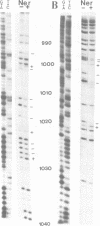
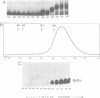
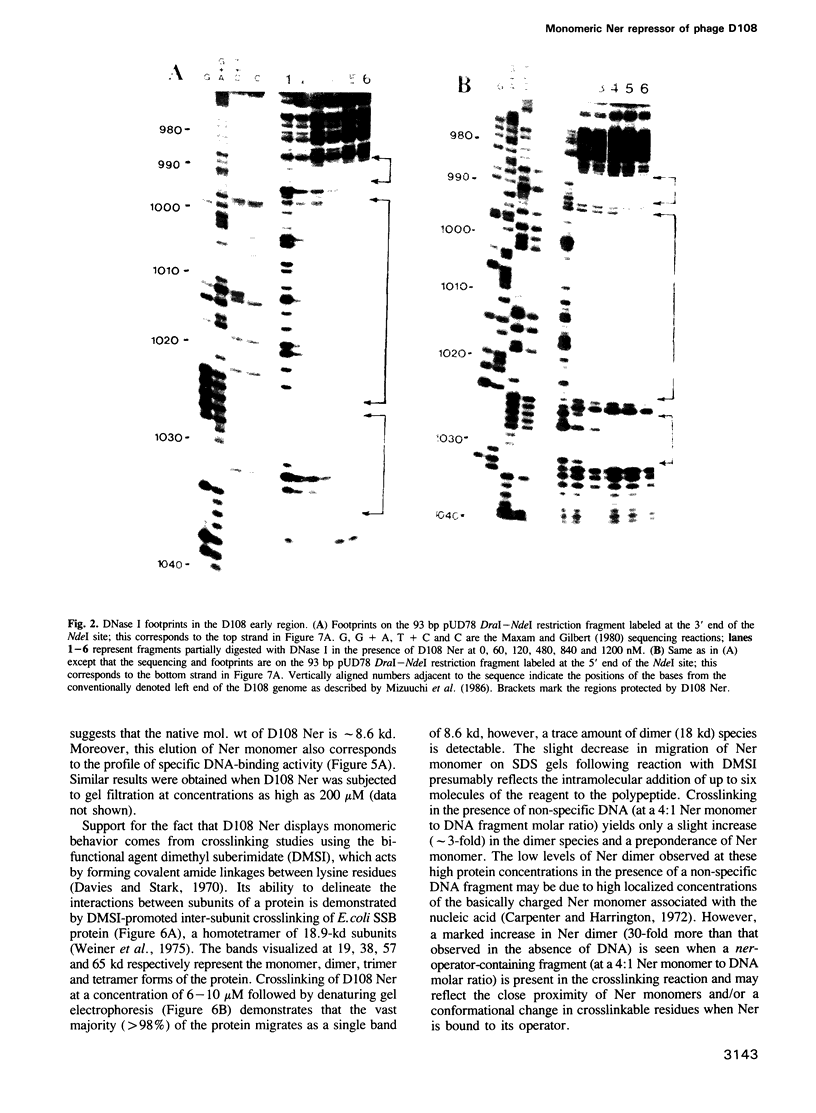
Abstract
We have purified the 8.6 kd ner gene product (a lambda Cro-like protein which negatively regulates transcription from two divergent and overlapping promoters) from the Mu-like transposable bacteriophage D108. Chemical and enzymatic protection experiments show the D108 ner-operator to contain two perfect 11 bp (5'-CCG-TGAGCTAC-3') inverted repeats separated by an 8 bp AT-rich region. Ner makes base-specific contacts in the major groove spanning the 11 bp repeats and also interacts with regions flanking these sites such that its operator comprises five turns of the DNA helix. Furthermore, gel filtration chromatography and dimethyl suberimidate crosslinking experiments indicate that D108 Ner (at concentrations exceeding 5 microM) is a monomer in solution, yet crosslinks as a dimer when bound to its operator site. As a small (73 amino acids) monomeric protein, Ner does not display strong homology with any known DNA-binding proteins. By virtue of the interactions with its operator it appears to bind DNA in a markedly different manner from other known prokaryotic repressors thus adding to the growing catalog of protein motifs used for specific binding to DNA.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abremski K., Gottesman S. Purification of the bacteriophage lambda xis gene product required for lambda excisive recombination. J Biol Chem. 1982 Aug 25;257(16):9658–9662. [PubMed] [Google Scholar]
- Aggarwal A. K., Rodgers D. W., Drottar M., Ptashne M., Harrison S. C. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science. 1988 Nov 11;242(4880):899–907. doi: 10.1126/science.3187531. [DOI] [PubMed] [Google Scholar]
- Allet B., Payton M., Mattaliano R. J., Gronenborn A. M., Clore G. M., Wingfield P. T. Purification and characterization of the DNA-binding protein Ner of bacteriophage Mu. Gene. 1988 May 30;65(2):259–268. doi: 10.1016/0378-1119(88)90462-3. [DOI] [PubMed] [Google Scholar]
- Anderson J. E., Ptashne M., Harrison S. C. Structure of the repressor-operator complex of bacteriophage 434. 1987 Apr 30-May 6Nature. 326(6116):846–852. doi: 10.1038/326846a0. [DOI] [PubMed] [Google Scholar]
- Anderson W. F., Ohlendorf D. H., Takeda Y., Matthews B. W. Structure of the cro repressor from bacteriophage lambda and its interaction with DNA. Nature. 1981 Apr 30;290(5809):754–758. doi: 10.1038/290754a0. [DOI] [PubMed] [Google Scholar]
- Carpenter F. H., Harrington K. T. Intermolecular cross-linking of monomeric proteins and cross-linking of oligomeric proteins as a probe of quaternary structure. Application to leucine aminopeptidase (bovine lens). J Biol Chem. 1972 Sep 10;247(17):5580–5586. [PubMed] [Google Scholar]
- Craig N. L., Nash H. A. E. coli integration host factor binds to specific sites in DNA. Cell. 1984 Dec;39(3 Pt 2):707–716. doi: 10.1016/0092-8674(84)90478-1. [DOI] [PubMed] [Google Scholar]
- Davies G. E., Stark G. R. Use of dimethyl suberimidate, a cross-linking reagent, in studying the subunit structure of oligomeric proteins. Proc Natl Acad Sci U S A. 1970 Jul;66(3):651–656. doi: 10.1073/pnas.66.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd I. B., Egan J. B. Systematic method for the detection of potential lambda Cro-like DNA-binding regions in proteins. J Mol Biol. 1987 Apr 5;194(3):557–564. doi: 10.1016/0022-2836(87)90681-4. [DOI] [PubMed] [Google Scholar]
- Drlica K., Rouviere-Yaniv J. Histonelike proteins of bacteria. Microbiol Rev. 1987 Sep;51(3):301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G. S., Hull R. C., Curtiss R., 3rd Mutator bacteriophage D108 and its DNA: an electron microscopic characterization. J Virol. 1981 Jan;37(1):420–430. doi: 10.1128/jvi.37.1.420-430.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goosen N., van Heuvel M., Moolenaar G. F., van de Putte P. Regulation of Mu transposition. II. The escherichia coli HimD protein positively controls two repressor promoters and the early promoter of bacteriophage Mu. Gene. 1984 Dec;32(3):419–426. doi: 10.1016/0378-1119(84)90017-9. [DOI] [PubMed] [Google Scholar]
- Goosen N., van de Putte P. Role of ner protein in bacteriophage Mu transposition. J Bacteriol. 1986 Aug;167(2):503–507. doi: 10.1128/jb.167.2.503-507.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull R. A., Gill G. S., Curtiss R., 3rd Genetic characterization of Mu-like bacteriophage D108. J Virol. 1978 Sep;27(3):513–518. doi: 10.1128/jvi.27.3.513-518.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. D., Poteete A. R., Lauer G., Sauer R. T., Ackers G. K., Ptashne M. lambda Repressor and cro--components of an efficient molecular switch. Nature. 1981 Nov 19;294(5838):217–223. doi: 10.1038/294217a0. [DOI] [PubMed] [Google Scholar]
- Jordan S. R., Pabo C. O. Structure of the lambda complex at 2.5 A resolution: details of the repressor-operator interactions. Science. 1988 Nov 11;242(4880):893–899. doi: 10.1126/science.3187530. [DOI] [PubMed] [Google Scholar]
- Krause H. M., Higgins N. P. Positive and negative regulation of the Mu operator by Mu repressor and Escherichia coli integration host factor. J Biol Chem. 1986 Mar 15;261(8):3744–3752. [PubMed] [Google Scholar]
- Krause H. M., Rothwell M. R., Higgins N. P. The early promoter of bacteriophage Mu: definition of the site of transcript initiation. Nucleic Acids Res. 1983 Aug 25;11(16):5483–5495. doi: 10.1093/nar/11.16.5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukolj G., Tolias P. P., DuBow M. S. Purification and characterization of the Ner repressor of bacteriophage Mu. FEBS Lett. 1989 Feb 27;244(2):369–375. doi: 10.1016/0014-5793(89)80565-4. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Levin D. B., DuBow M. S. Cloning and localization of the repressor gene (c) of the Mu-like transposable phage D108. FEBS Lett. 1987 Sep 28;222(1):199–203. doi: 10.1016/0014-5793(87)80219-3. [DOI] [PubMed] [Google Scholar]
- Levin D. B., DuBow M. S. Regulation of repressor and early gene expression in Mu-like transposable bacteriophage D108. Mol Gen Genet. 1989 Jun;217(2-3):392–400. doi: 10.1007/BF02464909. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McKay D. B., Steitz T. A. Structure of catabolite gene activator protein at 2.9 A resolution suggests binding to left-handed B-DNA. Nature. 1981 Apr 30;290(5809):744–749. doi: 10.1038/290744a0. [DOI] [PubMed] [Google Scholar]
- Mizuuchi M., Weisberg R. A., Mizuuchi K. DNA sequence of the control region of phage D108: the N-terminal amino acid sequences of repressor and transposase are similar both in phage D108 and in its relative, phage Mu. Nucleic Acids Res. 1986 May 12;14(9):3813–3825. doi: 10.1093/nar/14.9.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey J. H. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal Biochem. 1981 Nov 1;117(2):307–310. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Schleif R. DNA binding by proteins. Science. 1988 Sep 2;241(4870):1182–1187. doi: 10.1126/science.2842864. [DOI] [PubMed] [Google Scholar]
- Schumann W., Westphal C., Bade E. G., Holzer L. Origin and binding specificity of protein(s) coded for by Mu prophages. Mol Gen Genet. 1979 Jun 7;173(2):189–196. doi: 10.1007/BF00330310. [DOI] [PubMed] [Google Scholar]
- Tolias P. P., DuBow M. S. The cloning and characterization of the bacteriophage D108 regulatory DNA-binding protein ner. EMBO J. 1985 Nov;4(11):3031–3037. doi: 10.1002/j.1460-2075.1985.tb04040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolias P. P., Dubow M. S. The overproduction and characterization of the bacteriophage Mu regulatory DNA-binding protein ner. Virology. 1986 Jan 30;148(2):298–311. doi: 10.1016/0042-6822(86)90327-2. [DOI] [PubMed] [Google Scholar]
- Toussaint A., Faelen M., Desmet L., Allet B. The products of gene A of the related phages Mu and D108 differ in their specificities. Mol Gen Genet. 1983;190(1):70–79. doi: 10.1007/BF00330326. [DOI] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Hydroxyl radical "footprinting": high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leerdam E., Karreman C., van de Putte P. Ner, a cro-like function of bacteriophage Mu. Virology. 1982 Nov;123(1):19–28. doi: 10.1016/0042-6822(82)90291-4. [DOI] [PubMed] [Google Scholar]
- Vershon A. K., Liao S. M., McClure W. R., Sauer R. T. Bacteriophage P22 Mnt repressor. DNA binding and effects on transcription in vitro. J Mol Biol. 1987 May 20;195(2):311–322. doi: 10.1016/0022-2836(87)90652-8. [DOI] [PubMed] [Google Scholar]
- Vershon A. K., Youderian P., Susskind M. M., Sauer R. T. The bacteriophage P22 arc and mnt repressors. Overproduction, purification, and properties. J Biol Chem. 1985 Oct 5;260(22):12124–12129. [PubMed] [Google Scholar]
- Weiner J. H., Bertsch L. L., Kornberg A. The deoxyribonucleic acid unwinding protein of Escherichia coli. Properties and functions in replication. J Biol Chem. 1975 Mar 25;250(6):1972–1980. [PubMed] [Google Scholar]
- Wijffelman C., Gassler M., Stevens W. F., van de Putte P. On the control of transcription of bacteriophage Mu. Mol Gen Genet. 1974;131(2):85–96. doi: 10.1007/BF00266145. [DOI] [PubMed] [Google Scholar]
- Wolberger C., Dong Y. C., Ptashne M., Harrison S. C. Structure of a phage 434 Cro/DNA complex. Nature. 1988 Oct 27;335(6193):789–795. doi: 10.1038/335789a0. [DOI] [PubMed] [Google Scholar]
- Yin S., Bushman W., Landy A. Interaction of the lambda site-specific recombination protein Xis with attachment site DNA. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1040–1044. doi: 10.1073/pnas.82.4.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meeteren R., van de Putte P. Transcription of bacteriophage Mu. I. Hybridization analysis of RNA made in vitro. Mol Gen Genet. 1980;179(1):177–183. doi: 10.1007/BF00268461. [DOI] [PubMed] [Google Scholar]
- van de Putte P., Giphart-Gassler M., Goosen N., Goosen T., van Leerdam E. Regulation of integration and replication functions of bacteriophage Mu. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):347–353. doi: 10.1101/sqb.1981.045.01.048. [DOI] [PubMed] [Google Scholar]