering the progress of peptide-based siRNA vectors emphasizes that new strategies must be developed to improve peptide-mediated transfection. Accordingly, we propose that membrane-disrupting peptides carrying a net positive charge could provide an unexplored alternative for efficient siRNA transfection due to their dual functionality to both complex siRNA and disrupt endosomal compartments.
Acid activatable melittin has previously been utilized to improve endosomal escape of hepatocyte targeted chol-siRNA resulting in a 500-fold improvement in protein knockdown.33 In contrast, our work focuses on the development of melittin-derived peptides as an siRNA vector, not just as an excipient for endosomal escape. While our melittin derivatives are expected to improve upon existing peptide-mediated siRNA delivery by initiating endosomal escape, additional molecular mechanisms resulting in successful siRNA transfection remain to be identified. For example, recent work by van Asbeck et al. concludes that sensitivity to decomplexation by poly-anionic macromolecules contributes to improved transfection, but the role decomplexation plays in siRNA delivery to the cytoplasm was not established.34 Furthermore, pH-responsive fusogenic peptides from the influenza virus have previously been leveraged to augment peptide-mediated transfection, but their ability to improve siRNA transfection may be attributable to increased siRNA packaging or uptake and not endosomal escape.29 CPP/siRNA nanoparticles have been well characterized from a physico-chemical perspective; however, the mechanisms involved in peptide/siRNA nanocomplex transfection that contribute to successful bypass of endosomal entrapment and subsequent induction of RNAi have yet to be elucidated. Additional studies regarding the intracellular processing of peptide/siRNA nanocomplexes and the mechanism of siRNA release to the cytoplasm are required to further develop peptides for siRNA transfection.
We have previously reported that melittin derivative, p5RHH, is capable of siRNA transfection with an IC50 as low as 25nM without significant cytotoxicity at all tested doses.35 In the current work, this peptide is employed for the delivery of p65 and p100/52 siRNA for simultaneous knockdown of both canonical and non-canonical NFkB signaling pathways in a murine model of Human T-Lymphotropic Virus-1 (HTLV-1) induced adult T-cell leukemia/lymphoma (ATLL). For enhanced stability, we show here that an albumin-coated formulation of p5RHH exhibits remarkable transfection efficiency attributable to pH triggered nanoparticle disassembly. Detailed studies regarding the mechanism of action reveal that exposure to endosomal pH triggers both nanoparticle disassembly and endosomal escape. Moreover, it is clear from comparisons with nonfunctioning melittin derivatives that endosomal disruption alone does not result in successful induction of RNAi but requires concurrent siRNA release from the vector. Our results offer general parameters that yield efficient siRNA delivery into the cytoplasm by peptide vectors, which may aid the development noncovalent peptide/siRNA nanocomplexes for siRNA therapeutics.
RESULTS AND DISCUSSION
To formulate p5RHH/siRNA nanoparticles, p5RHH (10mM stock in DI H2O) is dissolved 1:200 in Dulbecco’s Phosphate Buffered Saline, vortexed for 30 seconds followed by addition of the appropriate amount of siRNA (100μM stock in 1x siRNA) and incubated at 37°C for 40 minutes (Figure 1a). 40 minute incubations were chosen based on the particle size as tracked by Deep-Etch Electron Microscopy. Electron micrographs (Supplemental Figure 1a) indicate that particles formed at this time point have not begun to exhibit further aggregation, allowing a platform for kinetic stabilization via albumin surface coating. Notably, 40 minute incubations also exhibit maximal transfection efficiency based on knockdown of green fluorescent protein (GFP) expression in B16–F10 melanoma cells (ATCC, Manassas, VA)(Supplemental Figure 1b).
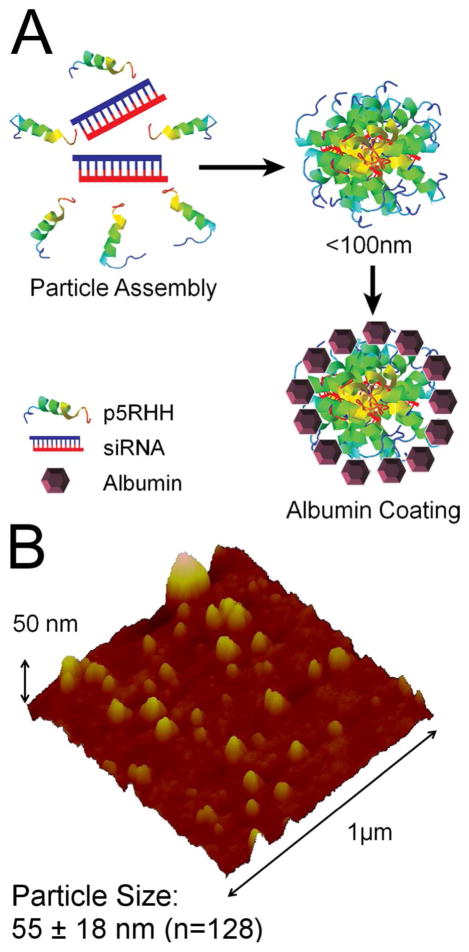
Figure 1.
(a) Scheme for formulation of albumin-stabilized p5RHH/siRNA nanoparticles. (b) Wet-mode AFM imaging of p5RHH/siRNA nanoparticles reveals an average particle size of ~55 ± 18 nm (n = 128).
Albumin is known to provide enhanced nanoparticle stability by coating nanoparticles to prevent flocculation.36 Albumin-stabilized formulations include a subsequent 30 minute incubation in the presence of 0.5mg/ml human serum albumin (50mg/ml stock in DI H2O) prior to use. The size of albumin stabilized p5RHH/siRNA nanoparticles 72 hours post formulation was measured to be ~55 ±18 nm (n = 128) by wet mode atomic force microscopy (Figure 1b), indicating that albumin prevents flocculation of p5RHH/siRNA nanoparticles. An analysis of time dependent particle size stability has previously been performed by DLS, and reveals a lack of particle aggregation after albumin coating during an initial overnight incubation.35 The AFM measurements provided here confirm a lack of aggregation while providing a more accurate assessment of particle diameter.
Considering the uncertainty surrounding the cellular entry of peptide/siRNA nanoparticles, uptake assays were performed to provide insight into the mechanism by which p5RHH achieves cytoplasmic delivery of siRNA.37 Flow cytometry assays depicting the uptake of Alexa488-labeled scrambled siRNA packaged with p5RHH provide a convenient experimental tool to determine the role of select endocytic pathways in p5RHH/siRNA nanoparticle uptake. Incubation of cells at 4°C causes near complete inhibition of p5RHH/siRNA uptake, thus rejecting the hypothesis that p5RHH mediates direct membrane translocation for cytoplasmic release of siRNA (Supplemental Figure 2). Instead, studies of p5RHH/siRNA uptake in the presence of endocytosis inhibitors indicate that macropinocytosis is the major pathway responsible for p5RHH/siRNA uptake (Figure 2a–d). The macropinocytosis inhibitor EIPA dramatically reduces p5RHH/siRNA uptake whereas caveolae inhibitor, filipin, and clathrin mediated endocytosis (CME) inhibitor, PAO, have no affect on p5RHH/siRNA uptake.
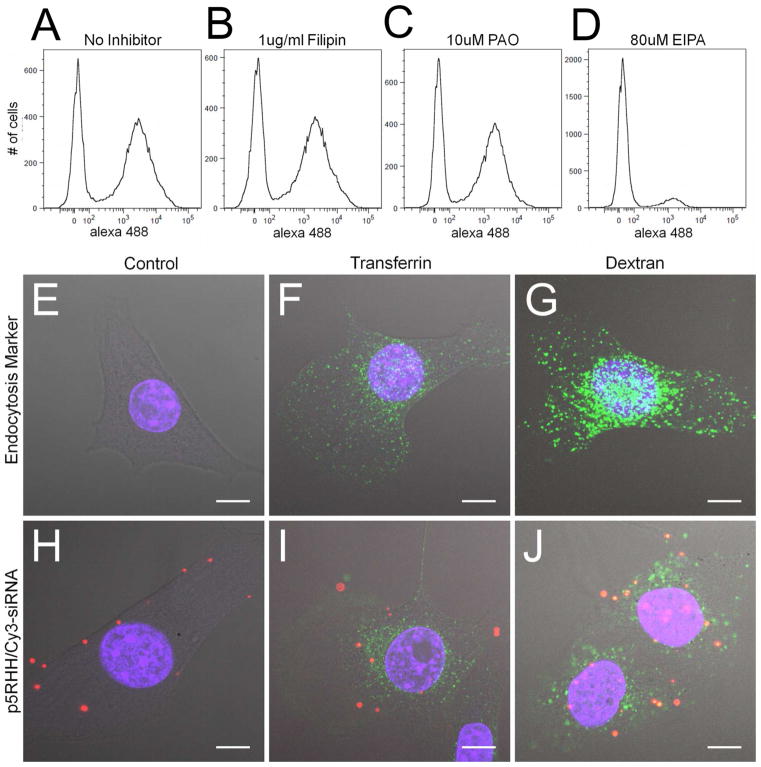
Figure 2.
(a–d) 40 minute uptake of p5RHH/Alexa488-siRNA nanoparticles shows that 60% of the treated cells take up (a) p5RHH/siRNA nanoparticles. The presence of endocytosis inhibitors indicates that (b) 100μg/mL filipin (caveolae inhibitor) and (c) 10μM PAO (clathrin mediated endocytosis inhibitor) do not inhibit p5RHH/siRNA nanoparticle uptake. Alternatively, treatment with (d) macropinocytosis inhibitor (EIPA, 80μM) nearly abolishes nanoparticle uptake. (e–j) Colocalization as determined by confocal microscopy shows that p5RHH/Cy-3 siRNA nanoparticles are taken up with FITC-70kDa dextran (j) but not FITC-transferrin (i). Scale bar 10μm.
The use of chemical endocytosis inhibitors is a common method for evaluating nanoparticle uptake. However, care must be taken to ensure the selectivity of those inhibitors.38–39 Consequently, uptake inhibition assays were performed for only 40 minutes at inhibitor concentrations that were determined to be specific to the expected pathway (Supplemental Figure 2), as demonstrated by inhibition of standard endosomal markers transferrin (CME) and 70kDa Dextran (macropinocytosis). B16 cells are known not to express caveolin-1, and not surprisingly uptake of caveolae marker Cholera Toxin B is not measureable in this cell type (unpublished observation).
Confocal microscopy confirms the flow cytometry data, illustrating strong colocalization of p5RHH/Cy3-siRNA with FITC-70kDa dextran (Figure 2j), but not with FITC-Transferrin (Figure 2i). Cells were incubated with uptake markers for only 40 minutes to minimize release of Cy-3 labeled siRNA into the cytoplasm, which could yield cytoplasmic or nuclear fluorescence that otherwise might confound the analysis, and thus cells exhibiting cytoplasmic release were not imaged to avoid these issues. Interestingly, the rapid (<1 hour uptake and release) of Cy-3 labeled siRNA confirms the rapid endosomal escape induced by p5RHH/siRNA nanoparticles (Supplemental Figure 3).
These results are in accordance with general rules regulating the cellular uptake of many positively charged peptides containing basic residues. Specifically, arginine residues can form bidentate ionic interactions with cell surface proteoglycans, which results in close association with the plasma membrane.40 Moreover, these nonspecific binding interactions can stimulate actin rearrangements that are required for fluid phase uptake by macropinocytosis. The robust uptake of positively charged peptides indicates that electrostatic association with the plasma membrane and subsequent fluid phase uptake is sufficient to achieve substantial peptide/siRNA uptake.
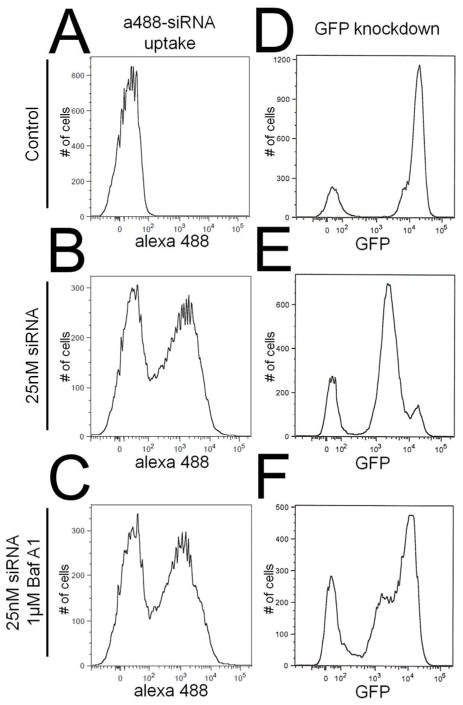
Proper siRNA trafficking subsequent to the initial endocytic event is also vitally important for successful siRNA transfection. In particular, the pH of endosomes and lysosomes is tightly controlled by acidification via membrane-bound vacuolar ATPases and can provide a trigger for environmentally sensitive siRNA release from p5RHH/siRNA nanoparticles.41–42 To determine if the low pH generated by these vacuolar ATPases is involved in siRNA release from endosomes, cells were incubated in the presence of bafilomycin A1 during the transfection. Compared to control cells transfected without bafilomycin A1 (Figure 3e), bafilomycin A1-treated cells (Figure 3f) led to a near complete loss of GFP knockdown as determined by flow cytometry. Since bafilomycin A1 could be slowing p5RHH/siRNA uptake, flow cytometric evaluation of the uptake of fluorescently-labeled siRNA in B16 cells was utilized to ensure that the concentration of bafilomycin A1 used in these assays did not impair p5RHH/siRNA uptake (Figure 3a–c). These data confirm the importance of endosomal acidification in the cytoplasmic release of siRNA when delivered to cells via p5RHH.
Figure 3.
(a–c) Bafilomycin A1 does not inhibit uptake of p5RHH/Alexa488-siRNA nanoparticles (c) compared to transfection in the absence of bafilomycin A1 (b). (d–f) On the other hand, bafilomycin A1 blocks knockdown of GFP (f) compared to transfection in the absence of bafilomycin A1 (e) indicating that endosomal acidification is crucial for p5RHH mediated siRNA transfection.
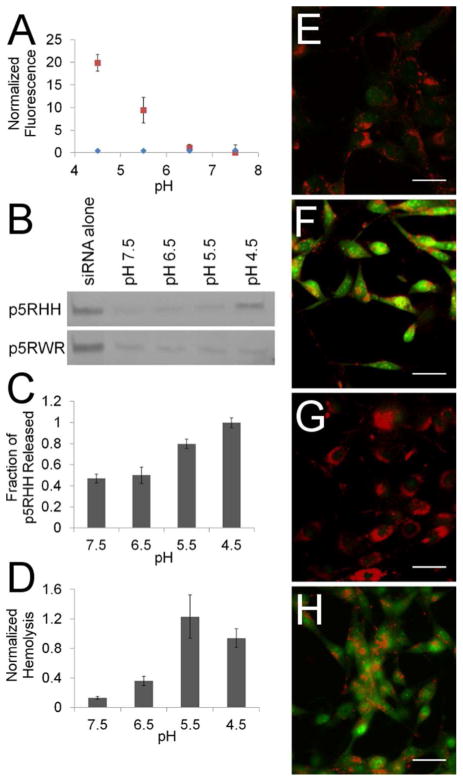
Because endosomal acidification is crucial to the ability of p5RHH to deliver siRNA to the cytoplasm, p5RHH/siRNA nanoparticles were incubated at low pH to ascertain how an increasingly acidic environment affects nanoparticle integrity. Dye-binding assays using the nucleic acid stain TOPRO3 reveal that siRNA becomes increasingly accessible at pH ≤ 5.5 as manifested by increased TOPRO3 fluorescence intensity (Figure 4a). To determine if increased dye accessibility was correlated with increased siRNA release, additional samples were run on a 20% polyacrylamide gel to resolve free siRNA (Figure 4b). Based on these data, it is apparent that siRNA does not become free to migrate into the gel until a pH of 4.5 is achieved. Taken together these assays imply a pH dependent mechanism for particle disassembly and siRNA release, with a lower pH (4.5) required for siRNA to be completely released than that required to initiate particle disassembly (pH 5.5). In contrast to p5RHH, p5RWR is unable to respond to pH as demonstrated by a lack of TOPRO3 fluorescence at pH ≤ 5.5 (Figure 4a), and a lack of siRNA release as measured by gel mobility (Figure 5b).
Figure 4.
(a) Fluorescence from TOPRO3 binding to siRNA increases dramatically at pH ≤ 5.5 when packaged via p5RHH (
 ), but not the non-functioning peptide p5RWR (
), but not the non-functioning peptide p5RWR (
 ). (b) Polyacrylamide gel electrophoresis confirms that p5RHH releases siRNA at pH 4.5 but p5RWR shows no pH-dependent release. (c) p5RHH is also released at low pH with an increase in p5RHH release at pH ≤ 5.5. (d) Freed p5RHH is capable of hemolysis, leading to increased hemoglobin release at pH ≤ 5.5. (e–h) Acridine orange release assays show that p5RHH/siRNA nanoparticles are able to disrupt endosomes (h) when tested in tissue culture, as exhibited by dye release similar to that of 100μM chloroquine (f), whereas p5RWR cannot (g). Scale bar 50μm.
). (b) Polyacrylamide gel electrophoresis confirms that p5RHH releases siRNA at pH 4.5 but p5RWR shows no pH-dependent release. (c) p5RHH is also released at low pH with an increase in p5RHH release at pH ≤ 5.5. (d) Freed p5RHH is capable of hemolysis, leading to increased hemoglobin release at pH ≤ 5.5. (e–h) Acridine orange release assays show that p5RHH/siRNA nanoparticles are able to disrupt endosomes (h) when tested in tissue culture, as exhibited by dye release similar to that of 100μM chloroquine (f), whereas p5RWR cannot (g). Scale bar 50μm.
Figure 5.
(a) Knockdown of GFP in B16 GFP cells reveals that only p5RHH can successfully deliver GFP siRNA to the cytoplasm, whereas p5RWR can not even with endosomal escape induced by chloroquine. (b) Flow cytometry reveals both p5RWR and p5RHH deliver similar amounts of alexa 488-labeled siRNA. Untreated control (
 ); 50nM a488 siRNA/p5RWR (
); 50nM a488 siRNA/p5RWR (
 ); 50nM a88 siRNA/p5RWR + chloroquine (
); 50nM a88 siRNA/p5RWR + chloroquine (
 ); 50nM a488 siRNA/p5RHH (
); 50nM a488 siRNA/p5RHH (
 ); 50nM a88 siRNA/p5RHH + chloroquine (
); 50nM a88 siRNA/p5RHH + chloroquine (
 ). Confocal microscopy (scale bar 10μm) reveals that p5RWR (c) delivers siRNA by remains in punctate vesicles whereas p5RHH achieves cytoplasmic distribution (d). Simultaneous incubation with chloroquine is required to release siRNA to the cytoplasm when transfected by p5RWR (e) but has no effect on p5RHH-mediated transfection (f).
). Confocal microscopy (scale bar 10μm) reveals that p5RWR (c) delivers siRNA by remains in punctate vesicles whereas p5RHH achieves cytoplasmic distribution (d). Simultaneous incubation with chloroquine is required to release siRNA to the cytoplasm when transfected by p5RWR (e) but has no effect on p5RHH-mediated transfection (f).
To corroborate particle disassembly, pH-dependent p5RHH release from p5RHH/siRNA nanoparticles was quantified after dialysis through a 10K dialysis membrane. These assays reveal that approximately 40% of p5RHH remained free after particle assembly, and a strong release of p5RHH occurred at pH ≤ 5.5 (Figure 4c). This pH dependence matches the pH dependence seen for siRNA dye binding confirming that pH does indeed trigger nanoparticle disassembly and subsequent release of both p5RHH and siRNA. The lytic capacity of liberated p5RHH can be quantified in vitro with red blood cell (RBC) hemolysis assays. When incubated at decreasing pH, the ability of p5RHH/siRNA nanoparticles to lyse RBC is enhanced, due to the release of free p5RHH at pH ≤ 5.5 (Figure 4d). These assays were performed at 4°C to decrease the rate of auto-hemolysis observed at higher temperatures. RBC hemolysis indicates that free p5RHH is capable of lysing membrane-bound structures and could potentially disrupt endosomal membranes in intact cells. A more complete characterization p5RHH’s hemolytic properties is provided in Supplementary Figure 5.
Endosomal disruption in living cells was observed by acridine orange staining. Cells were first loaded with acridine orange (10μM, 15 minutes), which fluoresces red at low pH in the endosome but green at cytoplasmic pH.43 Endosomal disruption can be visualized by an increase in cytoplasmic green fluorescence in the presence of 100μM chloroquine (Figure 4f). Similarly, cells transfected with p5RHH/siRNA also released acridine orange from cytoplasmic endosomal vesicles, confirming efficient endosomal disruption, whereas cells transfected with p5RWR/siRNA nanoparticles did not exhibit endosomal disruption (Figure 4h,g). These results highlight the importance of nanoparticle disassembly and release of membrane-active peptide measured in vitro for endosomal disruption in a cellular context. While p5RHH/siRNA nanoparticles are pH responsive and release p5RHH for endosomal disruption, p5RWR/siRNA nanoparticles do not disassemble and do not alter endosomal integrity.
One potential mechanism for the pH-responsive properties of p5RHH/siRNA nanoparticles appears to be protonation of histidine residues. With a pKa of 6, histidine likely provides the critical trigger for particle disassembly because increased siRNA dye binding and p5RHH release are recorded at pH < pKa of histidine. Traditionally, protonation of histidine has often been used as a trigger for siRNA delivery in the context of the proton sponge effect, in which the buffering capacity of histidine-containing polymers leads to the accumulation of Cl− counterions and ultimately osmotic rupture of the endosome.30, 44–48 In comparison to methods relying on endosomal buffering for osmotic rupture, the presence of only two histidine residues in our peptide suggests that these proposed modifications to melittin likely do not yield adequate buffering capacity to achieve the proton-sponge effect for endosomal escape. As an example, Lo and Wang have shown that TAT must be augmented by at least ten histidine residues for successful nucleic acid release into the cytoplasm.30 While we cannot completely rule out some contribution of the proton-sponge effect to the endosomolysis by p5RHH observed for acridine orange release, the need for only two histidine residues is an indication that pH triggers particle disassembly and subsequent release of the membrane lytic peptide. Moreover, based on our hemolysis studies, free p5RHH causes RBC disruption at concentrations greater than 100μM (Supplementary Figure 5b), well below the endosomal osmoticity required for osmotic disruption by chloroquine (>5mM).49 Consequently, osmotic rupture likely plays only a minor role if any.
When examining the ability of p5RHH to deliver GFP siRNA to B16-GFP cells, a strong decrease in GFP expression at 50nM siRNA is observed by western blotting 24 hours after transfection (Figure 5a). Moreover, transfection of cells in the presence of 50μM chloroquine, a known endosomolytic agent, does not improve knockdown.50 The lack of additional knockdown by chloroquine verifies that p5RHH itself is able to fully and efficiently release siRNA from the endosomal compartment, a finding that is visualized by confocal microscopy (Figure 5d,f).
Despite nearly equal uptake of p5RWR/siRNA nanoparticles (Figure 5b), p5RWR is unable to induce GFP knockdown even in the presence of chloroquine (Figure 5a). Confocal microscopy reveals a high degree of endosomal entrapment, suggesting p5RWR/siRNA nanoparticles do not reach the cytoplasm (Figure 5c), unless treated with chloroquine (Figure 5e). The fact that GFP knockdown remains impaired despite endosomal release by chloroquine (Figure 5a) indicates that siRNA accessibility to the RNA induced silencing complex is impaired, reflecting the poor siRNA release from p5RWR based nanoparticles observed by TOPRO3 binding and gel mobility shift assays in vitro (Figure 4a,b).
These data highlight that the ability of p5RHH/siRNA nanoparticles to disassemble in response to low pH is crucial for siRNA delivery to the cytoplasm. Specifically, nanoparticle disassembly with siRNA release from the vector and concurrent endosomolysis by p5RHH is a coordinated event yielding access of free siRNA to the cytoplasmic compartment. The essential role of coordinated siRNA release and endosomal escape in successful siRNA transfection is well known.51 For example, premature siRNA release in the endosome allows siRNA degradation by endosomal hydrolases. On the other hand, peptides which bind too strongly to siRNA, are also hypothesized to prevent successful RNAi.52 Consequently, siRNA release from p5RHH/siRNA nanoparticles must be concurrent with endosomal escape for maximal mRNA degradation.
The therapeutic potential of albumin coated p5RHH/siRNA nanoparticles was demonstrated by the highly efficient transfection of siRNAs targeting both the canonical and non-canonical NFkB pathways in F8 cells, a murine model of HTLV-1 induced ATLL. The transcription factor NFkB was chosen as a therapeutic target due to its central role in ATLL, where it promotes resistance to chemotherapy by driving the expression of anti-apoptotic proteins.53–55 While small molecule proteasome inhibitors and inhibitors of the IKK complex can decrease NFkB activation in some ATLL disease models,56–58 questions regarding their specificity and ability to inactivate NFkB in vivo highlight the need for more specific therapeutics.59–61 The potential synergy provided by direct inhibition of both canonical and non-canonical NFkB pathways via siRNA may be the key to successful blockade of NFkB signaling required for therapeutic success.
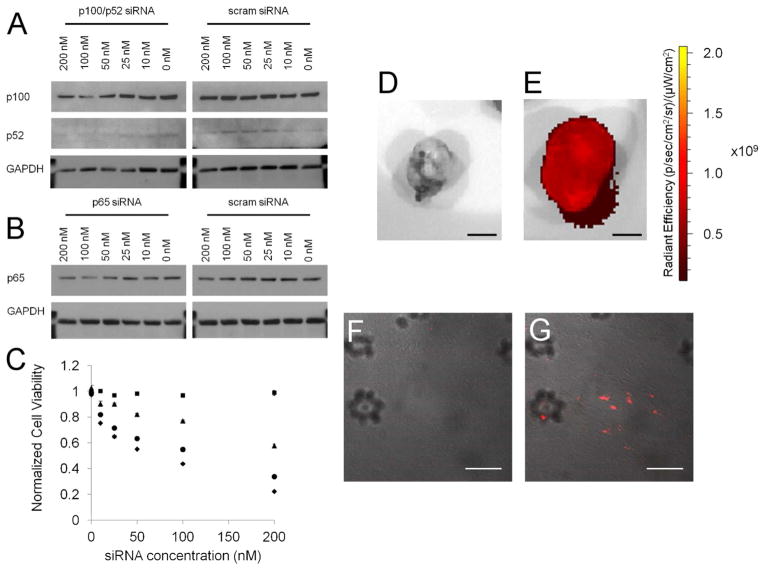
siRNAs were chosen to target the p65 subunit of the canonical pathway and p100/p52 subunit of the non-canonical pathway. Western blotting performed 24 hours after transfection revealed a dose-dependent decrease in the expression of both proteins that was not seen when cells were transfected with a scrambled siRNA control (Figure 6a,b). Alamar blue assays (Figure 6c) 48 hours after transfection demonstrate that knockdown of these pathways in vitro is therapeutically relevant as a strong decrease in cell viability is recorded with both p65 and p100/p52 siRNAs. Moreover, it is clear that blockade of the non-canonical NFkB pathway with p100/p52 siRNA (IC50~100nM) is superior to blockade of the canonical pathway (IC50~200nM) in this cell line. As reviewed by Rauch and Ratner, the non-canonical pathway plays a more prominent role in promoting anti-apoptotic protein expression than does the canonical pathway, labeling it as the more desirable target for modulating the proliferation of ATLL cells.53 Our data utilizing p5RHH-mediated siRNA delivery not only confirm this hypothesis but also reveal a synergistic response when targeting both the canonical and non-canonical NFkB pathways with a single p5RHH/siRNA formulation simultaneously packaging both p65 and p100/p52 siRNAs. Use of this dual targeted p5RHH/siRNA formulation improves NFkB blockade-mediated cell death, with an IC50 ~50nM. It is important to note that despite the ability to lyse RBC in vitro and endosomal membranes in vivo, transfection with scrambled siRNA does not result in any toxicity of F8 cells. Work in our lab has shown that N-terminal truncation of melittin decreases its lytic capacity by 2 orders of magnitude (unpublished observation), and while it appears that p5RHH is able to lyse endosomes at high concentration, p5RHH is safe after endosomal release and dilution in the cytoplasm.
Figure 6.
(a,b) Western blotting demonstrates a dose-dependent decrease in p100/p52 or p65 expression that is not seen when treating F8 cells with scrambled siRNA. (c) Alamar blue assays 48 hours post transfection reveals that scrambled siRNA (■) does not affect F8 cell viability. Knockdown of the canonical NFkB pathway with p65 siRNA (▲) has an IC50 of nearly 200nM. Targeting the non-canonical NFkB pathway with p100/p52 siRNA (●) yields an IC50 of 100nM. However, a nanoparticle formulation simultaneously carrying siRNA to block both canonical and non-canonical NFkB pathways (◆) improves the IC50 to 50nM. IVIS imaging (scale bar 5mm) reveals tumor localization of Cy5.5 labeled siRNA to the tumor of treated mice (e), and is confirmed by confocal microscopy (g) (scale bar 50μm). Non-treated controls shown for comparison (d, f)
Given the safety of p5RHH in tissue culture, pilot experiments were conducted to examine tumor localization of p5RHH/siRNA nanoparticles when delivered in vivo. IVIS imaging and confocal microscopy reveal delivery of Cy5.5 labeled scrambled siRNA to the tumor periphery (Figure 6d–g, Supplemental Figure 6) when introduced by tail-vein injection into mice carrying spontaneous ATLL tumors at a dose of 1mg/kg. IVIS imaging of resected organs reveals minimal uptake in traditional clearance organs such as the liver and spleen, but suggest renal clearance, as previously observed with some polyplexes and lipoplexes.62–63 The shift in clearance away from the liver and spleen suggests a potential role for albumin coating in protecting p5RHH/siRNA nanoparticles from opsonization in accordance with albumin’s previously described ability to act as a disopsonin.64–65
Prior attempts to target NFkB expression itself have focused on the use of naked antisense DNA oligonucleotides or lentiviral shRNA expression, which have limited therapeutic potential.66–67 The use of antisense oligonucleotides is inefficient, requiring an order of magnitude more oligonucleotide than our current siRNA formulation in vitro. On the other hand, viral vectors for shRNA expression present myriad challenges for human trials ranging from induction of cancer to toxicity associated with saturation effects.68–70 Due to the ability to simultaneously target both NFkB pathways, we believe that the current siRNA approach offers proof of concept that the use of p5RHH for highly efficient, low toxicity transfection of NFkB targeted siRNA reflects a synergistic strategy for the treatment of ATLL or other disease processes that are driven by NFkB induction.
CONCLUSION
In summary, membrane-lytic peptides can be recruited as endosomal escape agents to promote the cytoplasmic delivery of siRNA by preventing the siRNA entrapment associated with alternative PTD-mediated transfection. We report an albumin-stabilized p5RHH/siRNA formulation with a final size of ~55 ± 18 nm which enters cells via macropinocytosis. In this work, we have utilized flow cytometry to study both siRNA-mediated knockdown of GFP-PEST as well as endocytosis of fluorescently labeled siRNA-containing nanoparticles. The utility of concurrently quantifying both knockdown and uptake allows careful dissection of the cellular processing of siRNA containing nanoparticles exemplified in our studies revealing the importance of endosomal acidification for successful siRNA delivery to the cytoplasm by p5RHH. These methods can be applied generally to study additional steps in the uptake and processing or siRNA carrying nanoparticles. In the case of p5RHH, endosomal acidification provides a trigger for pH-mediated particle disassembly with concurrent siRNA release and endosomal escape brought on by release of free p5RHH. When utilized for the simultaneous transfection of p65 and p100/p52 siRNAs in a model of ATLL, p5RHH mediates a synergistic decrease in cell viability suggesting the potential of further in vivo studies. We believe that the unique ability of p5RHH/siRNA nanoparticles to efficiently coordinate peptide and siRNA release with endosomal escape portends potential for the use of p5RHH-mediated transfection in a variety of disease substrates. Furthermore, analysis of p5RHH’s mechanism of action provides insight that can guide the further development of future peptide vectors for siRNA transfection. We expect that the formulation methodology reported for p5RHH can be applied broadly to peptide/siRNA nanoparticles to prevent aggregation and potentially decrease opsonization.
MATERIALS AND METHODS
Preparation of peptide/siRNA nanoparticles and analysis
Melittin derivatives p5RHH (VLTTGLPALISWIRRRHRRHC) and p5RWR (VLTTGLPALISWIKRKRQQRWRRRR) were synthesized by Genscript (Piscataway, NJ), dissolved at 10mM in RNAse/DNAse free water (Sigma, St. Louis, MO) and stored in 4μl aliquots at −80°C before use. p5RHH/siRNA transfection complexes were prepared by diluting p5RHH 1:200 in Phosphate Buffered Saline (PBS, Gibco), vortexed for 30 seconds followed by addition of siRNA (stock concentration of 10μM in 1x siRNA buffer (Thermo Scientific, Waltham, MA)) to achieve a peptide to siRNA ratio of 100 to 1, and incubated for 40 minutes at 37°C with shaking in an Eppendorf Thermomixer R. For animal experiments, peptide and siRNA were incubated at a 10 fold higher concentration for 10 minutes on ice. Wet-mode atomic force microscopy was performed by ARC Technologies (White Bear Lake, MN).
Cell culture
B16–F10 (ATCC, Manassas, VA) cell lines were maintained under standard cell culture conditions (37°C and 5% CO2 in a humidified incubator) in DMEM (Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum (Gibco). F8 cells were generously provided by the Ratner lab and cultured in RPMI (Gibco) supplemented with 10% fetal bovine serum in accordance with previously publications.
Uptake Inhibition by Flow Cytometry
B16–F10 cells were incubated with alexa 488-labeled siRNA packaged with p5RHH (25nM), FITC-Transferrin (5μg/ml, Life Technologies) or 70kDa FITC-Dextran (100μg/ml, Sigma) in the presence or absence of endocytosis inhibitors for 40 minutes. After incubation, cells were washed 3x in PBS trypsinized and resuspended in FACS buffer (HBSS with 0.2% FBS and 0.5mM EDTA) for flow cytometry analysis. Inhibitors were used as follows: EIPA (80μM, Sigma), filipin (100μg/ml, Sigma), and PAO (10μM Sigma).
Confocal microscopy
B16–F10 cells were cultured on glass coverslips overnight before incubation with p5RHH nanoparticles and appropriate uptake markers for 40 minutes or 24 hours. p5RHH/Cy-3 siRNA nanoparticles were added at a final siRNA concentration of 200nM in the presence of either 70kDa FITC-Dextran (10mg/ml) or FITC-Transferrin (25μg/ml). After the incubation, cells were washed on ice 3x in PBS for 10 minutes and fixed in 4% paraformaldehyde before mounting on glass slides (Vectashield Mounting Medium with DAPI, Vector Labs, Burlingame, CA). Cells were imaged on a Zeiss Meta 510 (Thornwood, NY).
Analysis of GFP Knockdown
B16 GFP cells were plated at 150,000 cells/well in 6 well plates and transfected 12 hours later at a final concentration of 50nM siRNA in 1mL of 10% DMEM in the presence or absence of 1μM bafilomycin A1 (1mM stock in DMSO, Sigma). 24 hours after B16-GFP cells were transfected with p5RHH/siRNA nanoparticles containing GFP specific or scrambled siRNA, cells were trypsinized and resuspended in FACS buffer (0.2% FBS and 0.5mM EDTA) for analysis of GFP fluorescence. eGFP siRNA (Sense: 5′-GACGUAAACGGCCACAAGUUC-3′) was purchased from Sigma. Scrambled siRNA was purchased from Qiagen (Valencia, CA)
siRNA dye accessibility at low pH
Preformed p5RHH/siRNA nanoparticles were incubated in Hank’s Balanced Salt Solution (HBSS, Gibco) at the indicated pH for 30 minutes in the presence of TOPRO3 (Life Technologies) diluted 1 to 1000. TOPRO3 fluorescence was measured in a 96-well plate with excitation at 642 nm and emission at 661 nm. Fluorescence values were then normalized to siRNA only controls and presented as the average of 3 separate experiments.
pH dependent gel mobility assays
p5RHH/siRNA nanoparticles were incubated in HBSS at the indicated pH for 30 minutes before resolution on a 20% TBE-PAGE gel. siRNA was visualized by staining with SYBR GOLD in 1x TBE (IBIScientific) diluted 1 to 10000 for 15 minutes.
Acridine Orange Staining for Lysosomal Disruption
B16F10 cells plated on coverslips were loaded with acridine orange at 10μM for 15 minutes and washed for 3x in PBS before incubation in the presence of p5RHH/siRNA nanoparticles in 10% DMEM at a final siRNA concentration of 100nM for 12 hours. Alternatively, cells were exposed to chloroquine (Sigma) at 100μM for 15 minutes prior to imaging. Live cells were visualized by fluorescence microscopy on an Olympus BX610 (Tokyo, Japan).
RBC Hemolysis
Rabbit red blood cells (RBC) were isolated from whole blood by centrifugation and washed in PBS 3x before storage at 4°C. Prior to hemolysis studies, RBC were washed 3x in pH appropriate HBSS and diluted 1 to 5000. RBC in pH-specific buffer were then incubated with p5RHH/siRNA nanoparticles for 12 hours. The RBC remnants were pelleted by centrifugation and the hemoglobin content of the supernatant was measured by UV absorbance at 550 nm. Absorbance values were than normalized against maximum lysis by p5RHH only controls and presented as the average of 3 separate experiments.
Analysis of NFkB knockdown in F8 cells
F8 cells were plated in 6 well plates at 200,000 cells/well and transfected at varying siRNA concentrations in a final volume of 1 mL with the designated siRNA. siGENOME mouse NFkB (p65) siRNA 5 and siGENOME mouse NFKB2 (p100/p50) siRNA 1 were purchased from Dharmacon (Lafayette, CO). Scrambled siRNA was purchased from Qiagen (Valencia, CA). 24 hours after transfection, F8 cells were pelleted at 1000 rpm in a Precision AKR-1000. Cell pellets were then resuspended in 100μl RIPA buffer (10 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1.0% Igepal CA-630, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 1 mM EDTA, 5% glycerol) with 1 mM PMSF and Complete Protease Inhibitor Cocktail (Roche) and incubated on ice for 1 hour. Cell lysates were then centrifuged at 4°C for 5 minutes and supernatants stored at −20°C. Lysates were resolved on Nupage Bis-Tris gels (Life Technologies) and transferred to 0.22μm nitrocellulose before blocking in 5% bovine serum albumin (Sigma) in TBS-T. Primary antibodies used were rabbit anti-p65 (1:1000, Cell Signaling, Danvers, MA), rabbit anti-p100/p50 (1:1000, Cell Signaling). Secondary antibody: anti-Rabbit HRP (1:5000, Santa Cruz Biotechnology). Blots were developed using ECL Western Blotting Substrate (Pierce, Rockford, IL).
F8 cell Viability Measurements
F8 cells were plated in 24-well plates 12 hours before transfection at 20,000 cells/well in 400 μl and cultured under standard cell culture conditions. p5RHH/siRNA nanoparticles were prepared and incubated with cells for 48 hours in a final volume of 600 μl before viability measurements using Alamar Blue (Life Technologies. Briefly, Alamar Blue was diluted 1 to 10 into cell culture media and incubated with cells for 2–4 hours. Fluorescence was measured on a fluorescent plate reader with excitation at 570 nm and emission at 585 nm (Varian Cary Eclipse, Agilent Technologies, Santa Clara, CA).
Animal Experiments
The experimental animal protocols were approved by the Animal Care Committee of the Washington University School of Medicine. Transgenic mice with spontaneous tumors were a gift from the Ratner lab.71 Mice with advanced tumors were selected for pilot experiments and injected with a single dose at 1mg/kg 24 hours before sacrifice. Animals were perfused with saline, and tumors were excised for IVIS imaging and frozen sectioning.
Supplementary Material
Acknowledgments
We thank K. Boles for help producing the B16-GFP cell line used for siRNA screening and R. Roth for assistance with electron microscopy. Research described here was primarily supported by grants from the National Institutes of Health (U01 CA141541 to Dr. Robert Schreiber and RO1 HL073646-08 to SAW) as well as the Sigma Aldrich Predoctoral Fellowship.
Footnotes
Six Supplemental Files are available. Supplementary Figure 1 demonstrates characterization of p5RHH/siRNA nanoparticles by transfection efficiency and Deep-Etch Electron Microscopy. Supplementary Figure 2 confirms minimal p5RHH/siRNA uptake at 4°C. Supplementary Figure 3 reveals siRNA release within 1hr. Supplementary Figure 4 shows that endocytosis inhibitors are specific to the indicated pathway. Supplementary Figure 5 provides analysis of the hemolytic activity of p5RHH. Supplementary Figure 6 shows additional fields of view from confocal microscopy of tumors investigating Cy5.5 labeled siRNA delivery. Supplementary figures are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Shen H, Sun T, Ferrari M. Nanovector Delivery of siRNA for Cancer Therapy. Cancer Gene Ther. 2012;19:367–373. doi: 10.1038/cgt.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miele E, Spinelli GP, Miele E, Di Fabrizio E, Ferretti E, Tomao S, Gulino A. Nanoparticle-Based Delivery of Small Interfering RNA: Challenges for Cancer Therapy. Int J Nanomedicine. 2012;2012:3637–3657. doi: 10.2147/IJN.S23696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and Specific Genetic Interference by Double-Stranded RNA in Caenorhabditis Elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 4.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-Nucleotide RNAs Mediate RNA Interference in Cultured Mammalian Cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 5.Dominska M, Dykxhoorn DM. Breaking Down the Barriers: siRNA Delivery and Endosome Escape. J Cell Sci. 2010;123:1183–1189. doi: 10.1242/jcs.066399. [DOI] [PubMed] [Google Scholar]
- 6.Guzman-Villanueva D, El-Sherbiny IM, Herrera-Ruiz D, Vlassov AV, Smyth HDC. Formulation Approaches to Short Interfering RNA and MicroRNA: Challenges and Implications. J Pharm Sci. 2012;101:4046–4066. doi: 10.1002/jps.23300. [DOI] [PubMed] [Google Scholar]
- 7.Pecot CV, Calin GA, Coleman RL, Lopez-Berestein G, Sood AK. RNA Interference in the Clinic: Challenges and Future Directions. Nat Rev Cancer. 2011;11:59–67. doi: 10.1038/nrc2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Lu Z, Wientjes MG, Au JLS. Delivery of siRNA Therapeutics: Barriers and Carriers. AAPS J. 2010;12:492–503. doi: 10.1208/s12248-010-9210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rettig GR, Behlke MA. Progress Toward in Vivo Use of siRNAs-II. Mol Ther. 2012;20:483–512. doi: 10.1038/mt.2011.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alabi C, Vegas A, Anderson D. Attacking the Genome: Emerging siRNA Nanocarriers from Concept to Clinic. Curr Opin Pharm. 2012;12:427–433. doi: 10.1016/j.coph.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 11.Kesharwani P, Gajbhiye V, Jain NK. A Review of Nanocarriers for the Delivery of Small Interfering RNA. Biomaterials. 2012;33:7138–7150. doi: 10.1016/j.biomaterials.2012.06.068. [DOI] [PubMed] [Google Scholar]
- 12.Zhou J, Shum KT, Burnett J, Rossi J. Nanoparticle-Based Delivery of RNAi Therapeutics: Progress and Challenges. Pharmaceuticals. 2013;6:85–107. doi: 10.3390/ph6010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dokka S, Toledo D, Shi X, Castranova V, Rojanasakul Y. Oxygen Radical-Mediated Pulmonary Toxicity Induced by Some Cationic Liposomes. Pharm Res. 2000;17:521–525. doi: 10.1023/a:1007504613351. [DOI] [PubMed] [Google Scholar]
- 14.Filion MC, Phillips NC. Toxicity and Immunomodulatory Activity of Liposomal Vectors Formulated with Cationic Lipids toward Immune Effector Cells. Biochim Biophys Acta, Biomembr. 1997;1329:345–356. doi: 10.1016/s0005-2736(97)00126-0. [DOI] [PubMed] [Google Scholar]
- 15.Lv H, Zhang S, Wang B, Cui S, Yan J. Toxicity of Cationic Lipids and Cationic Polymers in Gene Delivery. J Controlled Release. 2006;114:100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 16.Soenen SJH, Brisson AR, De Cuyper M. Addressing the Problem of Cationic Lipid-Mediated Toxicity: The Magnetoliposome Model. Biomaterials. 2009;30:3691–3701. doi: 10.1016/j.biomaterials.2009.03.040. [DOI] [PubMed] [Google Scholar]
- 17.Akhtar S, Benter I. Toxicogenomics of Non-Viral Drug Delivery Systems for RNAi: Potential Impact on siRNA-Mediated Gene Silencing Activity and Specificity. Adv Drug Del Rev. 2007;59:164–182. doi: 10.1016/j.addr.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 18.Ballarin-Gonzalez B, Howard KA. Polycation-Based Nanoparticle Delivery of RNAi Therapeutics: Adverse Effects and Solutions. Adv Drug Del Rev. 2012;64:1717–1729. doi: 10.1016/j.addr.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 19.Abes R, Arzumanov AA, Moulton HM, Abes S, Ivanova GD, Iversen PL, Gait MJ, Lebleu B. Cell-Penetrating-Peptide-Based Delivery of Oligonucleotides: An Overview. Biochem Soc Trans. 2007;35:775–779. doi: 10.1042/BST0350775. [DOI] [PubMed] [Google Scholar]
- 20.Hassane FS, Saleh AF, Abes R, Gait MJ, Lebleu B. Cell Penetrating Peptides: Overview and Applications to the Delivery of Oligonucleotides. Cell Mol Life Sci. 2010;67:715–726. doi: 10.1007/s00018-009-0186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morris MC, Deshayes S, Heitz F, Divita G. Cell-Penetrating Peptides: From Molecular Mechanisms to Therapeutics. Biol Cell. 2008;100:201–217. doi: 10.1042/BC20070116. [DOI] [PubMed] [Google Scholar]
- 22.Meade BR, Dowdy SF. Enhancing the Cellular Uptake of siRNA Duplexes Following Noncovalent Packaging with Protein Transduction Domain Peptides. Adv Drug Del Rev. 2008;60:530–536. doi: 10.1016/j.addr.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Endoh T, Ohtsuki T. Cellular siRNA Delivery Using Cell-Penetrating Peptides Modified for Endosomal Escape. Adv Drug Del Rev. 2009;61:704–709. doi: 10.1016/j.addr.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 24.Laufer SD, Detzer A, Sczakiel G, Restle T. Selected Strategies for the Delivery of siRNA in Vitro and in Vivo. In: Erdmann VA, Barciszewski J, editors. RNA Technologies and Their Applications. Springer; Berlin Heidelberg: 2010. pp. 29–58. [Google Scholar]
- 25.Hoyer J, Neundorf I. Peptide Vectors for the Nonviral Delivery of Nucleic Acids. Acc Chem Res. 2012;45:1048–1056. doi: 10.1021/ar2002304. [DOI] [PubMed] [Google Scholar]
- 26.Nakase I, Akita H, Kogure K, Gräslund A, Langel Ü, Harashima H, Futaki S. Efficient Intracellular Delivery of Nucleic Acid Pharmaceuticals Using Cell-Penetrating Peptides. Acc Chem Res. 2012;45:1132–1139. doi: 10.1021/ar200256e. [DOI] [PubMed] [Google Scholar]
- 27.Veldhoen S, Laufer SD, Restle T. Recent Developments in Peptide-Based Nucleic Acid Delivery. Int J Mol Sci. 2008;9:1276–1320. doi: 10.3390/ijms9071276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trabulo S, Cardoso AL, Mano M, de Lima MCP. Cell-Penetrating Peptides—Mechanisms of Cellular Uptake and Generation of Delivery Systems. Pharmaceuticals. 2010;3:961–993. doi: 10.3390/ph3040961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lundberg P, El-Andaloussi S, Sütlü T, Johansson H, Langel Ü. Delivery of Short Interfering RNA Using Endosomolytic Cell-Penetrating Peptides. FASEB J. 2007;21:2664–2671. doi: 10.1096/fj.06-6502com. [DOI] [PubMed] [Google Scholar]
- 30.Lo SL, Wang S. An Endosomolytic TAT Peptide Produced by Incorporation of Histidine and Cysteine Residues as a Nonviral Vector for DNA Transfection. Biomaterials. 2008;29:2408–2414. doi: 10.1016/j.biomaterials.2008.01.031. [DOI] [PubMed] [Google Scholar]
- 31.Chiu YL, Ali A, Chu C-y, Cao H, Rana TM. Visualizing a Correlation between siRNA Localization, Cellular Uptake, and RNAi in Living Cells. Chem Biol. 2004;11:1165–1175. doi: 10.1016/j.chembiol.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 32.Shiraishi T, Nielsen PE. Enhanced Delivery of Cell-Penetrating Peptide–Peptide Nucleic Acid Conjugates by Endosomal Disruption. Nat Protocols. 2006;1:633–636. doi: 10.1038/nprot.2006.92. [DOI] [PubMed] [Google Scholar]
- 33.Wooddell CI, Rozema DB, Hossbach M, John M, Hamilton HL, Chu Q, Hegge JO, Klein JJ, Wakefield DH, Oropeza CE, et al. Hepatocyte-Targeted RNAi Therapeutics for the Treatment of Chronic Hepatitis B Virus Infection. Mol Ther. 2013;21:973–985. doi: 10.1038/mt.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Asbeck AH, Beyerle A, McNeill H, Bovee-Geurts PHM, Lindberg S, Verdurmen WPR, Hällbrink M, Langel Ü, Heidenreich O, Brock R. Molecular Parameters of siRNA–Cell Penetrating Peptide Nanocomplexes for Efficient Cellular Delivery. ACS Nano. 2013;7:3797–3807. doi: 10.1021/nn305754c. [DOI] [PubMed] [Google Scholar]
- 35.Hou KK, Pan H, Lanza GM, Wickline SA. Melittin Derived Peptides for Nanoparticle Based siRNA Transfection. Biomaterials. 2013;34:3110–3119. doi: 10.1016/j.biomaterials.2013.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicolás P, Saleta M, Troiani H, Zysler R, Lassalle V, Ferreira ML. Preparation of Iron Oxide Nanoparticles Stabilized with Biomolecules: Experimental and Mechanistic Issues. Acta Biomater. 2013;9:4754–4762. doi: 10.1016/j.actbio.2012.09.040. [DOI] [PubMed] [Google Scholar]
- 37.Veldhoen S, Laufer SD, Trampe A, Restle T. Cellular Delivery of Small Interfering RNA by a Non-Covalently Attached Cell-Penetrating Peptide: Quantitative Analysis of Uptake and Biological Effect. Nucleic Acids Res. 2006;34:6561–6573. doi: 10.1093/nar/gkl941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ivanov AI. Pharmacological Inhibition of Endocytic Pathways: Is It Specific Enough to Be Useful? In: Ivanov AI, editor. Exocytosis and Endocytosis. Humana Press; 2008. pp. 15–33. [DOI] [PubMed] [Google Scholar]
- 39.Vercauteren D, Vandenbroucke RE, Jones AT, Rejman J, Demeester J, De Smedt SC, Sanders NN, Braeckmans K. The Use of Inhibitors to Study Endocytic Pathways of Gene Carriers: Optimization and Pitfalls. Mol Ther. 2010;18:561–569. doi: 10.1038/mt.2009.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakase I, Takeuchi T, Tanaka G, Futaki S. Methodological and Cellular Aspects That Govern the Internalization Mechanisms of Arginine-Rich Cell-Penetrating Peptides. Adv Drug Del Rev. 2008;60:598–607. doi: 10.1016/j.addr.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 41.Xiao YT, Xiang LX, Shao JZ. Vacuolar H+-Atpase. Int J Biochem Cell Biol. 2008;40:2002–2006. doi: 10.1016/j.biocel.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 42.Merion M, Schlesinger P, Brooks RM, Moehring JM, Moehring TJ, Sly WS. Defective Acidification of Endosomes in Chinese Hamster Ovary Cell Mutants “Cross-Resistant” To Toxins and Viruses. Proc Natl Acad Sci US A. 1983;80:5315–5319. doi: 10.1073/pnas.80.17.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eissenberg LG, Goldman WE, Schlesinger PH. Histoplasma Capsulatum Modulates the Acidification of Phagolysosomes. J Exp Med. 1993;177:1605–1611. doi: 10.1084/jem.177.6.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chou ST, Leng Q, Scaria P, Woodle M, Mixson AJ. Selective Modification of HK Peptides Enhances siRNA Silencing of Tumor Targetsin Vivo. Cancer Gene Ther. 2011;18:707–716. doi: 10.1038/cgt.2011.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langlet-Bertin B, Leborgne C, Scherman D, Bechinger B, Mason AJ, Kichler A. Design and Evaluation of Histidine-Rich Amphipathic Peptides for siRNA Delivery. Pharm Res. 2010;27:1426–1436. doi: 10.1007/s11095-010-0138-2. [DOI] [PubMed] [Google Scholar]
- 46.Pichon C, Roufaï MB, Monsigny M, Midoux P. Histidylated Oligolysines Increase the Transmembrane Passage and the Biological Activity of Antisense Oligonucleotides. Nucleic Acids Res. 2000;28:504–512. doi: 10.1093/nar/28.2.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanaka K, Kanazawa T, Ogawa T, Takashima Y, Fukuda T, Okada H. Disulfide Crosslinked Stearoyl Carrier Peptides Containing Arginine and Histidine Enhance siRNA Uptake and Gene Silencing. Int J Pharm. 2010;398:219–224. doi: 10.1016/j.ijpharm.2010.07.038. [DOI] [PubMed] [Google Scholar]
- 48.Yu W, Pirollo KF, Yu B, Rait A, Xiang L, Huang W, Zhou Q, Ertem GZ, Chang EH. Enhanced Transfection Efficiency of a Systemically Delivered Tumor-targeting Immunolipoplex by Inclusion of a pH-sensitive Histidylated Oligolysine Peptide. Nucleic Acids Res. 2004;32:e48–e48. doi: 10.1093/nar/gnh049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krogstad DJ, Schlesinger PH. A Perspective on Antimalarial Action: Effects of Weak Bases on Plasmodium Falciparum. Biochem Pharmacol. 1986;35:547–552. doi: 10.1016/0006-2952(86)90345-x. [DOI] [PubMed] [Google Scholar]
- 50.Caron NJ, Quenneville SP, Tremblay JP. Endosome Disruption Enhances the Functional Nuclear Delivery of TAT-Fusion Proteins. Biochem Biophys Res Commun. 2004;319:12–20. doi: 10.1016/j.bbrc.2004.04.180. [DOI] [PubMed] [Google Scholar]
- 51.Alabi CA, Love KT, Sahay G, Stutzman T, Young WT, Langer R, Anderson DG. FRET-Labeled siRNA Probes for Tracking Assembly and Disassembly of siRNA Nanocomplexes. ACS Nano. 2012;6:6133–6141. doi: 10.1021/nn3013838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Geoghegan JC, Gilmore BL, Davidson BL. Gene Silencing Mediated by siRNA-Binding Fusion Proteins Is Attenuated by Double-Stranded RNA-Binding Domain Structure. Mol Ther — Nucleic Acids. 2012;1:e53. doi: 10.1038/mtna.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rauch DA, Ratner L. Targeting HTLV-1 Activation of NFκB in Mouse Models and ATLL Patients. Viruses. 2011;3:886–900. doi: 10.3390/v3060886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Keutgens A, Robert I, Viatour P, Chariot A. Deregulated NF-κB Activity in Haematological Malignancies. Biochem Pharmacol. 2006;72:1069–1080. doi: 10.1016/j.bcp.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 55.Horie R. NF-κB in Pathogenesis and Treatment of Adult T-Cell Leukemia/Lymphoma. Int Rev Immunol. 2007;26:269–281. doi: 10.1080/08830180701703364. [DOI] [PubMed] [Google Scholar]
- 56.Dewan MZ, Terashima K, Taruishi M, Hasegawa H, Ito M, Tanaka Y, Mori N, Sata T, Koyanagi Y, Maeda M, et al. Rapid Tumor Formation of Human T-Cell Leukemia Virus Type 1-Infected Cell Lines in Novel Nod-Scid/Γcnull Mice: Suppression by an Inhibitor against NF- B. J Virol. 2003;77:5286–5294. doi: 10.1128/JVI.77.9.5286-5294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mori N, Yamada Y, Ikeda S, Yamasaki Y, Tsukasaki K, Tanaka Y, Tomonaga M, Yamamoto N, Fujii M. Bay 11-7082 Inhibits Transcription Factor Nf-κB and Induces Apoptosis of HTLV-I–Infected T-Cell Lines and Primary Adult T-Cell Leukemia Cells. Blood. 2002;100:1828–1834. doi: 10.1182/blood-2002-01-0151. [DOI] [PubMed] [Google Scholar]
- 58.Shu ST, Nadella MVP, Dirksen WP, Fernandez SA, Thudi NK, Werbeck JL, Lairmore MD, Rosol TJ. A Novel Bioluminescent Mouse Model and Effective Therapy for Adult T-Cell Leukemia/Lymphoma. Cancer Res. 2007;67:11859–11866. doi: 10.1158/0008-5472.CAN-07-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mitra-Kaushik S, Harding JC, Hess JL, Ratner L. Effects of the Proteasome Inhibitor PS-341 on Tumor Growth in HTLV-1 Tax Transgenic Mice and Tax Tumor Transplants. Blood. 2004;104:802–809. doi: 10.1182/blood-2003-11-3967. [DOI] [PubMed] [Google Scholar]
- 60.Hajj HE, El-Sabban M, Hasegawa H, Zaatari G, Ablain J, Saab ST, Janin A, Mahfouz R, Nasr R, Kfoury Y, et al. Therapy-Induced Selective Loss of Leukemia-Initiating Activity in Murine Adult T Cell Leukemia. J Exp Med. 2010;207:2785–2792. doi: 10.1084/jem.20101095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watanabe M, Ohsugi T, Shoda M, Ishida T, Aizawa S, Maruyama-Nagai M, Utsunomiya A, Koga S, Yamada Y, Kamihira S, et al. Dual Targeting of Transformed and Untransformed HTLV-1–Infected T Cells by DHMEQ, a Potent and Selective Inhibitor of NF-κB, as a Strategy for Chemoprevention and Therapy of Adult T-Cell Leukemia. Blood. 2005;106:2462–2471. doi: 10.1182/blood-2004-09-3646. [DOI] [PubMed] [Google Scholar]
- 62.Shi B, Keough E, Matter A, Leander K, Young S, Carlini E, Sachs AB, Tao W, Abrams M, Howell B, et al. Biodistribution of Small Interfering RNA at the Organ and Cellular Levels after Lipid Nanoparticle-Mediated Delivery. J Histochem Cytochem. 2011;59:727–740. doi: 10.1369/0022155411410885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zuckerman JE, Choi CHJ, Han H, Davis ME. Polycation-siRNA Nanoparticles Can Disassemble at the Kidney Glomerular Basement Membrane. Proc Natl Acad Sci US A. 2012;109:3137–3142. doi: 10.1073/pnas.1200718109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thiele L, Diederichs JE, Reszka R, Merkle HP, Walter E. Competitive Adsorption of Serum Proteins at Microparticles Affects Phagocytosis by Dendritic Cells. Biomaterials. 2003;24:1409–1418. doi: 10.1016/s0142-9612(02)00525-2. [DOI] [PubMed] [Google Scholar]
- 65.Ogawara KI, Furumoto K, Nagayama S, Minato K, Higaki K, Kai T, Kimura T. Pre-Coating with Serum Albumin Reduces Receptor-Mediated Hepatic Disposition of Polystyrene Nanosphere: Implications for Rational Design of Nanoparticles. J Controlled Release. 2004;100:451–455. doi: 10.1016/j.jconrel.2004.07.028. [DOI] [PubMed] [Google Scholar]
- 66.Bernal-Mizrachi L, Lovly CM, Ratner L. The Role of NF-κB-1 and NF-κB-2-Mediated Resistance to Apoptosis in Lymphomas. Proc Natl Acad Sci US A. 2006;103:9220–9225. doi: 10.1073/pnas.0507809103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kitajima I, Shinohara T, Bilakovics J, Brown D, Xu X, Nerenberg M. Ablation of Transplanted HTLV-I Tax-Transformed Tumors in Mice by Antisense Inhibition of NF-Kappa B. Science. 1992;258:1792–1795. doi: 10.1126/science.1299224. [DOI] [PubMed] [Google Scholar]
- 68.Raper SE, Chirmule N, Lee FS, Wivel NA, Bagg A, Gao GP, Wilson JM, Batshaw ML. Fatal Systemic Inflammatory Response Syndrome in a Ornithine Transcarbamylase Deficient Patient Following Adenoviral Gene Transfer. Mol Genet Metab. 2003;80:148–158. doi: 10.1016/j.ymgme.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 69.Hacein-Bey-Abina S, Kalle CV, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, Morillon E, et al. LMO2-Associated Clonal T Cell Proliferation in Two Patients after Gene Therapy for Scid-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 70.Ahn M, Witting SR, Ruiz R, Saxena R, Morral N. Constitutive Expression of Short Hairpin RNA in Vivo Triggers Buildup of Mature Hairpin Molecules. Hum Gene Ther. 2011;22:1483–1497. doi: 10.1089/hum.2010.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rauch D, Gross S, Harding J, Niewiesk S, Lairmore M, Piwnica-Worms D, Ratner L. Imaging Spontaneous Tumorigenesis: Inflammation Precedes Development of Peripheral NK Tumors. Blood. 2009;113:1493–1500. doi: 10.1182/blood-2008-07-166462. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.