Abstract
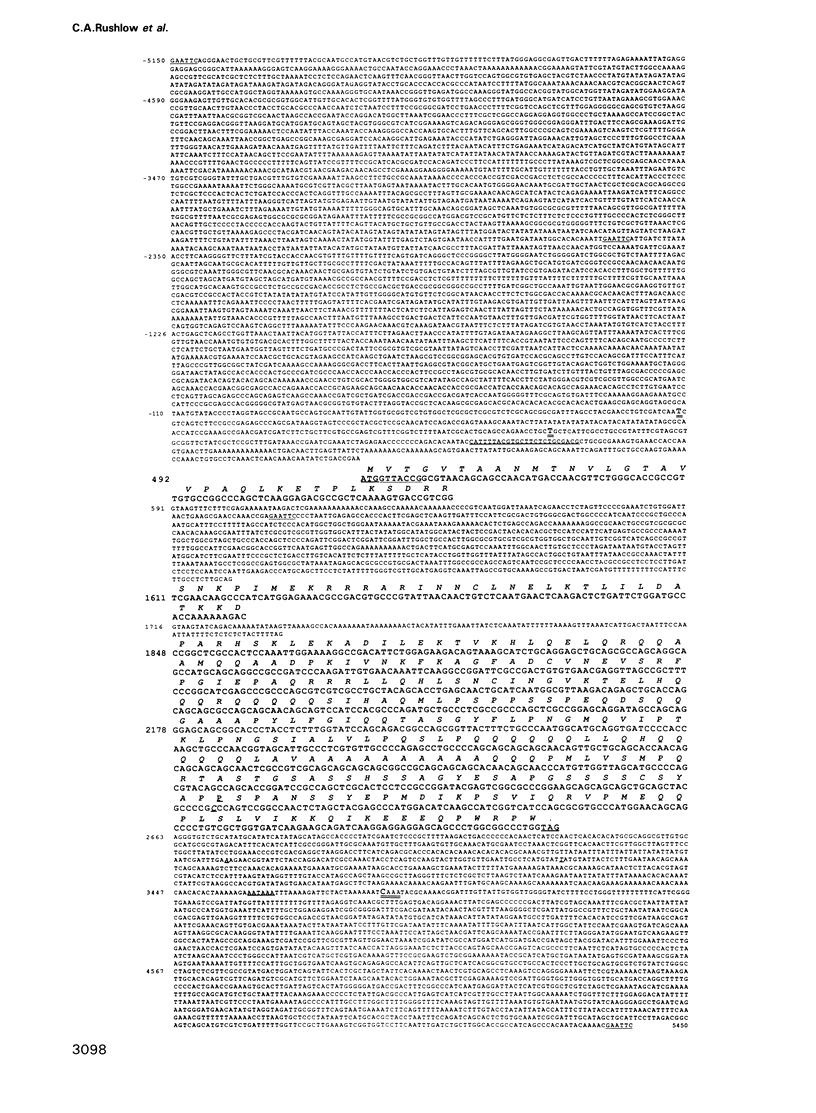
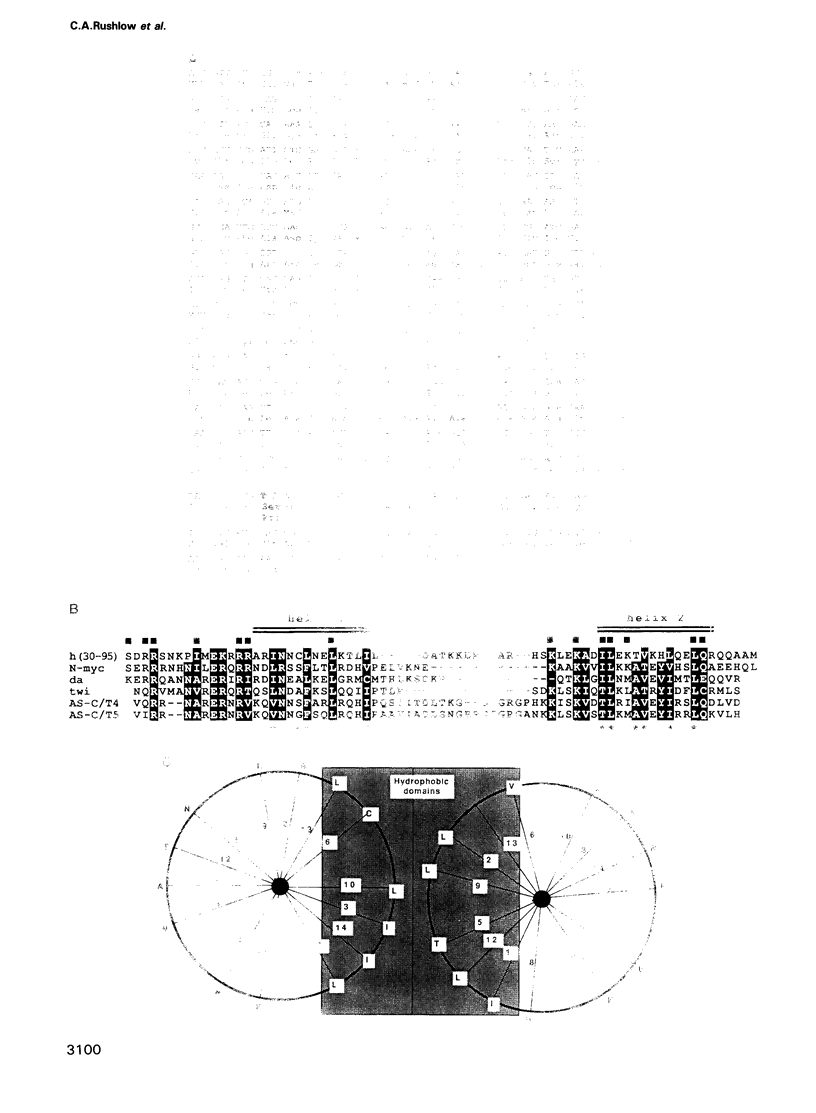
The Drosophila segmentation gene, hairy (h), acts to regulate embryonic segmentation and bristle pattern. We present the DNA sequence of the h gene and of h cDNAs, thereby deducing the organization of the h transcripts. The h gene encodes a 337 amino acid protein that acts in both embryonic segmentation and adult bristle patterning. The h protein includes a domain that shows extensive similarity to a domain of the proto-oncogene N-myc that may be involved in DNA binding and/or protein dimerization. We discuss mechanisms of h action as a transcriptional regulator.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akam M. The molecular basis for metameric pattern in the Drosophila embryo. Development. 1987 Sep;101(1):1–22. [PubMed] [Google Scholar]
- Alonso M. C., Cabrera C. V. The achaete-scute gene complex of Drosophila melanogaster comprises four homologous genes. EMBO J. 1988 Aug;7(8):2585–2591. doi: 10.1002/j.1460-2075.1988.tb03108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoist C., O'Hare K., Breathnach R., Chambon P. The ovalbumin gene-sequence of putative control regions. Nucleic Acids Res. 1980 Jan 11;8(1):127–142. doi: 10.1093/nar/8.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Spliced early mRNAs of simian virus 40. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1274–1278. doi: 10.1073/pnas.75.3.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Bopp D., Burri M., Baumgartner S., Frigerio G., Noll M. Conservation of a large protein domain in the segmentation gene paired and in functionally related genes of Drosophila. Cell. 1986 Dec 26;47(6):1033–1040. doi: 10.1016/0092-8674(86)90818-4. [DOI] [PubMed] [Google Scholar]
- Botas J., Moscoso del Prado J., García-Bellido A. Gene-dose titration analysis in the search of trans-regulatory genes in Drosophila. EMBO J. 1982;1(3):307–310. doi: 10.1002/j.1460-2075.1982.tb01165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campuzano S., Balcells L., Villares R., Carramolino L., García-Alonso L., Modolell J. Excess function hairy-wing mutations caused by gypsy and copia insertions within structural genes of the achaete-scute locus of Drosophila. Cell. 1986 Jan 31;44(2):303–312. doi: 10.1016/0092-8674(86)90764-6. [DOI] [PubMed] [Google Scholar]
- Carroll S. B., Laughon A., Thalley B. S. Expression, function, and regulation of the hairy segmentation protein in the Drosophila embryo. Genes Dev. 1988 Jul;2(7):883–890. doi: 10.1101/gad.2.7.883. [DOI] [PubMed] [Google Scholar]
- Carroll S. B., Scott M. P. Zygotically active genes that affect the spatial expression of the fushi tarazu segmentation gene during early Drosophila embryogenesis. Cell. 1986 Apr 11;45(1):113–126. doi: 10.1016/0092-8674(86)90543-x. [DOI] [PubMed] [Google Scholar]
- Caudy M., Vässin H., Brand M., Tuma R., Jan L. Y., Jan Y. N. daughterless, a Drosophila gene essential for both neurogenesis and sex determination, has sequence similarities to myc and the achaete-scute complex. Cell. 1988 Dec 23;55(6):1061–1067. doi: 10.1016/0092-8674(88)90250-4. [DOI] [PubMed] [Google Scholar]
- Cavener D. R. Comparison of the consensus sequence flanking translational start sites in Drosophila and vertebrates. Nucleic Acids Res. 1987 Feb 25;15(4):1353–1361. doi: 10.1093/nar/15.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman K. G., Poole S. J., Weir M. P., Soeller W. C., Kornberg T. The invected gene of Drosophila: sequence analysis and expression studies reveal a close kinship to the engrailed gene. Genes Dev. 1987 Mar;1(1):19–28. doi: 10.1101/gad.1.1.19. [DOI] [PubMed] [Google Scholar]
- Dente L., Cesareni G., Cortese R. pEMBL: a new family of single stranded plasmids. Nucleic Acids Res. 1983 Mar 25;11(6):1645–1655. doi: 10.1093/nar/11.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierks P., van Ooyen A., Cochran M. D., Dobkin C., Reiser J., Weissmann C. Three regions upstream from the cap site are required for efficient and accurate transcription of the rabbit beta-globin gene in mouse 3T6 cells. Cell. 1983 Mar;32(3):695–706. doi: 10.1016/0092-8674(83)90055-7. [DOI] [PubMed] [Google Scholar]
- Dingwall C., Laskey R. A. Protein import into the cell nucleus. Annu Rev Cell Biol. 1986;2:367–390. doi: 10.1146/annurev.cb.02.110186.002055. [DOI] [PubMed] [Google Scholar]
- Driever W., Nüsslein-Volhard C. A gradient of bicoid protein in Drosophila embryos. Cell. 1988 Jul 1;54(1):83–93. doi: 10.1016/0092-8674(88)90182-1. [DOI] [PubMed] [Google Scholar]
- Driever W., Nüsslein-Volhard C. The bicoid protein is a positive regulator of hunchback transcription in the early Drosophila embryo. Nature. 1989 Jan 12;337(6203):138–143. doi: 10.1038/337138a0. [DOI] [PubMed] [Google Scholar]
- Frigerio G., Burri M., Bopp D., Baumgartner S., Noll M. Structure of the segmentation gene paired and the Drosophila PRD gene set as part of a gene network. Cell. 1986 Dec 5;47(5):735–746. doi: 10.1016/0092-8674(86)90516-7. [DOI] [PubMed] [Google Scholar]
- Garcia-Bellido A., Merriam J. R. Genetic analysis of cell heredity in imaginal discs of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1971 Sep;68(9):2222–2226. doi: 10.1073/pnas.68.9.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halazonetis T. D., Georgopoulos K., Greenberg M. E., Leder P. c-Jun dimerizes with itself and with c-Fos, forming complexes of different DNA binding affinities. Cell. 1988 Dec 2;55(5):917–924. doi: 10.1016/0092-8674(88)90147-x. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Holmgren R. Cloning sequences from the hairy gene of Drosophila. EMBO J. 1984 Mar;3(3):569–573. doi: 10.1002/j.1460-2075.1984.tb01849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard K., Ingham P. Regulatory interactions between the segmentation genes fushi tarazu, hairy, and engrailed in the Drosophila blastoderm. Cell. 1986 Mar 28;44(6):949–957. doi: 10.1016/0092-8674(86)90018-8. [DOI] [PubMed] [Google Scholar]
- Howard K., Ingham P., Rushlow C. Region-specific alleles of the Drosophila segmentation gene hairy. Genes Dev. 1988 Aug;2(8):1037–1046. doi: 10.1101/gad.2.8.1037. [DOI] [PubMed] [Google Scholar]
- Hunt T. False starts in translational control of gene expression. Nature. 1985 Aug 15;316(6029):580–581. doi: 10.1038/316580a0. [DOI] [PubMed] [Google Scholar]
- Ingham P. W., Pinchin S. M., Howard K. R., Ish-Horowicz D. Genetic Analysis of the Hairy Locus in DROSOPHILA MELANOGASTER. Genetics. 1985 Nov;111(3):463–486. doi: 10.1093/genetics/111.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham P. W. The molecular genetics of embryonic pattern formation in Drosophila. Nature. 1988 Sep 1;335(6185):25–34. doi: 10.1038/335025a0. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D., Howard K. R., Pinchin S. M., Ingham P. W. Molecular and genetic analysis of the hairy locus in Drosophila. Cold Spring Harb Symp Quant Biol. 1985;50:135–144. doi: 10.1101/sqb.1985.050.01.019. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D., Pinchin S. M. Pattern abnormalities induced by ectopic expression of the Drosophila gene hairy are associated with repression of ftz transcription. Cell. 1987 Nov 6;51(3):405–415. doi: 10.1016/0092-8674(87)90636-2. [DOI] [PubMed] [Google Scholar]
- Klämbt C., Knust E., Tietze K., Campos-Ortega J. A. Closely related transcripts encoded by the neurogenic gene complex enhancer of split of Drosophila melanogaster. EMBO J. 1989 Jan;8(1):203–210. doi: 10.1002/j.1460-2075.1989.tb03365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T., Ziff E. The role of the leucine zipper in the fos-jun interaction. Nature. 1988 Dec 15;336(6200):646–651. doi: 10.1038/336646a0. [DOI] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986 Jan 31;44(2):283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988 Jun 24;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Laughon A., Scott M. P. Sequence of a Drosophila segmentation gene: protein structure homology with DNA-binding proteins. Nature. 1984 Jul 5;310(5972):25–31. doi: 10.1038/310025a0. [DOI] [PubMed] [Google Scholar]
- Marshall R. D. Glycoproteins. Annu Rev Biochem. 1972;41:673–702. doi: 10.1146/annurev.bi.41.070172.003325. [DOI] [PubMed] [Google Scholar]
- McCormick F., Clark B. F., la Cour T. F., Kjeldgaard M., Norskov-Lauritsen L., Nyborg J. A model for the tertiary structure of p21, the product of the ras oncogene. Science. 1985 Oct 4;230(4721):78–82. doi: 10.1126/science.3898366. [DOI] [PubMed] [Google Scholar]
- McGinnis W., Levine M. S., Hafen E., Kuroiwa A., Gehring W. J. A conserved DNA sequence in homoeotic genes of the Drosophila Antennapedia and bithorax complexes. 1984 Mar 29-Apr 4Nature. 308(5958):428–433. doi: 10.1038/308428a0. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Kingsbury R. Transcriptional control signals of a eukaryotic protein-coding gene. Science. 1982 Jul 23;217(4557):316–324. doi: 10.1126/science.6283634. [DOI] [PubMed] [Google Scholar]
- Moore C. L., Sharp P. A. Site-specific polyadenylation in a cell-free reaction. Cell. 1984 Mar;36(3):581–591. doi: 10.1016/0092-8674(84)90337-4. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozer B., Marlor R., Parkhurst S., Corces V. Characterization and developmental expression of a Drosophila ras oncogene. Mol Cell Biol. 1985 Apr;5(4):885–889. doi: 10.1128/mcb.5.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriel W. J., Cole J., Lehmann A. R. Molecular analysis of ouabain-resistant mutants of the mouse lymphoma cell line L5178Y. Mutagenesis. 1987 Sep;2(5):383–389. doi: 10.1093/mutage/2.5.383. [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989 Mar 10;56(5):777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Nakabeppu Y., Ryder K., Nathans D. DNA binding activities of three murine Jun proteins: stimulation by Fos. Cell. 1988 Dec 2;55(5):907–915. doi: 10.1016/0092-8674(88)90146-8. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980 Oct 30;287(5785):795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Odenwald W. F., Garbern J., Arnheiter H., Tournier-Lasserve E., Lazzarini R. A. The Hox-1.3 homeo box protein is a sequence-specific DNA-binding phosphoprotein. Genes Dev. 1989 Feb;3(2):158–172. doi: 10.1101/gad.3.2.158. [DOI] [PubMed] [Google Scholar]
- Parkhurst S. M., Corces V. G. Interactions among the gypsy transposable element and the yellow and the suppressor of hairy-wing loci in Drosophila melanogaster. Mol Cell Biol. 1986 Jan;6(1):47–53. doi: 10.1128/mcb.6.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman D., Halvorson H. O. A putative signal peptidase recognition site and sequence in eukaryotic and prokaryotic signal peptides. J Mol Biol. 1983 Jun 25;167(2):391–409. doi: 10.1016/s0022-2836(83)80341-6. [DOI] [PubMed] [Google Scholar]
- Poole S. J., Kauvar L. M., Drees B., Kornberg T. The engrailed locus of Drosophila: structural analysis of an embryonic transcript. Cell. 1985 Jan;40(1):37–43. doi: 10.1016/0092-8674(85)90306-x. [DOI] [PubMed] [Google Scholar]
- Rushlow C., Doyle H., Hoey T., Levine M. Molecular characterization of the zerknüllt region of the Antennapedia gene complex in Drosophila. Genes Dev. 1987 Dec;1(10):1268–1279. doi: 10.1101/gad.1.10.1268. [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi P., Ransone L. J., Lamph W. W., Verma I. M. Direct interaction between fos and jun nuclear oncoproteins: role of the 'leucine zipper' domain. Nature. 1988 Dec 15;336(6200):692–695. doi: 10.1038/336692a0. [DOI] [PubMed] [Google Scholar]
- Soeller W. C., Poole S. J., Kornberg T. In vitro transcription of the Drosophila engrailed gene. Genes Dev. 1988 Jan;2(1):68–81. doi: 10.1101/gad.2.1.68. [DOI] [PubMed] [Google Scholar]
- Stanton L. W., Schwab M., Bishop J. M. Nucleotide sequence of the human N-myc gene. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1772–1776. doi: 10.1073/pnas.83.6.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steller H., Pirrotta V. A transposable P vector that confers selectable G418 resistance to Drosophila larvae. EMBO J. 1985 Jan;4(1):167–171. doi: 10.1002/j.1460-2075.1985.tb02332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villares R., Cabrera C. V. The achaete-scute gene complex of D. melanogaster: conserved domains in a subset of genes required for neurogenesis and their homology to myc. Cell. 1987 Jul 31;50(3):415–424. doi: 10.1016/0092-8674(87)90495-8. [DOI] [PubMed] [Google Scholar]
- Vincent A., Colot H. V., Rosbash M. Sequence and structure of the serendipity locus of Drosophila melanogaster. A densely transcribed region including a blastoderm-specific gene. J Mol Biol. 1985 Nov 5;186(1):149–166. doi: 10.1016/0022-2836(85)90265-7. [DOI] [PubMed] [Google Scholar]
- Vogt P. K., Bos T. J., Doolittle R. F. Homology between the DNA-binding domain of the GCN4 regulatory protein of yeast and the carboxyl-terminal region of a protein coded for by the oncogene jun. Proc Natl Acad Sci U S A. 1987 May;84(10):3316–3319. doi: 10.1073/pnas.84.10.3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton K. A., Yedvobnick B., Finnerty V. G., Artavanis-Tsakonas S. opa: a novel family of transcribed repeats shared by the Notch locus and other developmentally regulated loci in D. melanogaster. Cell. 1985 Jan;40(1):55–62. doi: 10.1016/0092-8674(85)90308-3. [DOI] [PubMed] [Google Scholar]