Abstract
Limb-girdle muscular dystrophy primarily affects the muscles of the hips and shoulders (the “limb-girdle” muscles), although it is a heterogeneous disorder that can present with varying symptoms; there is currently no cure. We sought to identify the genetic basis of limb-girdle muscular dystrophy type 1 in an American family of Northern European descent using exome sequencing. Exome sequencing was performed on DNA samples from two affected siblings and one unaffected sibling and resulted in the identification of eleven candidate mutations that co-segregated with the disease. Notably, this list included a previously reported mutation in DNAJB6, p.Phe89Ile, which was recently identified as a cause of limb-girdle muscular dystrophy type 1D. Additional family members were Sanger sequenced and the mutation in DNAJB6 was only found in affected individuals. Subsequent haplotype analysis indicated that this DNAJB6 p.Phe89Ile mutation likely arose independently of the previously reported mutation. Since other published mutations are located close by in the G/F domain of DNAJB6, this suggests that the area may represent a mutational hotspot. Exome sequencing provided an unbiased and effective method for identifying the genetic etiology of limb-girdle muscular dystrophy type 1 in a previously genetically uncharacterized family. This work further confirms the causative role of DNAJB6 mutations in limb-girdle muscular dystrophy type 1D.
Keywords: limb-girdle muscular dystrophy, DNAJB6, exome sequencing
Introduction
Autosomal dominant limb-girdle muscular dystrophies (LGMD1) are a genetically and clinically heterogeneous group of disorders. LGMD1 often presents with progressive proximal muscle weakness of the upper and lower extremities; however several forms may present with primarily distal weakness, and clinical heterogeneity may exist even within the same family [1]. To date, mutations in five causative genes have been associated with LGMD1, including the MYOT gene encoding myotilin in LGMD1A [2], the LMNA gene encoding lamin A/C in LGMD1B [3], the CAV3 gene which encodes caveolin in LGMD1C [4]. More recently, mutations in the DNAJB6 gene have been described in LGMD1D patients [5–7] and mutations in Transportin 3 (TNPO3) gene have been linked to LGMD1F [8, 9]. Additional loci linked to LGMD1 have been mapped, but causative genes have not been identified [1].
Using exome sequencing, we sought to determine the genetic basis for LGMD1 in an American family of Northern European descent that presented with an autosomal dominant pattern of inheritance. Exome sequencing has been used to great effect to identify the underlying mutations in a number of diseases, including recently in LGMD1 [5, 8].
PATIENTS AND METHODS
All aspects of this study were approved by the Stanford University Institutional Review Board and written informed consent was received.
Proband (III-3, Figure 1A arrowhead) presented at 55 years of age to clinic, with a history of slow running and difficulty climbing stairs, beginning shortly after high school. He began using a cane in his late 30s, two canes in his 40s, and at 56 years of age, developed acute worsening of upper and lower extremity strength owing to severe cervical spinal cord stenosis and subsequently became wheelchair dependent. A left biceps muscle biopsy at 32 years of age revealed marked variation in fiber size, scattered fibers with single or multiple rimmed vacuoles (Fig 2A, B), as well as several angular atrophic fibers characteristic of neurogenic atrophy (Fig. 2C). There were normal amounts of connective and adipose tissue and no increase in central nuclei or fiber splitting. Examination showed marked weakness of shoulder abduction, elbow flexion, hip flexion and extension, and ankle plantar flexion. Ankle dorsiflexion strength was normal, but there was moderately severe weakness of intrinsic hand muscles and deep finger flexors. An EKG and echocardiogram at 54 years of age were normal. He has mild dyspnea and sleep-disordered breathing for which he required positive airway pressure support, with a forced vital capacity that was 42% of predicted. Genetic testing for LGMD1 genes MYOT, CAV3 and LMNA was negative.
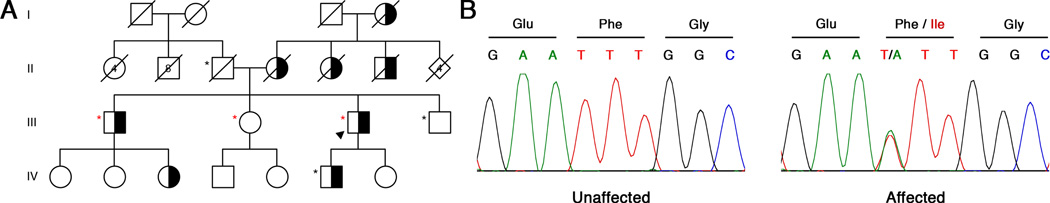
Figure 1. Mutations in DNAJB6 Cause LGMD1D.
(A) An American family of Northern European descent with LGMD1. Exome sequencing was performed in individuals marked with a red asterisk, Sanger sequencing in those with a black asterisk. Proband is indicated by black arrowhead. Affected individuals are colored black and white to indicate dominant inheritance.
(B) A c.265T>A mutation was discovered only in affected patients in the DNAJB6 gene, resulting in a p.Phe89Ile amino acid substitution.
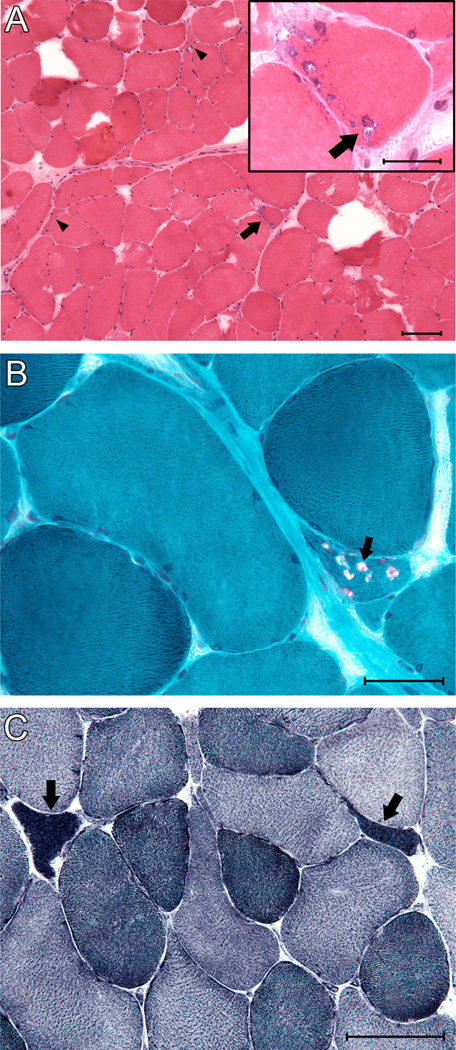
Figure 2.
Histopathological examination of LGMD1D muscle biopsy. (A) Low magnification photomicrograph of hematoxylin and eosin stained cryosection of biceps muscle biopsy, showing marked variation in fiber sizes, and a fiber with several rimmed vacuoles (arrow), highlighted in modified Gomori trichrome stain (inset, arrow), and scattered angular atrophic fibers (arrowheads). Hematoxylin and eosin, scale bar = 100 microns. Inset: High magnification view of region indicated by arrow, scale bar = 25 microns. (B) High magnification view of biceps muscle biopsy cryosection showing moderate variation in fiber size, a minimal increase in endomysial connective tissue, and a fiber containing numerous rimmed vacuoles (arrow). Modified Gomori trichrome, scale bar = 50 microns. (C) Higher power photomicrograph showing overly dark angular atrophic fibers typical of neurogenic atrophy (arrows). NADH enzyme histochemistry, scale bar = 100 microns.
The proband’s son (IV-6, Figure 1A) was evaluated by a neuromuscular specialist at 28 years of age due to a history of difficulty climbing stairs and rising from a sitting position beginning around age 16, although slow running was noted as early as age 8. Physical examination revealed proximal greater than distal weakness affecting the lower extremities more than the upper, with prominent atrophy of the thigh adductors and the medial head of gastrocnemius, scapular winging, and Achilles tendon contractures (summarized in Table 1). He does not use assistive devices, and an EKG was normal at time of presentation.
Table 1.
Clinical Summary of the Patients
| Clinical Presentation | III-3 | IV-6 |
|---|---|---|
| Proximal atrophy | + | + |
| DTR patella/ankle | 0/0 | trace/0 |
| Deltoid (L/R) | 1–2/1–2 | 4+/4+ |
| Biceps (L/R) | 4−/4− | 4/4 |
| Triceps (L/R) | 4−/4− | 4+/4+ |
| FDI (L/R) | 4/4 | 5/5 |
| EIP (L/R) | 4/4 | Not Tested |
| Hip flexors (L/R) | 0–1/0–1 | 4−/4− |
| Hip extensors (L/R) | 0–1/0–1 | 2/2 |
| Knee flexors (L/R) | 0/0 | 3+/3 |
| Knee extensors (L/R) | 1/1 | 4/4 |
| Ankle dorsiflexion (L/R) | 5/5 | Limited by tight heel cords |
| Ankle plantarflexion (L/R) | 4−/4− | 4/4 |
| Scapula winging | + | + |
| FVC (% predicted) | 42 | 85 |
| MIP (cm H2O) | −60 | −87 |
| Creatine Kinase level (U/L) | 181 | 3000 |
Proband pedigree analysis was consistent with autosomal dominant inheritance (Figure 1).
Exome Sequencing
Genomic DNA samples were isolated from three family members using Oragene•DISCOVER (OGR-500) saliva collection kits; the three remaining DNA samples were sent from Prevention Genetics (Marshfield, WI). Exomes were generated for three family members using the SureSelect Human All Exon 50Mb kit (Agilent, Santa Clara, CA). Sequencing was performed with 250bp paired-end reads on an Illumina MiSeq platform (Illumina Inc., San Diego, CA). Reads were aligned to the human reference genome (UCSC hg19, GRCh37, Feb. 2009 release) using bowtie2 [10] and SAMtools [11]. We applied GATK base quality score recalibration, indel realignment, duplicate removal, and performed coverage calculations, SNP and INDEL discovery and genotyping across each sample using standard hard filtering parameters or variant quality score recalibration [12]. Variants were filtered against dbSNPv137, 1000 genomes and ESP 6500 databases and were then annotated using ANNOVAR [13]. We assessed segregation of candidate mutations by Sanger sequencing using standard methods in all six family members for which DNA was available.
Haplotype Analysis
For each microsatellite marker, amplification of short tandem repeats (STRs) was performed using primers sequences available on the UniSTS database (http://www.ncbi.nlm.nih.gov/unists). Forward primers were tagged with 6-FAM and fragment analysis was then performed. Size characterization was performed using Peak Scanner software (Applied Biosystems).
Results
Exome Sequencing
Exome sequencing was performed for two affected brothers (Figure 1A red asterisks; III-1 and III-3) and one unaffected sister (III-2); average coverage depth was 60X. Eleven novel variants shared between the two affected brothers (III-1 and III-3) but not present in the unaffected sister (III-2) were identified (Supplemental Table 1). Included in the list of disease-segregating variants was a variation in the DNAJB6 gene, c.265T>A, producing a p.Phe89Ile substitution, which is located in the G/F domain of the protein (Figure 1B). We subsequently focused on DNAJB6 since this same mutation, as well as other mutations in DNAJB6, have been previously associated with LGMD1 [1, 5–7]. To confirm the association of this mutation with disease in this family, we performed Sanger sequencing on DNA from the remaining three family members for which a DNA sample was available (Figure 1A, black asterisks). The c.265T>A mutation in DNAJB6 was found in all affected and none of the unaffected family members. Combined with the previously reported mutations in DNAJB6 causing LGMD1D [1, 5–7], we determined that the causative mutation in this family was the c.265T>A/p.Phe89Ile mutation in DNAJB6.
Haplotype Analysis
Since a previous study identified the same mutation, p.Phe89Ile, in two other US families (DUK1047 and DUK1701 [6, 14]), we wondered if the mutation arose independently in our family or if the three families shared a common origin for the mutation. We performed a haplotype analysis of this region of chromosome 7 to compare the family reported in this study to the two in which this mutation was previously reported (Supplemental Table 2, [6, 14]). Microsatellite repeat length is different between this family and the two previously reported families, ruling out a common ancestry, but hinting at a potential mutational hotspot (Supplemental Table 2, [14]).
Pathology
Muscle pathology in DNAJB6 dystrophy is characterized by fiber size variation, regenerating fibers, rimmed vacuoles, cytoplasmic inclusions, and disorganized myofibrils, commonly referred to as moth-eaten fibers on NADH-tetrazolium staining [5, 6, 15]. Rimmed basophilic vacuoles are the key finding on muscle biopsy; however they may also be found in other autosomal dominant LGMDs, specifically LGMD1A [16]. It should be noted that these myopathological changes are consistent with those seen in myofibrillar myopathy (MFM), a clinically and genetically heterogeneous group of muscle disorders with shared muscle pathology characterized by disorganized myofibrils and cytoplasmic desmin-positive inclusions [17]. Upon examination of a biceps muscle biopsy of patient III-3 taken at age 32, we noted key pathological features previously linked to DNAJB6 dystrophy, including rimmed basophilic vacuoles (Figure 2). These pathological features combined with our genetic data led us to conclude that the p.Phe89Ile mutation in the DNAJB6 gene is causative for LGMD1D in this family.
Discussion
Using the unbiased technique of exome sequencing, we identified a mutation in DNAJB6 in a family with LGMD1. LGMD1D was originally described in several Finnish families with linkage to chromosome 7q36 and recently three studies in Finnish, Italian, American, and Japanese families revealed the causative gene to be DNAJB6 [5–7, 15, 18]. The nomenclature of LGMD associated with DNAJB6 mutations is confusing, with some groups utilizing the HUGO Gene Nomenclature Committee’s locus designation of LGMD1D, while some use LGMD1E [19] and others use LGMD1D/1E [5]; herein we have used the HUGO gene nomenclature “LGMD1D”.
DNAJB6 dystrophy is primarily associated with proximal limb weakness [1, 5–7], however distal predominant weakness has also been reported [5]. Onset may occur as early as age 13, but more commonly occurs in the fourth decade of life, and progresses to wheelchair dependence 20–30 years after symptoms begin [1, 5–7]. Bulbar, cardiac and pulmonary involvements have been uncommon in reported cases[1, 5, 6]. Consistent with prior descriptions of DNAJB6 dystrophy, all patients in this series presented with slowly progressive lower extremity weakness that was predominantly proximal, though the proband also had significant weakness of ankle plantar flexion and some weakness of intrinsic hand muscles. Age of onset was early in all patients reported here, with symptoms present as early as 8 years of age. The proband’s markedly reduced forced vital capacity raises the important possibility that ventilatory function can be impaired in LGMD1D.
DNAJB6 is a member of the DNAJ/HSP40 chaperone family and is known to play an important role in suppressing protein aggregation and toxicity of polyglutamine proteins commonly found in various neurodegenerative diseases [20]; Udan-Johns, 2013}. DNAJB6 has three domains: an N-terminal J domain, a variable C-terminal domain, and a G/F domain in which all of the mutations linked to LGMD1D (p.Phe89Ile, p.Phe93Leu, p.Phe93Ile, and p.Pro96Arg) have been found[5–7, 20]. We identified the previously reported c.265T>A base pair change that resulted in a p.Phe89Ile amino acid substitution in the G/F domain of the protein in all affected members of this pedigree (Figure 1; Table S1) [6].
We note that we also identified a sequence variation in TTN, an exceptionally large gene with mutations causing LGMD2J, a recessive form of LGMD [21], in both affected siblings (III-1 and III-3) but not the unaffected sister (III-2, Table S1). We performed Sanger sequencing and found this variant also segregates with the disease in the six family members for which we had DNA available. Unfortunately, no further DNA samples were available in this pedigree. Patients with some heterozygous TTN mutations typically present with tibial muscular dystrophy [21], which presents with a phenotype distinct from that described here. Furthermore, the TTN sequence variant that we identified is located in one of the 132 Fibronectin type 3 (FN3) domains, within the highly repetitive region of Ig and FN3 domains, far from any known disease causing mutations [22]. Given that this family presents with pathology and symptoms very similar to previously reported patients with DNAJB6 mutations and that heterozygous TTN mutations generally cause a distinct phenotype, we hypothesize that this TTN variation is not responsible for the disease [5–7]. However, we cannot rule out the possibility that the TTN sequence variant is playing a role in the clinical presentation in this family, for example in the respiratory symptoms reported in the proband, as TTN mutations can cause hereditary myopathy with early respiratory failure (HMERF) [23]. These findings also highlight the utility of performing exome sequencing compared to candidate gene targeted sequencing.
Our studies corroborate previously published work showing that mutations in DNAJB6 are associated with LGMD1D. We identified a point mutation in DNAJB6, c.265T>A, which results in an amino acid change that was previously shown to be associated with LGMD1D, p.Phe89Ile [6], in an American family of Northern European descent. We performed haplotype analysis of the region surrounding DNAJB6 and determined that the mutation reported in this study arose independently of the two previously reported families with p.Phe89Ile mutations, and combined with the other mutations reported in the area, this suggests that there may be a mutational hotspot in the G/F domain of DNAJB6 [5–7]. This work emphasizes the clinical utility of unbiased approaches like exome sequencing for identifying the causative mutations in inherited diseases.
Supplementary Material
Acknowledgements
We thank patients and their family for participating in this study. Supported by NIH Director’s New Innovator Award DP2OD004417 (A.D.G.), NIH grants NS065317 (A.D.G.) and NS073660 (A.D.G.). A.D.G. is a Pew Scholar in the Biomedical Sciences, supported by The Pew Charitable Trusts, and a Rita Allen Foundation Scholar. A.R.R. is supported by an Alzheimer’s Disease Research grant from the BrightFocus Foundation. J.D.B. is supported by the NIH-NHGRI Stanford Genome Science training grant. We thank Namita Goyal, MD for clinical data, including patient history and physical exam.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Flanigan K. The muscular dystrophies. Semin Neurol. 2012;32:255–263. doi: 10.1055/s-0032-1329199. [DOI] [PubMed] [Google Scholar]
- 2.Hauser M, Horrigan S, Salmikangas P, Torian U, Viles K, Dancel R, et al. Myotilin is mutated in limb girdle muscular dystrophy 1A. Hum Mol Genet. 2000;9:2141–2147. doi: 10.1093/hmg/9.14.2141. [DOI] [PubMed] [Google Scholar]
- 3.Muchir A, Bonne G, van der Kooi A, van Meegen M, Baas F, Bolhuis P, et al. Identification of mutations in the gene encoding lamins A/C in autosomal dominant limb girdle muscular dystrophy with atrioventricular conduction disturbances (LGMD1B) Hum Mol Genet. 2000;9:1453–1459. doi: 10.1093/hmg/9.9.1453. [DOI] [PubMed] [Google Scholar]
- 4.Minetti C, Sotgia F, Bruno C, Scartezzini P, Broda P, Bado M, et al. Mutations in the caveolin-3 gene cause autosomal dominant limb-girdle muscular dystrophy. Nat Genet. 1998;18:365–368. doi: 10.1038/ng0498-365. [DOI] [PubMed] [Google Scholar]
- 5.Harms M, Sommerville R, Allred P, Bell S, Ma D, Cooper P, et al. Exome sequencing reveals DNAJB6 mutations in dominantly-inherited myopathy. Ann Neurol. 2012;71:407–416. doi: 10.1002/ana.22683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sarparanta J, Jonson P, Golzio C, Sandell S, Luque H, Screen M, et al. Mutations affecting the cytoplasmic functions of the co-chaperone DNAJB6 cause limb-girdle muscular dystrophy. Nat Genet. 2012;44:450. doi: 10.1038/ng.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sato T, Hayashi Y, Oya Y, Kondo T, Sugie K, Kaneda D, et al. DNAJB6 myopathy in an Asian cohort and cytoplasmic/nuclear inclusions. Neuromuscul Disord. 2013;23:269–276. doi: 10.1016/j.nmd.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 8.Torella A, Fanin M, Mutarelli M, Peterle E, Del Vecchio Blanco F, Rispoli R, et al. Next-Generation Sequencing Identifies Transportin 3 as the Causative Gene for LGMD1F. PloS one. 2013;8:e63536. doi: 10.1371/journal.pone.0063536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melia MJ, Kubota A, Ortolano S, Vilchez JJ, Gamez J, Tanji K, et al. Limb-girdle muscular dystrophy 1F is caused by a microdeletion in the transportin 3 gene. Brain : a journal of neurology. 2013;136:1508–1517. doi: 10.1093/brain/awt074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Langmead B, Salzberg S. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DePristo M, Banks E, Poplin R, Garimella K, Maguire J, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38 doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Speer MC, Vance JM, Grubber JM, Lennon Graham F, Stajich JM, Viles KD, et al. Identification of a new autosomal dominant limb-girdle muscular dystrophy locus on chromosome 7. American journal of human genetics. 1999;64:556–562. doi: 10.1086/302252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sandell S, Huovinen S, Sarparanta J, Luque H, Raheem O, Haapasalo H, et al. The enigma of 7q36 linked autosomal dominant limb girdle muscular dystrophy. J Neurol Neurosurg Psychiatry. 2010;81:834–839. doi: 10.1136/jnnp.2009.192351. [DOI] [PubMed] [Google Scholar]
- 16.Olive M, Goldfarb L, Shatunov A, Fischer D, Ferrer I. Myotilinopathy: refining the clinical and myopathological phenotype. Brain : a journal of neurology. 2005;128:2315–2326. doi: 10.1093/brain/awh576. [DOI] [PubMed] [Google Scholar]
- 17.De Bleecker J, Engel A, Ertl B. Myofibrillar myopathy with abnormal foci of desmin positivity. II. Immunocytochemical analysis reveals accumulation of multiple other proteins. J Neuropathol Exp Neurol. 1996;55:563–577. doi: 10.1097/00005072-199605000-00009. [DOI] [PubMed] [Google Scholar]
- 18.Hackman P, Sandell S, Sarparanta J, Luque H, Huovinen S, Palmio J, et al. Four new Finnish families with LGMD1D; refinement of the clinical phenotype and the linked 7q36 locus. Neuromuscul Disord. 2011;21:338–344. doi: 10.1016/j.nmd.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 19.Bushby K, Beckmann J. The 105th ENMC sponsored workshop: pathogenesis in the non-sarcoglycan limb-girdle muscular dystrophies, Naarden, April 12–14, 2002. Neuromuscul Disord. 2003;13:80–90. doi: 10.1016/s0960-8966(02)00183-9. [DOI] [PubMed] [Google Scholar]
- 20.Hageman J, Rujano M, van Waarde M, Kakkar V, Dirks R, Govorukhina N, et al. A DNAJB chaperone subfamily with HDAC-dependent activities suppresses toxic protein aggregation. Mol Cell. 2010;37:355–369. doi: 10.1016/j.molcel.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 21.Hackman J, Vihola A, Udd A. The role of titin in muscular disorders. Ann Med. 2003;35:434–441. doi: 10.1080/07853890310012797. [DOI] [PubMed] [Google Scholar]
- 22.Kontrogianni-Konstantopoulos A, Ackermann MA, Bowman AL, Yap SV, Bloch RJ. Muscle giants: molecular scaffolds in sarcomerogenesis. Physiological reviews. 2009;89:1217–1267. doi: 10.1152/physrev.00017.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toro C, Olive M, Dalakas MC, Sivakumar K, Bilbao JM, Tyndel F, et al. Exome sequencing identifies titin mutations causing hereditary myopathy with early respiratory failure (HMERF) in families of diverse ethnic origins. BMC neurology. 2013;13:29. doi: 10.1186/1471-2377-13-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.