Abstract
The human liver fatty acid binding protein (L-FABP) T94A variant, the most common in the FABP family, has been associated with elevated liver triglyceride (TG) levels. How this amino acid substitution elicits these effects is not known. This issue was addressed with human recombinant wild-type (WT, T94T) and T94A variant L-FABP proteins as well as cultured primary human hepatocytes expressing the respective proteins (genotyped as TT, TC, and CC). T94A substitution did not or only slightly alter L-FABP binding affinities for saturated, monounsaturated, or polyunsaturated long chain fatty acids (LCFA), nor did it change the affinity for intermediates in TG synthesis. Nevertheless, T94A substitution markedly altered the secondary structural response of L-FABP induced by binding LCFA or intermediates of TG synthesis. Finally, T94A substitution markedly diminished polyunsaturated fatty acid, eicosapentaenoic acid (EPA) or docosahexaenoic acid (DHA), induction of peroxisome proliferator-activated receptor alpha (PPARα) - regulated proteins such as L-FABP, fatty acid transport protein 5 (FATP5), and PPARα itself in cultured primary human hepatocytes. Thus, while T94A substitution did not alter the affinity of human L-FABP for LCFAs, it significantly altered human L-FABP structure and stability as well as conformational and functional response to these ligands.
Keywords: L-FABP, fatty acids, PUFA, TG, liver, human
INTRODUCTION
Liver fatty acid binding protein (L-FABP) comprises 2–7% of cytosolic protein (0.2–0.7mM), representing 80–90% of cytosolic long chain fatty acids (LCFA) and LCFA-CoA binding capacity [1;2]. L-FABP is also thought to be a potential modifier protein promoting an early adaptive response to hepatocyte stress by which potentially lipotoxic LCFAs are partitioned into stable intracellular triglyceride (TG) stores [3]. There is significant evidence that a LCFA-CoA binding protein such as L-FABP is required for optimal activity of transacylase enzymes in the murine TG synthesis pathway. First, free unbound LCFA-CoA but not L-FABP-bound LCFA-CoA inhibits transacylase enzymes mediating the first two steps in TG synthesis [4–7]. In fact, L-FABP-bound LCFA-CoA stimulates these enzymes [7–10] by removing the inhibitory LCFA-CoA and presenting it for utilization [4;5;7;11]. Hepatic L-FABP is upregulated in human non-alcoholic fatty liver disease (NAFLD) and in NAFLD animal models [12–15]. In contrast, L-FABP ablation decreases hepatic TG accumulation [16–21]. Second, L-FABP also prevents LCFA lipotoxicity by binding/linking with oxidized and reactive LCFA species [22–29], which depletes the L-FABP pool as NAFLD progresses to non-alcoholic steatohepatitis (NASH) [14;23–27]. Third, in the longer term L-FABP may also prevent/ameliorate lipotoxicity by enhancing LCFA β-oxidation. Murine and/or human L-FABP enhance LCFA uptake [1;30–33;33–36], target/cotransport n-3 polyunsaturated fatty acids (PUFA) such as eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) [37] as well as other LCFAs [35;38–41] into the nucleus, directly bind peroxisome proliferator-activated receptor alpha (PPARα) [42–46], and activate LCFA-induced PPARα-transcription of LCFA β-oxidative enzymes [37;47].
A recently identified polymorphism in the human L-FABP gene, resulting in a T94A substitution, occurs with high frequency (26–38% minor allele freq.; 8.3+1.9% homozygous) in the human population and represents the most frequent polymorphism in the FABP family (MAF for 1000 genomes in NCBI dbSNP database; ALFRED database) [48–54]. However, direct structural and functional studies of the impact of the L-FABP T94A variant are lacking or contradictory. For example, overexpression of human L-FABP T94A in ‘Chang liver cells’ did not enhance LCFA uptake and decreased TG accumulation [55]. In contrast, murine L-FABP overexpression increased [30–33] while L-FABP ablation [1;34–36] or antisense treatment [33] decreased LCFA uptake and decreased hepatic TG [16–20]. This initially suggested that the L-FABP T94A variant does not bind LCFA and represents a ‘loss-of-function’ variant analogous to the L-FABP null mouse [55]. This conclusion, however, is at variance with the finding that expression of the L-FABP T94A variant is associated with elevated plasma triglycerides [49;56], increased LDL cholesterol [49;53], atherothrombotic cerebral infarction [51], and NAFLD in human subjects [53]. It is not clear if the discrepancy may be due to ‘Chang liver cells’ being derived from human cervical cancer cells and thus not necessarily recapitulating these aspects of hepatocyte function [57–59].
In summary, a more complete understanding of the structure, LCFA binding properties, and function of the human L-FABP T94A variant in LCFA-induced PPARα transcriptional activity in human hepatocytes is needed. To address these issues, structural and LCFA binding studies were initiated with purified recombinant human WT and T94A variant L-FABP proteins. While the human T94A variant L-FABP did not differ much in affinity for LCFA and LCFA metabolites, its structural response to these ligands differed markedly from that of the human WT T94T L-FABP. Furthermore, T94A diminished n-3 PUFA-mediated induction of PPARα transcriptional activity in human hepatocytes.
RESULTS
Key amino acids differentiating human wild-type and T94A variant L-FABP from rat L-FABP
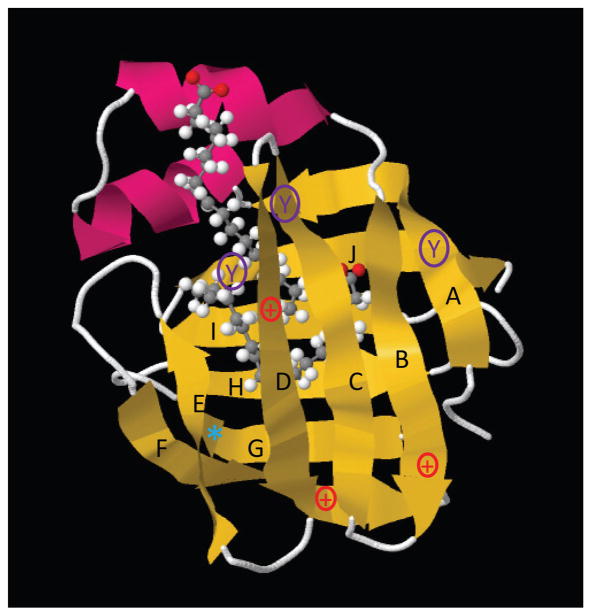
The common ribbon structure of both human and murine L-FABP demonstrates the classic β-barrel comprised of β-sheets A-J enclosing the ligand binding pocket (Fig. 1 yellow) plus two α-helices near the opening of the binding pocket (Fig. 1, red) (RCSB Protein Data Bank ID code: 2LKK) [60]. While the amino acid sequence of human T94T L-FABP is 82.7% identical and 89.8% similar to the rat L-FABP, these proteins differ significantly in charge, aromatic amino acid residues, ligand binding cavity volume, and in the case of the human L-FABP T94A variant alanine (smaller non-polar) substituted for threonine (larger polar) [61–64].
Figure 1. Key amino acids differentiating human wild-type and T94A variant L-FABP from rat L-FABP.
Ribbon structure of L-FABP (RCSB Protein Data Bank ID code: 2LKK) demonstrating the classic β-barrel (yellow β-sheets A–J) and α-helices (red) common to both human and rat L-FABPs [60]. Shown within the β-barrel are two bound oleic acids [60]. Key amino acids differentiating human and rat L-FABPs are designated as follows: 1) tyrosine (Y) in β-sheet A is found in both human and rat L-FABPs; 2) tyrosines (Y) in β-sheets C and J are found only in rat L-FABP; 3) three positively charged amino acids (+) in β-sheets B, D, and F in rat L-FABP are replaced by neutral amino acids in human L-FABPs; 4) threonine (T), a polar non-charged amino acid, is found at position 94 (*) in β-sheet G of both wild-type human and rat L-FABP, but is replaced by the smaller non-polar alanine (A) in the human L-FABP T94A variant.
The human L-FABPs have three neutral amino acids where the rat L-FABP has three positively-charged amino acids (Fig. 1, +). This results in significantly lower isoelectric point values, pI=6.60 for human L-FABP T94T and T94A as compared to the pI = 7.79 for rat L-FABP. While neither human nor rat L-FABPs contain tryptophan, the human L-FABP contains only a single tyrosine while the rat L-FABP has three tyrosines (Fig. 1, Y). The tyrosine residue common to both the human and rat L-FABPs (Fig. 1, Y in β-sheet A) is outside the ligand binding pocket. In contrast, the two additional tyrosine residues in the rat L-FABP (Fig. 1, Y in β-sheets C and J) are within/near the ligand binding site. Consequently, the tyrosine emission of the rat, but not human, L-FABP is more sensitive to occupancy of the ligand binding pocket (not shown). The threonine (T) at position 94 (Fig. 1, *) in β-sheet G of both wild-type human and rat L-FABP is replaced with alanine (A) in the human L-FABP T94A variant. While threonine is a polar non-charged amino acid, alanine is non-polar, uncharged, and significantly smaller—occupying 24% less volume and 18% less surface area [65;66]. Finally, the ligand binding cavity volume of rat L-FABP is 1.3–2.1 fold larger than that of other FABP family members, and that of the human wild-type L-FABP T94T is larger than that of the rat L-FABP [62–64].
The effects of these differences on the structure, stability, specificity for binding LCFAs and intermediates in TG synthesis, conformational responsiveness to ligand binding, and function are addressed in the following sections.
SDS-PAGE and Western analysis of rat, T94T and T94A L-FABPs
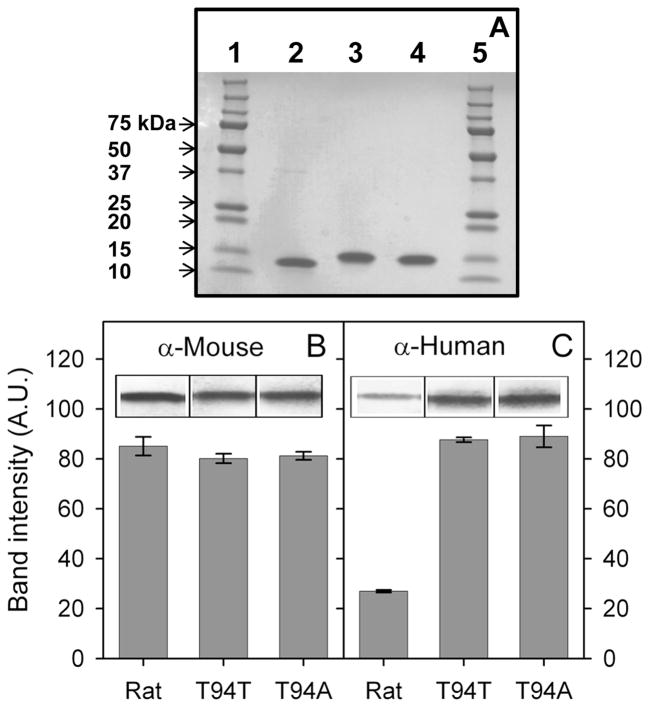
Purified rat L-FABP as well as human T94T and T94A L-FABP proteins were detected as single bands near 14 kDa on SDS-PAGE gels (Figs 2A). MALDI-TOF analysis of the final, his-tag removed T94T and T94A variant were consistent with molecular weights of these two proteins based on amino acid sequence (not shown). Western analyses showed that while the anti-mouse L-FABP antibody reacted equally well with both human and rat L-FABP (Fig. 2B), the anti-human L-FABP antibody reacted better with human than rat L-FABP (Fig. 2C). These findings suggested that the differences in amino acid sequence/composition between rat and human L-FABP were sufficient to significantly alter the antigenic epitopes. In contrast, the human L-FABP T94A mutation did not elicit any additional differential response to anti-mouse or anti-human L-FABP antibodies.
Figure 2. SDS-PAGE and Western blot analysis of rat, T94T WT, and T94A variant human L-FABPs.
Panel A: SDS-PAGE analysis of rat, T94T WT, and T94A variant human L-FABPs (3 μg each lane). Lane 1 and 5: Precision Plus Protein Standard (10 μL, Bio-Rad Labs); lane 2: rat L-FABP; lane 3: T94T WT human L-FABP; lane 4: T94A variant human L-FABP. Panel B: Western blot analysis with anti-mouse L-FABP antibody. Panel C: Western blot analysis with anti-human L-FABP antibody.
Tyrosine fluorescence of rat, T94T and T94A L-FABP in denaturation conditions
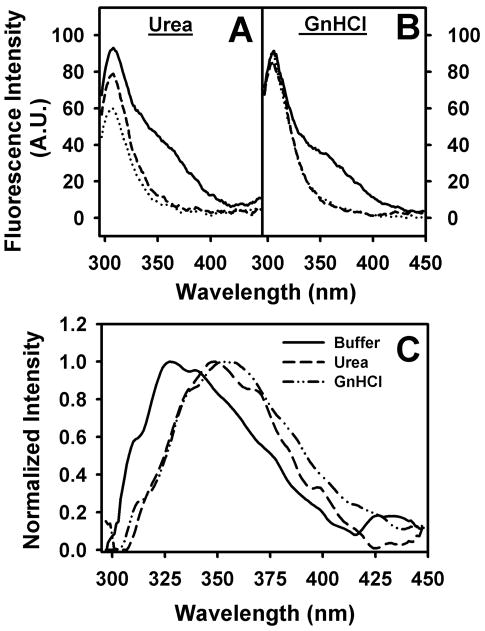
To examine the impact of the differences in the respective amino acid sequence on L-FABP stability, tyrosine fluorescence spectra were recorded under increasing denaturation conditions using 8M Urea (Fig. 3A) and 6M Guanidinium hydrochloride (GnHCl, Fig. 3B). While partial unfolding by urea increased the aqueous exposure of tyrosine residues in both rat and human L-FABPs, the tyrosines in rat L-FABP appeared more resistant to aqueous quenching upon urea-induced denaturation than that of human L-FABPs (Fig. 3A&B, solid line vs. dotted and dash line). Among the human L-FABPs, the T94A variant appeared more resistant to urea-induced unfolding than T94T (Fig. 3A, dashed line vs dotted line). GnHCl, a stronger protein unfolding agent, abolished these differences between the various types of L-FABPs (Fig. 3B).
Figure 3. L-FABP Tyrosine fluorescence in denaturation conditions.
Tyrosine fluorescence spectra of rat, Human T94T WT and T94A variant in 8M urea (Panel A) and 6M Guanidinium chloride (Panel B) were recorded as described in Experimental Procedures (solid line, rat L-FABP; dotted line, T94T; dash line; T94A). Panel C, normalized difference spectra of (normalized rat L-FABP - normalized T94T) tyrosine fluorescence emission (solid line, buffer; ---, 8M urea; -··-, 6M GnHCl). Representative spectra were shown.
Interestingly, qualitative comparison of the rat and human L-FABP tyrosine emission spectra suggested a broad shoulder at longer emission wavelength in the rat but not human L-FABP (Fig 3A&B). This was quantitatively confirmed when the tyrosine emission spectra of the human wild-type L-FABP T94T were normalized to those of the rat L-FABP and subtracted. The rat L-FABP exhibited an additional emission maximum near 330 nm in the folded-state (i.e. buffer only) but shifted to 350 nm in the presence of unfolding agents urea or GnHCl (Fig. 3C).
Since neither rat nor human L-FABP contain a tryptophan residue, the additional emission peak was likely due to tyrosinate-like emission. Tyrosinate-like fluorescence emission can occur in proteins where one or more of the tyrosines are hydrogen bonded through the phenolic hydroxyls, especially to neighboring basic amino acid residue(s), glutamic acid (E) or aspartic acid (D) [67]. While the rat and human forms of L-FABP contain a common tyrosine, Y7, the rat L-FABP contains two additional tyrosines, Y54 and Y120. Since the emission is not evident in the human L-FABP proteins, it would seem that these additional residues would be the most likely candidates for producing the tyrosinate-like emission in the recombinant rat L-FABP. However, examination of the lowest energy minimized solution-state structures of rat L-FABP reveals that only the hydroxyl of the Y7 tyrosine residue is involved in hydrogen bonding with no evidence in the human L-FABP structures. In the rat apo-L-FABP structure (RCSB Protein Data Bank ID code: 2JU3), hydrogen bonding occurs between Y7 and F3, but interestingly in the rat holo-L-FABP (RCSB Protein Data Bank ID code: 2JU7), hydrogen bonding was evident between Y7 and an aspartic acid residue, D107 (RCSB Protein Data Bank ID code: 2LKK)[68]. However, since D107 appeared quite flexible and located in a solvent-exposed region of a β-turn [68], it was unclear why the interaction was only subtly affected by denaturation in 8M urea and 6M GnHCl.
Rat, T94T and T94A L-FABP binding to 1-anilinonaphthalene-8-sulfonic acid (ANS)
Since the human and rat L-FABPs differ significantly in amino acid sequence, charge, and ligand binding cavity volume [61–64], a displacement assay was developed to examine the impact of these variations on binding affinity and specificity of these proteins for LCFAs and intermediates in TG synthesis. ANS was chosen for this assay because it is a synthetic fluorophore previously used to examine the binding sites in FABPs [45;69–71].
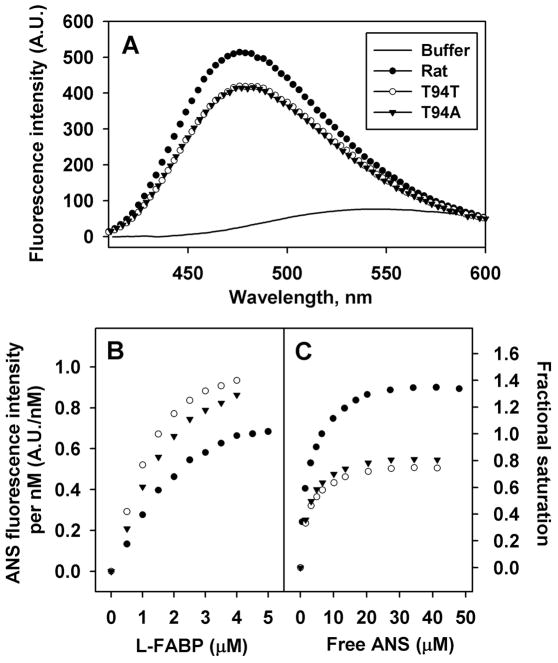
While the fluorescence of ANS was very weak in buffer (Fig. 4A solid line), it differentially increased upon binding to L-FABPs with the highest emission intensity obtained with rat L-FABP (●). Emission spectra of ANS bound to human wild-type L-FABP T94T (○) and T94A variant (▼) were similar at lower intensity than for rat L-FABP (Fig. 4A). Quantitative analysis of multiple spectra established that ANS maximal emission intensity was 3 nm blue-shifted and about 1.3-fold higher (p<0.05) when bound to rat than human L-FABPs (Table 1). This suggested that when bound to rat L-FABP the ANS was localized in a more hydrophobic environment and/or more ANS was bound per protein molecule.
Figure 4. ANS binding to Rat, T94T, T94A L-FABPs.
Panel A, representative ANS (35μM) emission spectra when bound to L-FABPs (500nM). Panel B, representative reverse titration curves. ANS (100nM) were titrated with increasing amount of the L-FABP. Panel C, representative forward titration curves. L-FABP 500nM were titrated with increasing amount of ANS. Solid line, buffer only, no proteins; ●, rat L-FABP; ○, T94T; ▼, T94A.
Table 1.
ANS binding to rat, T94T and T94A L-FABP
| Rat | T94T | T94A | |
|---|---|---|---|
| Fluorescence emission Maximum (nm) | 477 | 480 | 480 |
| A.U./nM when fully bound | 0.78 ± 0.02 | 1.03 ± 0.01 * | 0.98 ± 0.04* |
| Kd (μM) | 2.5 ± 0.2 | 2.3 ± 0.1 | 2.4 ± 0.1 |
| Bmax | 1.42 ± 0.01 | 0.80 ± 0.02 * | 0.86 ± 0.01 *# |
Data are presented as Mean ± SE (n=3 to 5).
P<0.05, human vs. rat L-FABP;
P<0.05, T94A vs. T94T.
To distinguish these possibilities, reverse titrations were carried out in order to obtain the fluorescence efficiency (fluorescence intensity/nM ANS) when ANS was fully bound to L-FABP. The fluorescence efficiency was in turn used in forward titrations to determine Kd’s as described in Methods. In reverse titration, ANS (100nM) was titrated with increasing amount of L-FABP proteins and ANS fluorescence intensity per nM ANS was plotted against L-FABP concentrations (Fig. 4B). Curve fitting of the latter data showed that the ANS fluorescence intensity/nM ANS fully bound to the L-FABPs was 0.8 for rat L-FABP and 1.0 for both human L-FABPs (Table 1). Next, the forward titration was carried out by titrating 500nM L-FABP with increasing amount of ANS. From the ANS fluorescence intensity/nM and total ANS concentration, the fractional saturation and free ANS concentration was calculated and the ANS binding curves plotted (Fig. 4C).
Quantitative analysis of multiple binding curves showed that the ANS binding affinities (Kds) of human and rat L-FABPs were similar (Table 1). In contrast, the Bmax values were significantly higher for rat (Bmax = 1.4) than human L-FABPs (Bmax = 0.8) (Table 1). This was consistent with rat L-FABP having two binding sites for ANS, while human L-FABPs had only one. Since ANS is negatively charged molecule, its binding to proteins depends on both hydrophobicity and positive charges of the binding sites. As indicated by the ANS spectral shift above, the human L-FABP ligand binding site was less hydrophobic environment and human L-FABP has three fewer positively charged amino acid residues than the rat L-FABP (Fig. 1, +).
Binding of long chain fatty acid (LCFA) to rat and human L-FABPs – ANS displacement assays
Since changes in even a single amino acid can significantly alter LCFA binding affinity and/or specificity of L-FABP and other FABPs [72–76], the specificity of rat and human L-FABPs for LCFAs was examined.
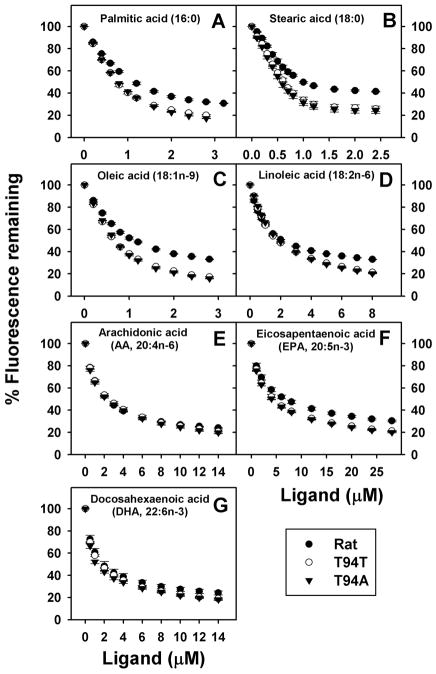
A variety of LCFAs effectively displacement ANS bound to both rat and human L-FABPs including: saturated palmitic and stearic acids (Fig. 5A,B), monounsaturated oleic acid (Fig. 5C), diunsaturated linoleic acid (Fig. 5D), n-6 polyunsaturated arachidonic acid (Fig. 5E), and n-3 polyunsaturated eicosapentaenoic and docosahexaenoic acids (Fig. 5F,G). In almost all cases the LCFAs more completely displaced ANS from the human L-FABPs than from the rat L-FABP, consistent with the nearly 2-fold higher quantity of ANS bound to the latter. The displacement curves of T94T and T94A were essentially superimposable and indicated displacement of the ANS from its single binding site therein.
Figure 5. ANS displacement from L-FABP by long chain fatty acids (LCFAs).
L-FABP (500nM) was incubated with ANS 35μM for 10min. ANS displacement were done by titration with small aliquots of ligands (Panel A, palmitic acid; Panel B, stearic acid; Panel C, oleic acid; Panel D, linoleic acid; Panel E, arachidonic acid; Panel F, eicosapentaenoic acid; Panel G, docosahexaenoic acid.) ●, rat L-FABP; ○, T94T; ▼, T94A.
Quantitative analysis of multiple ANS displacement curves revealed that rat L-FABP bound LCFAs with the following order of binding affinities (Table 2): saturated (palmitic, stearic acid) and monounsaturated (oleic acid) LCFAs > diunsaturated (linoleic acid) and polyunsaturated (DHA > AA > EPA) LCFAs. As compared to rat L-FABP, human L-FABPs bound some LCFAs (palmitic acid, oleic acid) more strongly, but bound others slightly more weakly (linoleic acid) or with similar affinities (stearic acid, AA, EPA, DHA) (Table 2). For all LCFAs, there were only small differences in binding affinities between human T94T and T94A L-FABPs with the T94A variant binding palmitic acid and linoleic acid slightly more weakly (Table 2). Taken together, the ANS displacement studies suggested that all three L-FABPs effectively bound LCFAs with some preference for saturated and monounsaturated LCFAs. However, there was little difference between the human wild-type L-FABP T94T and T94A variant in affinities for the respective LCFAs.
Table 2.
Impact of human L-FABP T94A variant on binding of LCFA and intermediates in triglyceride synthesis.
| ANS Displacement – Ki (μM)a | |||
|---|---|---|---|
|
| |||
| Ligand | Rat | T94T | T94A |
|
| |||
| Fatty Acids | |||
|
| |||
| Palmitic acid (16:0) | 0.048 ± 0.001 | 0.038 ± 0.001 * | 0.043 ± 0.001 *# |
| Stearic acid (18:0) | 0.037 ± 0.004 | 0.029 ± 0.002 | 0.028 ± 0.001 |
| Oleic acid (18:1n-9) | 0.043 ± 0.002 | 0.035 ± 0.001 * | 0.039 ± 0.001 |
| Linoleic acid(18:2n-6) | 0.081 ± 0.005 | 0.113 ± 0.005 * | 0.132 ± 0.003 *# |
| Arachidonic acid (AA, 20:4n-6) | 0.110 ± 0.006 | 0.113 ± 0.006 | 0.108 ± 0.006 |
| Eicosapentaenoic acid (EPA, 20:5n-3) | 0.24 ± 0.03 | 0.20 ± 0.03 | 0.19 ± 0.01 |
| Docosahexaenoic acid (DHA, 22:6n-3) | 0.081 ± 0.015 | 0.072 ± 0.005 | 0.065 ± 0.009 |
|
| |||
| Intermediates in TG Synthesis | |||
|
| |||
| Oleoyl CoA (18:1) | 2.41 ± 0.12 | 1.11 ± 0.04 * | 1.03 ± 0.02 * |
| LPA(18:1) | 99 ± 10 | 0.052 ± 0.007* | 0.036 ± 0.004* |
| 2-OMG(18:1) | ND | ND | ND |
| PODG (16:0, 18:1) | ND | ND | ND |
| POPA (16:0, 18:1) | 9.1 ± 0.8 | 2.8 ± 0.3 * | 2.8 ± 0.2 * |
Values represent mean ± SE, (n=3–5); ND: no displacement; LPA binding to rat L-FABP was measured by Tyrosine quenching rather than ANS displacement.
P<0.05, human vs. rat L-FABP;
P<0.05, T94A vs. T94T.
Binding of intermediates in triglyceride (TG) synthesis by rat and human L-FABPs – ANS displacement assays
To avoid the toxicity elicited by accumulation of excess LCFAs and their active metabolites (LCFA-CoAs), these lipids rapidly oxidized or incorporated into triglycerides for storage or secretion [7;10;77;78]. However, rat and human L-FABPs differ significantly in amino acid sequence [61] and changes in even a single amino acid can significantly alter the affinity and/or specificity of L-FABP and other FABPs for larger ligands (e.g. LCFA-CoA, lysophosphatidic acid, cholesterol than LCFA [73;79–81]. Therefore, the ability of the rat and human L-FABPs to binding intermediates in the synthesis of triglycerides was examined.
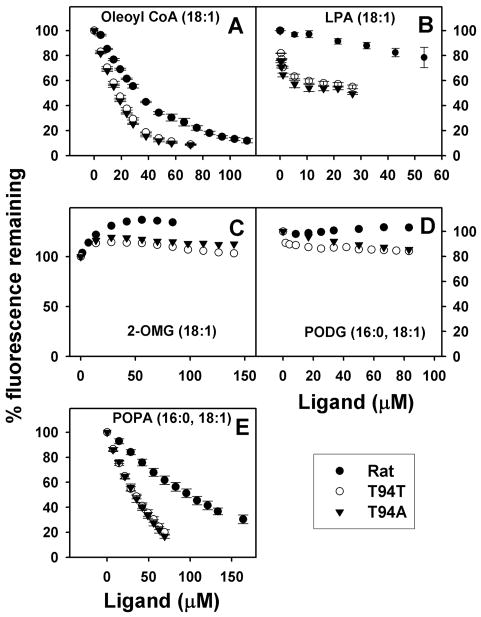
In the ANS displacement assays, only some intermediates in the synthesis of triglycerides effectively bound to rat and human L-FABPs including oleoyl-CoA (Fig. 6A), lysophosphatidic acid (Fig. 6B), and palmitoyl-oleoyl-phosphatidic acid (Fig. 6E), but not mono- or di-glycerides (Fig. 6C,D). Ligands that did not displace ANS may have much weaker binding affinity than ANS. Quantitative analysis of multiple binding curves showed that these ligands were in general more weakly bound than LCFAs by the three L-FABPs in the order: LCFAs ≈ lysophosphatidic acid > oleoyl-CoA > palmitoyl-oleoyl-phosphatidic acid ⋙ mono- or di-glycerides (Table 2). Both human L-FABPs bound oleoyl-CoA, lysophosphatidic acid, and palmitoyl-oleoyl-phosphatidic acid more strongly than did the rat L-FABP (Table 2). However, the human L-FABP T94A variant did not significantly differ from the wild-type L-FABP T94T in affinity for these ligands (Table 2).
Figure 6. ANS displacement from L-FABP by intermediates in triglyceride (TG) synthesis.
L-FABP (500nM) was incubated with ANS 35μM for 10min. ANS displacement were done by titration with small aliquots of ligands (Panel A, Oleoyl CoA; Panel B, LPA (for rat L-FABP, the curved was obtained by tyrosine quenching); Panel C, 2-OMG; Panel D, POPG; Panel E, POPA). ●, rat L-FABP; ○, T94T; ▼, T94A.
Effects of ligand binding on rat, human WT T94T, and human T94A variant L-FABP protein secondary structure-circular dichroism (CD)
Ligand-induced conformational changes in L-FABP may significantly impact the transfer of ligand to and the interaction with target proteins [43–45;82]. To resolve the impact of ligand binding on the secondary structures of the L-FABPs, circular dichroic spectra were obtained for each protein in buffer with or without ligand followed by secondary structure analysis as described in Methods.
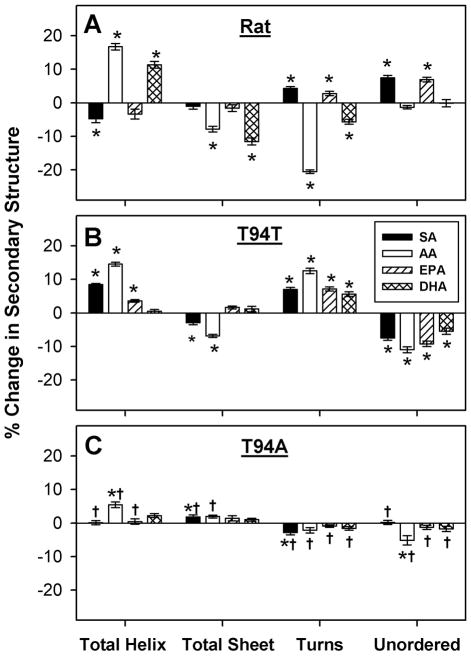
The secondary structure of the rat L-FABP was sensitive to LCFA binding and highly specific for the type of LCFA bound (Fig. 7A). There was no obvious pattern common to all LCFAs with regards to changes in total proportions of helix, sheet, turn and unordered secondary structure elicited by LCFA binding to rat L-FABP (Fig. 7A, Table S1). Each was distinctive.
Figure 7. Changes in secondary structure of rat and human L-FABP upon interaction with stearic acid, arachidonic acid, EPA, or DHA.
L-FABP (0.5 μM) was examined by CD spectroscopy and subsequent secondary structure analysis in the absence or presence of 5 μM ligand as described in Experimental Procedures. The data were presented as % change in secondary structure (L-FABP/ligand – L-FABP only) of rat (panel A), T94T (panel B), and T94A (panel C) L-FABP upon interaction with stearic acid (SA, black bar), arachidonic acid (AA, white bar), EPA (hatched bar), and DHA (cross hatched bar). *, P < 0.05 for L-FABP/ligand secondary structure vs. L-FABP only secondary structure; †, P < 0.05 for T94A/ligand secondary structure vs. T94T/ligand secondary structure.
LCFA-induced changes in wild-type human T94T secondary structure were overall somewhat smaller and differed from those induced by the same LCFA in rat L-FABP (Fig. 7B vs. Fig. 7A). Each specific LCFA elicited a different pattern of secondary structure change on binding human WT L-FABP T94T (Fig. 7B; Table S1) than in rat L-FABP (Fig. 7A, Table S1).
In marked contrast to the rat and human wild-type L-FABPs, the secondary structure of the human L-FABP T94A variant was relatively insensitive to LCFA binding. Secondary structure changes upon binding arachidonic acid, for example, were all <8% with most being only 1–5% (Fig. 7C, AA; Table S1). Similar considerations held for the other LCFAs examined. Further, human L-FABP T94A variant structural changes upon binding SA, AA, EPA and DHA were significantly different from those upon binding to human wild-type L-FABP T94T (Fig. 7C, †).
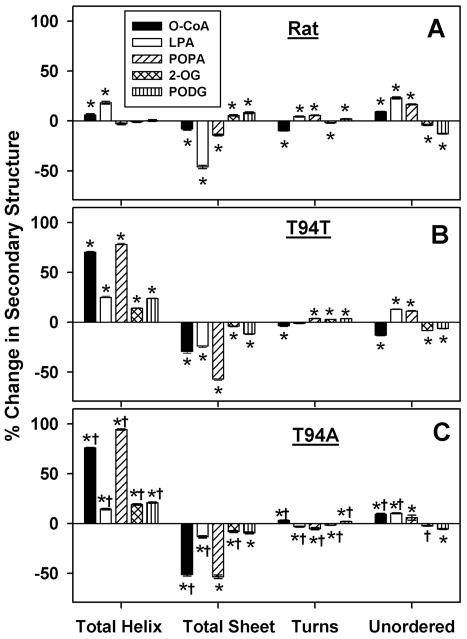
Intermediates of TG synthesis elicited much larger changes than LCFAs in secondary structure of all three L-FABPs (Fig. 8 vs Fig. 7; Table S2 vs Table S1). Furthermore, the conformational changes induced by binding of TG synthesis intermediates to the L-FABPs were in the opposite order: human L-FABP T94A variant > human wild-type L-FABP T94T > rat L-FABP. Ligands which did not bind or for which a Kd could not accurately be resolved (2-OG, PODG) elicited no or only small changes in L-FABP secondary structure.
Figure 8. Changes in secondary structure of rat and human L-FABP upon interaction with O-CoA, LPA, POPA, 2-OMG or PODG.
L-FABP (0.5 μM) was examined by CD spectroscopy and subsequent secondary structure analysis in the absence or presence of 5 μM ligand as described in Experimental Procedures. The data were presented as % change in secondary structure (L-FABP/ligand – L-FABP only) of rat (panel A), T94T (panel B), and T94A (panel C) L-FABP upon interaction with Oleoyl CoA (black bar), LPA (white bar), POPA (hatched bar), and 2-OMG (cross hatched bar), and PODG (vertical hatched bar). *, P < 0.05 for L-FABP/ligand secondary structure vs. L-FABP only secondary structure; †, P < 0.05 for T94A/ligand secondary structure vs. T94T/ligand secondary structure.
Regarding the pattern of secondary structure changes upon binding these ligands, the rat and human L-FABPs differed markedly. The pattern of secondary structure changes induced by ligands (especially O-CoA, LPA, POPA) in rat L-FABP was again somewhat dependent on the type of ligand (Fig. 8A; Table S2). In contrast, both human L-FABPs showed an overall similar qualitative pattern of secondary structure changes upon binding these ligands (Fig. 8B,C; Table S2). However, secondary structural responses of human wild-type L-FABP T94T and T94A variant upon binding the respective ligands were significantly different (Fig. 8C, †; Talbe S2, †).
Functional significance of the human L-FABP T94A mutation: impact on LCFA-mediated transcription of PPARα-regulated proteins in cultured primary human hepatocytes
Both murine and human wild-type L-FABP T94T directly interact with the respective PPARα [43–46;82] to facilitate ligand-induction of PPARα transcription of genes in LCFA metabolism [42;47;82–84]. While human wild-type L-FABP T94T and T94A variant did not differ in binding affinity for LCFAs such as EPA and DHA (Table 2), they differed markedly in secondary structural response to these ligands (Fig. 7). Both exogenous n-3 polyunsaturated LCFAs (EPA, DHA) as well as de novo LCFA synthesis (from glucose) activate PPARα [47;85], the functional significance of the T94A substitution on L-FABP ability to induce ligand-mediated PPARα transcriptional activity was examined in cultured primary human hepatocytes expressing wild-type L-FABP T94T, heterozygous, or homozygous L-FABP T94A variant as described in Methods.
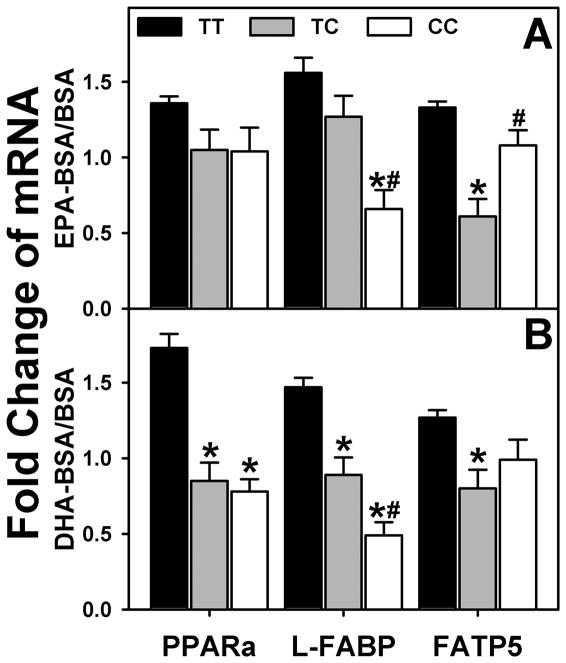
In hepatocytes expressing the wild-type L-FABP T94T (genotyped as TT, Fig. 9A,B; black bars), both EPA and DHA induced transcription of all PPARα regulated genes examined: PPARα itself; L-FABP, the key protein in cytoplasmic LCFA transport and nuclear targeting; FATP5, the key plasma membrane fatty acid translocase in LCFA uptake. In contrast, the expression of the human L-FABP T94A variant (genotyped as TC and CC), especially in hepatocytes homozygous for the T94A variant (genotyped as CC, Fig. 9A,B; open bars), significantly decreased or tended to impair the ability of EPA and DHA to induce PPARα transcription of PPARα, L-FABP, and FATP5.
Figure 9. Effect of human L-FABP T94A mutation on ligand-induced transcription of PPARα-regulated proteins.
Primary human hepatocytes were cultured overnight, and then incubated for 24h with fatty acid-free BSA (Alb) or EPA/BSA, or DHA/BSA (200 μM EPA or DHA) in 6 mM glucose-containing medium as we described in Experimental Procedures. rtPCR was used to determine human PPARα, L-FABP, and FATP5 mRNA levels normalized to an internal control (18S RNA). Values presented were the fold change induced by BSA/EPA or BSA/DHA complex relative to BSA only. Mean ± SEM, n=8–10 different samples in each group, *p<0.05, homozygous T94A (CC) and heterozygous (TC) variants were compared to wild-type (TT); #p<0.05, homozygous T94A (CC) were compared to heterozygous (TC) variants.
Thus, the L-FABP T94A variant impaired LCFA-mediated (i.e. EPA, DHA, de novo synthesized LCFA) signaling to PPARα. The altered L-FABP T94A function correlated with diminished ability of LCFA binding to alter the secondary structure of the L-FABP protein rather than any alteration in LCFA binding affinity.
DISCUSSION
The human L-FABP T94A variant is the most common coding polymorphism in the entire FABP family, occurring with 26–38% minor allele frequency and 8.3±1.9% homozygous in the human population (MAF for 1000 genomes in NCBI dbSNP database; ALFRED database) [48–54]. It has been associated with lipid dysregulation evidenced by elevated plasma triglycerides [49;56] and LDL cholesterol [49;53], atherothrombotic cerebral infarction [51], and non-alcoholic fatty liver disease (NAFLD) [53]. However, little is known about the mechanism(s) whereby expression of the L-FABP T94A variant elicits these effects. To our knowledge there have been no reports actually examining the impact of the T94A substitution on human L-FABP’s ability to bind LCFAs and/or intermediates in TG synthesis, structurally respond to binding of these ligands, or functionally respond to LCFA in primary hepatocytes. The studies described herein provide the following new insights beginning to address these issues:
First, the human L-FABP T94A variant was more resistant to unfolding by urea than the WT human L-FABP T94T. The functional significance of the T94As resistance to unfolding may relate to its ability to interact with target proteins such as PPARα in the nucleus [42–46], GPAT in the endoplasmic reticulum [4;5;7], CPT1A in mitochondria [86], and FATP at the plasma membrane [36]. It was previously shown that wt L-FABP was more stable to temperature unfolding [87]. Protein chemical unfolding and thermal unfolding could be different. For example, a study with human serum albumin showed guanidine and urea interacted with the protein by electrostatic forces, resulted in random coiled conformation, while thermal denaturation produces a molten globe state and the protein aggregation [88]. In a more recent study, it was reported that during chemical denaturation the protein conformations of the transition state were more extended than at high temperature, also the folding routes were different from thermal denaturation [89]. Therefore chemical and thermal denaturation measure different aspect of protein stability. It was shown thermal denaturation affected L-FABP α-helical structure more than β-sheet structure [87]. Interestingly, the human L-FABPs differed significantly from the rat L-FABP in antigenicity, spectral properties, and detergent stability. These species-dependent differences were attributed to the nearly 20% of the human WT L-FABP amino acid sequence being not only non-identical, nearly half of which are nonconservative replacements, as compared to rat L-FABP [61].
Second, the T94A substitution did not or only slightly altered the affinity but not specificity of ligand binding. While the human L-FABP T94A variant differed slightly from the WT L-FABP T94T in affinity for a few LCFAs (16:0, 18:2), it did not differ significantly in affinity for other LCFAs or the more complex ligand intermediates in TG synthesis. Our earlier report also found T94T and T94A have the same (or similar) Ki for phytanic acid, fenofibrate and fenofibric acid [87]. Human L-FABP apparently binds only one molecule of ANS while rat L-FABP binds two. Our fluorescence binding data showed rat L-FABP binds more than one molecules of ANS (Bmax = 1.4), which is in agreement with previous report of Velkov et al [70]. However our human L-FABP data showed it only bind one ANS molecule (Bmax = 0.8)—in agreement with an earlier fluorescence binding assay report wherein human L-FABP bound one ANS with Kd = 2.0 μM [90]. In contrast, an NMR study showed human L-FABP bound two molecules of ANS [91]. However, experiments performed therein utilized a different technique and under very different conditions than those described herein: 1). Concentrations of human L-FABP and ANS used for NMR experiments were much higher than those for fluorescence binding assay; 2) NMR experiments were performed at pH 5.5 while fluorescence binding assay was done at pH 7.4. Since ANS has a negative charged sulfonate group, electrostatic interactions influenced by pH play very important roles in ANS binding to proteins. For example, electrospray ionization mass spectrometry (ESIMS) of ANS binding to different proteins determined different amounts of ANS bound at different pH [92]. For apomyoglobin the maximal ANS binding was observed at pH 4.0 where each protein molecule contained one to six molecules of bound ANS while at neutral pH only a single molecule of ANS was bound [92]. Since L-FABP has more protonated amino acids at pH 5.5 than at pH 7.4, this may account for the additional ANS binding at pH 5.5 [92]; 3) The second ANS bound by L-FABP could be in a more aqueous environment detectable by NMR but not fluorescence.
Third, the human and rat L-FABPs differed significantly in binding affinity, but not specificity for LCFAs. The human L-FABP exhibited small, but significant, differences in the binding of the more saturated LCFAs (16:0, 18:1, 18:2) while its affinity for the polyunsaturated LCFAs (20:4, 20:5, 22:6) did not differ from that of the rat L-FABP. Although two earlier studies separately examined the LCFA binding profiles of rat [7;9;93] and human [94] wild-type L-FABPs, direct comparisons are difficult since different displacement assays were used in each of these studies and both differed from that presented herein. Both human and rat wild-type L-FABP bound the key ligand substrates for triglyceride synthesis: i) LCFA-CoA, consistent with earlier studies [7;9;93;94]; ii) lysophosphatidic, consistent with earlier studies [93;94]; iii) phosphatidic acid (POPA); and iv) L-FABP binding to monoacylglycerol is controversial depending on the technique used. Gel filtration chromatography of liver cytosol, solution NMR, Lipidex, and binding assay with fluorescent MG analog 12-(9-anthroyloxy)oleoyl-sn-1-glycerol (MG12AO) showed that L-FABP binds MG [95;96]. However, fluorescence displacement assays such as the ANS displacement used herein and DAUDA displacement described earlier [75] found little displacement. The exact reason for these differences is not clear. It is important to note that while the key enzyme in triglyceride synthesis from monoacylglycerides (i.e. MGAT) was first thought to be exclusively localized in human and rodent intestine, subsequent studies showed that MGAT was also significantly expressed in human (but not rodent) liver [97]. With respect to the other major intermediate in triglyceride synthesis, neither human nor rat L-FABPs bound diglycerides (PODG). In all cases, the present data demonstrated for the first time that human L-FABP bound several of these LCFA-derived intermediates in TG synthesis with 2–3 fold higher binding affinity than did the rat L-FABP.
Fourth, the L-FABP T94A variant exhibited diminished secondary structural response to LCFA binding. [56]. LCFAs altered L-FABP secondary structure in the order: rat L-FABP > human WT L-FABP T94T ≫ human L-FABP T94A variant. Ligand-induced conformational changes in L-FABP are thought to facilitate ligand transfer from L-FABP to bound PPARα [43–45]. A recent NMR study showed that human WT T94T L-FABP binding of GW7647, another PPARα selective drug, altered the ligand binding cavity and its portal region conformation to stabilize/optimize ligand entry/exit from the β-barrel [45]. Indeed, L-FABP T94A variant expressing human subjects are less responsive to fenofibrate lowering of elevated triglyceride to basal levels [56].
Fifth, binding of intermediates in triglyceride synthesis to the human L-FABP T94A variant altered secondary structure more than with the wild-type L-FABP. Intermediates in TG synthesis that were bound by L-FABP altered L-FABP secondary structure in the reverse order: human L-FABP T94A variant > human WT L-FABP T94T ≫ rat L-FABP. L-FABP is known to facilitate GPAT mediated incorporation of LCFA-CoA into lysophosphatidic acid, the key rate limiting step in de novo phophosphatic acid and triglyceride synthesis [4–7;11;98]. Furthermore, L-FABP conformation significantly determines the ability of GPAT to facilitate incorporation of LCFA-CoA into lysophosphatidic acid [6;7]. Thus, the greater conformational change of the human L-FABP T94A variant to LCFA-CoA binding suggests that this in turn may facilitate LPA (and thus PA and TG) synthesis to help account for the increased NAFLD in human subjects expressing the L-FABP T94A variant [53].
Sixth, human L-FABP T94A variant expressing human primary hepatocytes exhibited substitution diminished LCFA-mediated transcription of PPARα-regulated proteins involved in LCFA uptake (FATP5), intracellular transport (L-FABP), and PPARα itself. L-FABP directly interacts with FATP5 at the mouse hepatocyte plasma membrane [36], and PPARα in the nucleus [42–46;99]. These interactions in turn enhance LCFA uptake [1;34;36;100], LCFA β-oxidation [34;36;78;86], and PPARα transcriptional activity [47;83;84], respectively. Indeed, overexpression of human WT L-FABP T94T, but not the T94A variant, enhanced LCFA uptake in cultured transformed ‘Chang’ liver cells FABP [55]. Furthermore, fenofibrate (like EPA and DHA) was less effective in inducing PPARα transcription of LCFA β-oxidative enzymes (Fig. 9) and is less effective in lowering elevated plasma triglyceride to basal levels in L-FABP T94A variant human subjects [56]. Consequently, the T94A substitution elicited TG accumulation in human subjects expressing the T94A variant [53].
In summary, the T94A substitution in the human T94A variant L-FABP significantly altered the secondary structure and conformational response to LCFA binding. This in turn diminished the ability of the human L-FABP to facilitate LCFA-induction of PPARα transcriptional activity in cultured primary human hepatocytes. WT L-FABP is known to mediate LCFA transport uptake [1;34;36;100], transport through the cytoplasm [10;32;101], and targeting/cotransport into nuclei [35;37–40]. Conversely binding of EPA, DHA, or xenobiotic ligands induces L-FABP translocation into the nucleus [47;83;84]. Inside nuclei, L-FABP directly interacts with PPARα [43–45] to facilitate LCFA transfer [45] for inducing PPARα transcriptional activity [33;35;42;47;83;84]. The sensitivity of L-FABP activity to subtle conformational differences elicited by single amino acid substitutions or conformers is illustrated by studies with the murine L-FABP [5–7;9;86]. The clinical significance of these findings is supported by fenofibrate’s lessened effectiveness in lowering elevated serum triglycerides to basal levels in human subjects expressing the L-FABP T94A variant [56]. Finally, the finding of similar or equal binding affinities for LCFAs demonstrates for the first time that the human L-FABP T94A variant is an altered-function rather than functionless mutation analogous to L-FABP ablation. Loss of L-FABP reduces LCFA and LCFA-CoA binding by cytosolic proteins by 80–90% as well as abolishes L-FABP facilitation of ligand (PUFA, fibrate, TOFA, C57)-mediated induction of PPARα transcriptional activity in murine hepatocytes [47;83;84]. Expression of the T94A variant diminished the ability of PUFA (EPA, DHA) to induce transcription of a variety of proteins involved in LCFA uptake and metabolism in cultured primary human hepatocytes.
MATERIALS AND METHODS
Materials
Antibody against Human L-FABP (H-120), a rabbit polyclonal antibody raised against amino acids 7–126 mapping within an internal region of L-FABP of human origin was purchased from Santa Cruz Biotechnology (Dallas, TX). Antibody against mouse L-FABP was developed in our laboratory as described [102;103]. Mini-PROTEAN TGX Any kD precast polyacrylamide gels as well as Precision Plus Protein Dual Xtra Standards were purchased from Bio-Rad (Hercules, CA). SimplyBlue SafeStain was obtained from Invitrogen (Carlsbad, CA). ANS (1-anilinonaphthalene-8-sulfonic acid) was purchased from Life Technologies (Grand Island, NY). Stearic acid, palmitic acid, oleic acid, linoleic acid, arachidonic acid (AA), cis-5,8,11,14,17-eicosapentaenoic acid (EPA), cis-4,7,10,13,16,19-docosahexaenoic acid (DHA), Oleoyl CoA, and Oleoyl-L-α-lysophosphatidic acid sodium salt (LPA) were purchased from Sigma (St. Louis, MO). 2-oleoyl glycerol (2-OG), 1-palmitoyl-2-oleoyl-sn-glycerol (PODG), and 1-palmitoyl-2-Oleoyl-sn-Glycero-3-Phosphate monosodium salt (POPA) were obtained from Avanti Polar Lipids (Alabaster, Alabama). All reagents and solvents used were of the highest grade available.
Proteins
Recombinant rat, T94T wild-type (WT) human, and T94A mutant human liver fatty acid binding proteins (L-FABPs) were isolated, purified, and dilapidated as described [16;87;104]. Recombinant rat L-FABP as well as human WT T94T and T94A mutant L-FABP protein concentrations were analyzed by amino acid analysis and molecular weights confirmed by matrix-assisted laser desorption time-of-flight (MALDI-TOF) mass spectrometry (Dr. Larry Daggett, Protein Chemistry Laboratory, Texas A&M University, College Station, TX). The respective dilapidated, non-His-tagged recombinant proteins were shown to be >98% pure by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE, 3 μg protein/lane) utilizing a mini-PROTEAN TGX Any kD precast polyacrylamide gel followed by gel staining/detaining with SimplyBlue SafeStain according to the manufacturer’s instructions.
Western blotting
To determine if rat L-FABP, human wild-type L-FABP T94T, and human L-FABP T94A variant reacted equally well with rabbit polyclonal antisera against wild-type mouse or human L-FABP, western blotting was performed with the respective recombinant proteins and antibodies similarly as described previously by our lab [34;35;43].
Fluorescence Spectra and denaturation of Recombinant L-FABP Proteins
Since rat and human L-FABP contain 3 and 1 tyrosine residues, respectively, but no tryptophan, L-FABP was excited at 280 nm and fluorescence emission spectra of tyrosine residues were recorded between 295 and 420 nm using a Varian Cary Eclipse Fluorescence Spectrophotometer (Varian, Inc., Palo Alto, CA). Fluorescence emission intensities at equivalent quantities of tyrosine were obtained using 200nM rat L-FABP, 600nM WT human L-FABP T94T, and 600nm human T94A variant. Temperature was maintained at 24°C with a circulating water bath. The denaturation experiments were done by recording L-FABP tyrosine fluorescence spectra in 8M urea or 6M guanidinium chloride at room temperature.
Ligand Binding: ANS Displacement Assay. ANS Binding to rat and human L-FABPs
ANS is essentially nonfluorescent in buffer, but its fluorescence increases dramatically upon binding to L-FABP. ANS fluorescence emission spectra were obtained by scanning from 410–600nm with 380 nm excitation. In order to determine the Kd of ANS binding to L-FABP, forward titration (500nM L-FABP titrated with 0–48μM ANS), and reverse titrations (100nM ANS titrated with 0–4μM L-FABP) were performed. From the curve fitting of the reverse titration, the fluorescence intensity of ANS (per nM) when fully bound to L-FABP was calculated. This parameter was then used to calculate the fractional saturation and free ANS concentration in forward titration. Binding curved were constructed by plotting fractional saturation (Y) vs free ANS concentration (X), from which Kd and Bmax were determined by curve fitting.
LCFA and LCFA metabolite binding to rat and human L-FABPs: ANS displacement assay
A solution of L-FABP (500nM) and ANS (35μM) was titrated with increasing amount of LCFA, LCFA-CoA, or intermediates in triglyceride synthesis. Displacement curves were constructed by plotting % ANS fluorescence remaining vs ligand concentration. EC50 was obtained from the displacement curve. Ki was calculated from the Kd for ANS determined above and from the EC50 according to the following equation: .
For LPA binding to rat L-FABP, tyrosine quenching was used to construct the binding curve. Tyrosine emission spectra of Rat L-FABP (200nM) were recorded as described above with increasing amount of LPA. Kd was calculated by fitting the curve (100-% fluorescence remaining vs LPA concentration to a hyperbolic equation.
Circular Dichroism Spectroscopy
Circular dichroism (CD) spectroscopy measurements were performed utilizing a JASCO J-815 CD spectrometer (JASCO, Easton, MD) equipped with a Model PFD-425S Peltier Type FDCD attachment for temperature regulation. All temperature and ligand interaction CD experiments were done at 0.5 μM protein (determined by amino acid analysis as above) in a buffer containing 10 mM potassium phosphate (pH 7.4) with or without 1% ethanol. The 1% ethanol had no effect on protein CD spectra or secondary structure analyses (data not shown). Prior to CD scanning from 185 nm to 250 nm, samples were incubated with stirring (250 rpm) at 25 °C for 10 min. The final CD spectrum, representing an average of ten scans, was background-subtracted and mathematically smoothed by the Means-Movement method using a convolution width = 5. The CD manufacturer’s analysis software was used to perform secondary structure analysis using SDP (soluble and denatured protein) 48 as the reference set. While all spectra were analyzed by CONTIN, CDSSTR, and SELCON 3, the CONTIN analysis most consistently yielded the lowest root-mean-square deviation (RMSD) (data not shown).
Ligand interaction CD studies of rat and human L-FABPs (0.5 μM protein in 10 mM potassium phosphate, pH 7.4) were performed as follows: Long chain fatty acids or intermediates in triglyceride synthesis (5 μM) were added from stock solutions (500 μM in ethanol) such that the final ethanol concentration was 1%. Each sample was incubated with stirring at 25 °C for 10 min in the Fluorescence Detected Circular dichroism (FDCD) attachment prior to obtaining the CD spectrum. The final CD spectrum (average of ten scans) was again background subtracted (buffer/ligand/ethanol), mathematically smoothed, and secondary structure analyzed as described above. The percent change in secondary structure between samples was calculated using the following formula: .
PUFA-mediated induction of transcription of PPARα-regulated proteins in cultured primary human hepatocytes: Qrt-PCR
Commercially obtained (Life Technologies, Grand Island, NY) cryopreserved primary human hepatocytes from female (50+3 yrs old) Caucasian donors were genotyped to determine WT T94T (TT), heterozygous (TC), or T94A (CC) variant expression as in [49;50]. The hepatocytes were thawed, plated and cultured overnight according to the manufacturers’ instructions (Life Technologies, Grand Island, NY) and then incubated with 40μM BSA (fatty acid free) or BSA/DHA or BSA/EPA (1:5) complex for 24 hours in glucose free William’s E media (US Biological, Salem, MA) to which 6mM glucose, 100nM insulin, and 10nM dexamethasone had been added. Hepatocytes total mRNA was isolated with RN-easy kit (Qiagen Sciences, Maryland, USA) and RN-ase free DNase set (Qiagen GmbH, Hilden, Germany). All human mRNA analysis reagents (One-Step RT-PCR Master Mix, TaqMan Gene Expression Assays, TaqMan) were from Applied Biosystems (Life Technologies, Grand Island, NY). Human L-FABP, fatty acid transport protein-5 (FATP5), and peroxisome proliferator activated receptor-α (PPAR-α) mRNA levels were determined according to the procedures provided by the manufacturer.
Statistics
One-way analysis of variance (ANOVA) combined with the Newman-Keuls multiple-comparisons post-test (GraphPad Prism Version 3.03, San Diego, CA) was used for all statistical analyses. Data were expressed as means ± standard error of the mean (n = 4–6) with P indicated in the respective figure legends. SigmaPlot 2002 for Windows Version 8.02 (SPSS, Chicago, IL) was used for graphical analysis.
Supplementary Material
Table S1. L-FABP secondary structural changes (%) upon binding stearic acid (SA), arachidonic acid (AA), eicosapentaenoic acid (EPA), docosahexaenoic acid (DHA).
Table S2. L-FABP secondary structure changes (%) upon binding to intermediates in TG synthesis.
Acknowledgments
This work was supported in part by the USPHS National Institutes of Health Grants DK41402 (FS, ABK) and DK70965 (BPA).
Abbreviations
- AA
arachidonic acid
- ANS
1-anilinonaphthalene-8-sulfonic acid
- CD
circular dichroism
- DGAT
diacylglycerol acyltransferase
- DHA
cis-4,7,10,13,16,19-docosahexaenoic acid
- EPA
cis-5,8,11,14,17-eicosapentaenoic acid
- FATP5
Fatty Acid Transport Protein 5
- GPAT
glycerol-3-phosphate acyltransferase
- LCFA
long chain fatty acid
- L-FABP
liver fatty acid binding protein or FABP1
- LPA
Oleoyl-L-α-lysophosphatidic acid sodium salt
- MGAT
monoacylglycerol acyltransferase
- NAFLD
non-alcoholic fatty liver disease
- NASH
non-alcoholic steatohepatitis
- 2-OMG
2-oleoyl glycerol
- O-CoA
Oleoyl Coenzyme A
- PODG
1-palmitoyl-2-oleoyl-sn-glycerol
- POPA
1-palmitoyl-2-Oleoyl-sn-Glycero-3-Phosphate monosodium salt
- PPARα,-β/δ, or - γ
peroxisome proliferator-activated receptor alpha, beta/delta, or gamma
- PUFA
polyunsaturated fatty acids
- SNP
single nucleotide polymorphism
- T94T
wild type human L-FABP
- T94A
human L-FABP T94A variant
- TG
triglyceride
- WT
wild-type
Footnotes
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
References
- 1.Martin GG, Danneberg H, Kumar LS, Atshaves BP, Erol E, Bader M, Schroeder F, Binas B. Decreased liver fatty acid binding capacity and altered liver lipid distribution in mice lacking the liver fatty acid binding protein (L-FABP) gene. J Biol Chem. 2003;278:21429–21438. doi: 10.1074/jbc.M300287200. [DOI] [PubMed] [Google Scholar]
- 2.Martin GG, Huang H, Atshaves BP, Binas B, Schroeder F. Ablation of the liver fatty acid binding protein gene decreases fatty acyl CoA binding capacity and alters fatty acyl CoA pool distribution in mouse liver. Biochem. 2003;42:11520–11532. doi: 10.1021/bi0346749. [DOI] [PubMed] [Google Scholar]
- 3.Anstee QM, Daly AK, Day CP. Genetic modifiers of non-alcoholic fatty liver disease progression. Biochim Biophys Acta. 2011;1812:1557–1566. doi: 10.1016/j.bbadis.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 4.Bordewick U, Heese M, Borchers T, Robenek H, Spener F. Compartmentation of hepatic fatty-acid-binding protein in liver cells and its effect on microsomal phosphatidic acid biosynthesis. Biol Chem Hoppe-Seyler. 1989;370:229–238. doi: 10.1515/bchm3.1989.370.1.229. [DOI] [PubMed] [Google Scholar]
- 5.Jolly CA, Hubbell T, Behnke WD, Schroeder F. Fatty acid binding protein: stimulation of microsomal phosphatidic acid formation. Arch Biochem Biophys. 1997;341:112–121. doi: 10.1006/abbi.1997.9957. [DOI] [PubMed] [Google Scholar]
- 6.Jolly CA, Murphy EJ, Schroeder F. Differential influence of rat liver fatty acid binding protein isoforms on phospholipid fatty acid composition: phosphatidic acid biosynthesis and phospholipid fatty acid remodeling. Biochim Biophys Acta. 1998;1390:258–268. doi: 10.1016/s0005-2760(97)00186-0. [DOI] [PubMed] [Google Scholar]
- 7.Schroeder F, Jolly CA, Cho TH, Frolov AA. Fatty acid binding protein isoforms: structure and function. Chem Phys Lipids. 1998;92:1–25. doi: 10.1016/s0009-3084(98)00003-6. [DOI] [PubMed] [Google Scholar]
- 8.Hubbell T, Behnke WD, Woodford JK, Schroeder F. Recombinant liver fatty acid binding protein interactions with fatty Acyl-Coenzyme A. Biochemistry. 1994;33:3327–3334. doi: 10.1021/bi00177a025. [DOI] [PubMed] [Google Scholar]
- 9.Frolov A, Cho TH, Murphy EJ, Schroeder F. Isoforms of rat liver fatty acid binding protein differ in structure and affinity for fatty acids and fatty acyl CoAs. Biochemistry. 1997;36:6545–6555. doi: 10.1021/bi970205t. [DOI] [PubMed] [Google Scholar]
- 10.McArthur MJ, Atshaves BP, Frolov A, Foxworth WD, Kier AB, Schroeder F. Cellular uptake and intracellular trafficking of long chain fatty acids. J Lipid Res. 1999;40:1371–1383. [PubMed] [Google Scholar]
- 11.Jolly CA, Wilton DA, Schroeder F. Microsomal fatty acyl CoA transacylation and hydrolysis: fatty acyl CoA species dependent modulation by liver fatty acyl CoA binding proteins. Biochim Biophys Acta. 2000;1483:185–197. doi: 10.1016/s1388-1981(99)00170-5. [DOI] [PubMed] [Google Scholar]
- 12.Yang SY, He XY, Schulz H. Fatty acid oxidation in rat brain is limited by the low activity of 3-ketoacyl-coenzyme A thiolase. J Biol Chem. 1987;262:13027–13032. [PubMed] [Google Scholar]
- 13.Higuchi N, Kato M, Tanaka M, et al. Effects of insulin resistance and hepatic lipid accumulation on hepatic mRNA expression levels of apoB, MTP, and L-FABP in non-alcoholic fatty liver disease. Exp and Ther Med. 2011;2:1077–1081. doi: 10.3892/etm.2011.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charlton M, Viker K, Krishnan A, Sanderson S, Veldt B, Kaalsbeek AJ, Kendrick M, Thompson G, Que F, Swain J, Sarr M. Differential expression of lumican and fatty acid binding protein-1: new insights into the histologic spectrum of nonalcoholic fatty liver disease. Hepatology. 2009;49:1375–1384. doi: 10.1002/hep.22927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baumgardner JN, Shankar K, Hennings L, Badger TM, Ronis MJJ. A new model for nonalcoholic steatohepatitis in the rat utilizing total enteral nutrition to overfeed a high-polyunsaturated fat diet. Am J Physiol Gastrointest and Liver Phys. 2007;294:G27–G38. doi: 10.1152/ajpgi.00296.2007. [DOI] [PubMed] [Google Scholar]
- 16.Martin GG, Atshaves BP, Huang H, McIntosh AL, Williams BW, Pai P-J, Russell DH, Kier AB, Schroeder F. Hepatic phenotype of liver fatty acid binding protein (L-FABP) gene ablated mice. Am J Physiol. 2009;297:G1053–G1065. doi: 10.1152/ajpgi.00116.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newberry EP, Xie Y, Kennedy S, Buhman KK, Luo J, Gross RW, Davidson NO. Decreased hepatic triglyceride accumulation and altered fatty acid uptake in mice with deletion of the liver fatty acid binding protein gene. J Biol Chem. 2003;278:51664–51672. doi: 10.1074/jbc.M309377200. [DOI] [PubMed] [Google Scholar]
- 18.Newberry EP, Kennedy SM, Xie Y, Sternard BT, Luo J, Davidson NO. Diet-induced obesity and hepatic steatosis in L-FABP−/− mice is abrogated with SF, but not PUFA, feeding and attenuated after cholesterol supplementation. Am J Physiol Gastrointest and Liver Phys. 2008;294:G307–G314. doi: 10.1152/ajpgi.00377.2007. [DOI] [PubMed] [Google Scholar]
- 19.Newberry EP, Kennedy SM, Xie Y, Luo J, Stanley SE, Semenkovich CF, Crooke RM, Graham MJ, Davidson NO. Altered hepatic triglyceride content after partial hepatectomy without impaired liver regeneration in multiple muring genetic models. Hepatology. 2008;48:1097–1105. doi: 10.1002/hep.22473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie Y, Newberry EP, Kennedy SM, Luo J, Davidson NO. Increased susceptibility to diet-induced gallstones in liver fatty acid binding protein knockout mice. J Lipid Res. 2009;50:977–987. doi: 10.1194/jlr.M800645-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen A, Tang Y, Davis V, Hsu F-F, Kennedy SM, Song H, Turk J, Brunt EM, Newberry EP, Davidson NO. L-FABP modulates murine stellate cell activation and diet induced nonalcoholic fatty liver disease. Hepatology. 2013;57:2202–2212. doi: 10.1002/hep.26318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guzman C, Benet M, Pisonero-Vaquero S, Moya M, Garcia-Mediavilla MV, Martinez-Chantar ML, Gonzalez-Gallego J, Castell JV, Sanchez-Campos S, Jover R. The human liver fatty acid binding protein (FABP1) gene is activated by FOXA1 and PPARa; and repressed by C/EBPa: implicaiton in FABP1 down-regulation in nonalcoholic liver disease. Biochim Biophys Acta. 2013;1831:803–818. doi: 10.1016/j.bbalip.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 23.Rajaraman G, Wang GQ, Yan J, Jiang P, Gong Y, Burczynski FJ. Role of cytosolic liver fatty acid binding protein in hepatocellular oxidative stress: effect of dexamethasone and clofibrate treatment. Mol Cell Biochem. 2007;295:27–34. doi: 10.1007/s11010-006-9268-6. [DOI] [PubMed] [Google Scholar]
- 24.Wang G, Gong Y, Anderson J, Sun D, Minuk G, Robertes MS, Burczynski FJ. Antioxidative function of L-FABP in L-FABP stably transfected Chang liver cells. Hepatology. 2005;42:871–879. doi: 10.1002/hep.20857. [DOI] [PubMed] [Google Scholar]
- 25.Wang G, Shen H, Rajaraman G, Roberts MS, Gong Y, Jiang P, Burczynski FJ. Expression and antioxidant function of liver fatty acid binding protein in normal and bile-duct ligated rats. Eur J Pharm. 2007;560:61–68. doi: 10.1016/j.ejphar.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 26.Yan J, Gong Y, She Y-M, Wang G, Roberts MS, Burczynski FJ. Molecular mechanism of recombinant liver fatty acid binding protein’s antioxidant activity. J Lipid Res. 2009;50:2445–2454. doi: 10.1194/jlr.M900177-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan J, Gong Y, She YM, Wang G, Robertes MS, Burczynski FJ. Molecular mechanism of recombinant L-FABP’s antioxidant activity. J Lipid Res. 2010;50:2445–2454. doi: 10.1194/jlr.M900177-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smathers RL, Fritz KS, Galligan JJ, Shearn CT, Reigan P, Marks MJ, Petersen DR. Characterizationof 4-HNE modified L-FABP reveals alterations in structural and functional dynamics. PPAR Research. 2012;7:e38459. doi: 10.1371/journal.pone.0038459. doi:10/1371/journal.pone.0038459 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan W, Chen K, Zheng G, Wang W, Teng A, Liu A, Ming D, Yan P. Role of liver fatty acid binding protein in hepatocellular injury: Effect of CrPic treatment. J Inorganic Biochem. 2013;124:46–53. doi: 10.1016/j.jinorgbio.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 30.Murphy EJ, Prows DR, Jefferson JR, Schroeder F. Liver fatty acid binding protein expression in transfected fibroblasts stimulates fatty acid uptake and metabolism. Biochim Biophys Acta. 1996;1301:191–198. doi: 10.1016/0005-2760(96)00024-0. [DOI] [PubMed] [Google Scholar]
- 31.Prows DR, Murphy EJ, Schroeder F. Intestinal and liver fatty acid binding proteins differentially affect fatty acid uptake and esterification in L-Cells. Lipids. 1995;30:907–910. doi: 10.1007/BF02537481. [DOI] [PubMed] [Google Scholar]
- 32.Murphy EJ. L-FABP and I-FABP expression increase NBD-stearate uptake and cytoplasmic diffusion in L-cells. Am J Physiol. 1998;275:G244–G249. doi: 10.1152/ajpgi.1998.275.2.G244. [DOI] [PubMed] [Google Scholar]
- 33.Wolfrum C, Buhlman C, Rolf B, Borchers T, Spener F. Variation of liver fatty acid binding protein content in the human hepatoma cell line HepG2 by peroxisome proliferators and antisense RNA affects the rate of fatty acid uptake. Biochim Biophys Acta. 1999;1437:194–201. doi: 10.1016/s1388-1981(99)00008-6. [DOI] [PubMed] [Google Scholar]
- 34.Atshaves BP, McIntosh AL, Lyuksyutova OI, Zipfel WR, Webb WW, Schroeder F. Liver fatty acid binding protein gene ablation inhibits branched-chain fatty acid metabolism in cultured primary hepatocytes. J Biol Chem. 2004;279:30954–30965. doi: 10.1074/jbc.M313571200. [DOI] [PubMed] [Google Scholar]
- 35.McIntosh AL, Atshaves BP, Hostetler HA, Huang H, Davis J, Lyuksyutova OI, Landrock D, Kier AB, Schroeder F. Liver type fatty acid binding protein (L-FABP) gene ablation reduces nuclear ligand distribution and peroxisome proliferator activated receptor-alpha activity in cultured primary hepatocytes. Arch Biochem Biophys. 2009;485:160–173. doi: 10.1016/j.abb.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Storey SM, McIntosh AL, Huang H, Martin GG, Landrock KK, Landrock D, Payne HR, Kier AB, Schroeder F. Loss of intracellular lipid binding proteins differentially impacts saturated fatty acid uptake and nuclear targeting in mouse hepatocytes. Am J Physiol Gastrointest and Liver Phys. 2012;303:G837–G850. doi: 10.1152/ajpgi.00489.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McIntosh AL, Huang H, Atshaves BP, Wellburg E, Kuklev DV, Smith WL, Kier AB, Schroeder F. Fluorescent n-3 and n-6 very long chain polyunsaturated fatty acids: three photon imaging and metabolism in living cells overexpressing liver fatty acid binding protein. J Biol Chem. 2010;285:18693–18708. doi: 10.1074/jbc.M109.079897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lawrence JW, Kroll DJ, Eacho PI. Ligand dependent interaction of hepatic fatty acid binding protein with the nucleus. J Lipid Res. 2000;41:1390–1401. [PubMed] [Google Scholar]
- 39.Huang H, Starodub O, McIntosh A, Kier AB, Schroeder F. Liver fatty acid binding protein targets fatty acids to the nucleus: real-time confocal and multiphoton fluorescence imaging in living cells. J Biol Chem. 2002;277:29139–29151. doi: 10.1074/jbc.M202923200. [DOI] [PubMed] [Google Scholar]
- 40.Huang H, Starodub O, McIntosh A, Atshaves BP, Woldegiorgis G, Kier AB, Schroeder F. Liver fatty acid binding protein colocalizes with peroxisome proliferator receptor alpha and enhances ligand distribution to nuclei of living cells. Biochemistry. 2004;43:2484–2500. doi: 10.1021/bi0352318. [DOI] [PubMed] [Google Scholar]
- 41.Hostetler HA, McIntosh AL, Petrescu AD, Huang H, Atshaves BP, Murphy EJ, Kier AB, Schroeder F. Fluorescence methods to assess the impact of lipid binding proteins on ligand activated gene expression. In: Murphy EJ, Rosenberger TA, editors. Methods in Lipid-Mediated Signaling. CRC Press-Taylor and Francis Group, LLC; Boca Raton, FL: 2010. pp. 299–348. [Google Scholar]
- 42.Wolfrum C, Borrmann CM, Borchers T, Spener F. Fatty acids and hypolipidemic drugs regulate PPARalpha and PPARgamma gene expresion via L-FABP: a signaling path to the nucleus. Proc Natl Acad Sci. 2001;98:2323–2328. doi: 10.1073/pnas.051619898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hostetler HA, McIntosh AL, Atshaves BP, Storey SM, Payne HR, Kier AB, Schroeder F. Liver type Fatty Acid Binding Protein (L-FABP) interacts with peroxisome proliferator activated receptor-a in cultured primary hepatocytes. J Lipid Res. 2009;50:1663–1675. doi: 10.1194/jlr.M900058-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hostetler HA, Balanarasimha M, Huang H, Kelzer MS, Kaliappan A, Kier AB, Schroeder F. Glucose regulates fatty acid binding protein interaction with lipids and PPARa. J Lipid Res. 2010;51:3103–3116. doi: 10.1194/jlr.M005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Velkov T. Interactions between human liver fatty acid binding protein and peroxisome proliferator activated receptor drugs. PPAR Research. 2013;2013:1–14. doi: 10.1155/2013/938401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smathers RL, Galligan JJ, Shearn CT, Fritz KS, Mercer K, Ronis M, Orlicky DJ, Davidson NO, Petersen DR. Susceptibility of L-FABP −/− mice to oxidative stress in early-stage alcoholic liver. J Lipid Res. 2013;54:1335–1345. doi: 10.1194/jlr.M034892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petrescu AD, Huang H, Martin GG, McIntosh AL, Storey SM, Landrock D, Kier AB, Schroeder F. Impact of L-FABP and glucose on polyunsaturated fatty acid induction of PPARa regulated b-oxidative enzymes. Am J Physiol Gastrointest and Liver Phys. 2012;304:G241–G256. doi: 10.1152/ajpgi.00334.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robitaille J, Brouillette C, Lemieux S, Perusse L, Gaudet D, Vohl M-C. Plasma concentrations of apolipoprotein B are modulated by a gene-diet interaction effect between the L-FABP T94A polymorphism and dietary fat intake in French-Canadian men. Mol Gen and Metab. 2004;82:296–303. doi: 10.1016/j.ymgme.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 49.Fisher E, Weikert C, Klapper M, Lindner I, Mohlig M, Spranger J, Boeing H, Schrezenmeir J, Doring F. L-FABP T94A is associated with fasting triglycerides and LDL-cholesterol in women. Mol Gen and Metab. 2007;91:278–284. doi: 10.1016/j.ymgme.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 50.Weikert MO, Loeffelholz Cv, Roden M, Chandramouli V, Brehm A, Nowotny P, Osterhoff MA, Isken F, Spranger J, Landau BR, Pfeiffer A, Mohlig M. A Thr94Ala mutation in human liver fatty acid binding protein contributes to reduced hepatic glycogenolysis and blunted elevation of plasma glucsoe levels in lipid-exposed subjects. Am J Physiol Endocrinol Metab. 2007;293:E1078–E1084. doi: 10.1152/ajpendo.00337.2007. [DOI] [PubMed] [Google Scholar]
- 51.Yamada Y, Kato K, Oguri M, Yoshida T, Yokoi K, Watanabe S, Metoki N, Yoshida H, Satoh K, Ichihara S, Aoyagi Y, Yasunaga A, Park H, Tanaka M, Nozawa Y. Association of genetic variants with atherothrombotic cerebral infarction in Japanese individuals with metabolic syndrome. Int J Mol Med. 2008;21:801–808. [PubMed] [Google Scholar]
- 52.Bu L, Salto LM, De Leon KJ, De Leon M. Polymorphisms in fatty acid binding protein 5 show association with type 2 diabetes. Diabetes Res Clin Prac. 2011;92:82–91. doi: 10.1016/j.diabres.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peng X-E, Wu YL, Lu Q-Q, Ju Z-J, Lin X. Two genertic variants in FABP1 and susceptibility to non-alcoholic fatty liver disease in a Chinese population. Gene. 2012;500:54–58. doi: 10.1016/j.gene.2012.03.050. [DOI] [PubMed] [Google Scholar]
- 54.Mansego ML, Martinez F, Martinez-Larrad MT, Zabena C, Rojo G, Morcillo S, Soriguer F, Martin-Escudero JC, Serrano-Rios M, Redon J, Chaves FJ. Common variants of the liver fatty acid binding protein gene influence the risk of Type 2 Diabetes and insulin resistance in Spanish population. PLoS ONE. 2012;7:e31853. doi: 10.1371/journal.pone.0031853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao N, Qu X, Yan J, Huang Q, Yuan HY, Ouyang D-S. L-FABP T94A decreased fatty acid uptake and altered hepatic triglyceride and cholesterol accumulation in Chang liver cells stably transfected with L-FABP. Mol Cell Biochem. 2010;345:207–214. doi: 10.1007/s11010-010-0574-7. [DOI] [PubMed] [Google Scholar]
- 56.Brouillette C, Bose Y, Perusse L, Gaudet D, Vohl M-C. Effect of liver fatty acid binding protein (FABP) T94A missense mutation on plasma lipoprotein responsiveness to treatment with fenofibrate. J Hum Gen. 2004;49:424–432. doi: 10.1007/s10038-004-0171-2. [DOI] [PubMed] [Google Scholar]
- 57.Masters JR. HeLa cells 50 years on: the good, the bad and the ugly. Nature Rev Cancer. 2002;2:315–319. doi: 10.1038/nrc775. [DOI] [PubMed] [Google Scholar]
- 58.Nelson-Rees WA, Flandermeyer RR. HeLa cultures defined. Science. 1976;191:96–98. doi: 10.1126/science.1246601. [DOI] [PubMed] [Google Scholar]
- 59.Masters JR. Cell line misidentification: the beginning of the end. Nature Rev Cancer. 2010;10:441–448. doi: 10.1038/nrc2852. [DOI] [PubMed] [Google Scholar]
- 60.Cai J, Lucke C, Chen Z, Qiao Y, Klimtchuk E, Hamilton JA. Solution structure and backbone dynamics of human liver fatty acid binding protein: fatty acid binding revisited. Biophys J. 2012;102:2585–2594. doi: 10.1016/j.bpj.2012.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Betts MJ, Russell RB. Amino Acid Properties and Conseaquences of Substitutions. In: Barnes MR, Gray IC, editors. Bioinformatics for Geneticists. 2003. pp. 289–316. [Google Scholar]
- 62.Thompson J, Winter N, Terwey D, Bratt J, Banaszak L. The crystal structure of the liver fatty acid-binding protein. J Biol Chem. 1997;272:7140–7150. doi: 10.1074/jbc.272.11.7140. [DOI] [PubMed] [Google Scholar]
- 63.Cai J, Lucke C, Qiao Y, Klimtchuk E, Hamilton JA. Solution structure and backbone dynamics of human liver fatty acid binding protein. Biophys J. 2010;98:238a. doi: 10.1016/j.bpj.2012.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sharma A, Sharma A. Fatty acid induced remodeling within the human liver fatty acid binding protein. J Bio Chem. 2011:286. doi: 10.1074/jbc.M111.270165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zamyatin AA. Protein volume in solution. Prog Biophys Mol Biol. 1972;24:107–123. doi: 10.1016/0079-6107(72)90005-3. [DOI] [PubMed] [Google Scholar]
- 66.Chotia C. the nature of the accessible and buried surfaces of proteins. J Mol Biol. 1975;105:1–14. doi: 10.1016/0022-2836(76)90191-1. [DOI] [PubMed] [Google Scholar]
- 67.Ross JBA, Laws WR, Rousslang KW, Wyssbrod HR. Tyrosine Fluorescence and Phosphorescence from Proteins and Polypeptides. In: Lakowicz JR, editor. Topics in Fluorescence Spectroscopy. Plenum Press; New York: 1992. pp. 1–63. [Google Scholar]
- 68.He Y, Yang X, Wang H, Estephan R, Francis F, Kodukula S, Storch J, Stark RE. Solution-state molecular structure of apo and oleate-liganded liver fatty acid binding protein. Biochemistry. 2007;46:12543–12556. doi: 10.1021/bi701092r. [DOI] [PubMed] [Google Scholar]
- 69.Chuang S, Velkov T, Horne J, Wielens J, Chalmers DK, Porter CJH, Scanlon MJ. Probing fibrate binding specificity of rat liver fatty acid binding protein. J Med Chem. 2009;52:5344–5355. doi: 10.1021/jm801349e. [DOI] [PubMed] [Google Scholar]
- 70.Chuang S, Velkov T, Horne J, Porter CJH, Scanlon MJ. Characterization of the drug binding specificity of rat liver fatty acid binding protein. J Med Chem. 2008;51:3755–3764. doi: 10.1021/jm701192w. [DOI] [PubMed] [Google Scholar]
- 71.Carbone V, Velkov T. Interaction of phthalates and phonxy acid herbicide environmental pollutants with intestinal intracellular lipid binding proteins. Chem Res in Tox. 2013;26:1240–1250. doi: 10.1021/tx400170t. [DOI] [PubMed] [Google Scholar]
- 72.Baier LJ, Sacchettini JC, Knowler WC, Eads J, Paolisso G, Tataranni PA, Mochizuki H, Bennett PH, Bogardus C, Prochazka M. An amino acid substitution in the human intestinal fatty acid binding protein is associated with increased fatty acid binding, increased fat oxidation, and insulin resistance. J Clin Inv. 1995;95:1281–1287. doi: 10.1172/JCI117778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Woodford JK, Behnke WD, Schroeder F. Liver fatty acid binding protein enhances sterol transfer by membrane interaction. Mol Cell Biochem. 1995;152:51–62. doi: 10.1007/BF01076463. [DOI] [PubMed] [Google Scholar]
- 74.Schulenberg-Schell H, Schafer P, Keuper HJ, Stanislawski B, Hoffmann E, Ruterjans H, Spener F. Interactions of fatty acids with neutral fatty-acid-binding protein from bovine liver. Eur J Biochem. 1988;170:565–574. doi: 10.1111/j.1432-1033.1988.tb13735.x. [DOI] [PubMed] [Google Scholar]
- 75.Thumser AE, Voysey J, Wilton DC. Mutations of recombinant rat liver fatty acid-binding protein at residues 102 and 122 alter its structural integrity and affinity for physiological ligands. Biochem J. 1996;314:943–949. doi: 10.1042/bj3140943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jakoby MG, Miller KR, Joner JJ, Bauman A, Cheng L, Li E, Cistola DP. Ligand-protein electrostatic interactions govern the specificity of retinol- and fatty acid-binding proteins. Biochemistry. 1993;32:872–878. doi: 10.1021/bi00054a019. [DOI] [PubMed] [Google Scholar]
- 77.Powell GL, Tippett PS, Kiorpes TC, McMillin-Wood J, Coll KE, Schultz H, Tanaka K, Kang ES, Shrago E. Fatty acyl CoA as an effector molecule in metabolism. Fed Proc. 1985;44:81–84. [Google Scholar]
- 78.Atshaves BP, Martin GG, Hostetler HA, McIntosh AL, Kier AB, Schroeder F. Liver fatty acid binding protein (L-FABP) and Dietary Obesity. Journal of Nutritional Biochemisty. 2010;21:1015–1032. doi: 10.1016/j.jnutbio.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Evans C, Wilton DC. The chemical modification of cysteine-69 of rat liver fatty acid- binding protein (FABP): a fluorescence approach to FABP structure and function. Mol Cell Biochem. 1990;98:135–140. doi: 10.1007/BF00231377. [DOI] [PubMed] [Google Scholar]
- 80.Thumser AE, Evans C, Worrall AF, Wilton DC. Effect on ligand binding of arginine mutations in recombinant rat liver fatty acid-binding protein. Biochem J. 1994;297:103–107. doi: 10.1042/bj2970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hagan RM, Davies JK, Wilton DA. Effect of charge reversal mutations in the a-helical region of L_FABP on binding of fatty acyl CoAs, lysophospholipids, and bile acids. Mol Cell Biochem. 2002;239:55–60. [PubMed] [Google Scholar]
- 82.Schroeder F, Petrescu AD, Huang H, Atshaves BP, McIntosh AL, Martin GG, Hostetler HA, Vespa A, Landrock K, Landrock D, Payne HR, Kier AB. Role of fatty acid binding proteins and long chain fatty acids in modulating nuclear receptors and gene transcription. Lipids. 2008;43:1–17. doi: 10.1007/s11745-007-3111-z. [DOI] [PubMed] [Google Scholar]
- 83.Huang H, McIntosh AL, Martin GG, Petrescu AD, Landrock K, Landrock D, Kier AB, Schroeder F. Inhibitors of fatty acid synthesis induce PPARa-regulated fatty acid b-oxidative enzymes: synergistic roles of L-FABP and glucose. PPAR Research. 2013;2013:1–22. doi: 10.1155/2013/865604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Petrescu AD, McIntosh AL, Storey SM, Huang H, Martin GG, Landrock D, Kier AB, Schroeder F. High glucose potentiates liver fatty acid binding protein (L-FABP) mediated fibrate induction of PPARa in mouse hepatocytes. Biochim Biophys Acta. 2013;1831:1412–1425. doi: 10.1016/j.bbalip.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chakravarthy MV, Pan Z, Zhu Y, Tordjman K, Schneider JG, Coleman T, Turk J, Semenkovich CF. “New” hepatic fat activates PPARalpha to maintain glucose, lipid, and cholesterol homeostasis. Cell Metabolism. 2005;1:309–322. doi: 10.1016/j.cmet.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 86.Hostetler HA, Lupas D, Tan Y, Dai J, Kelzer MS, Martin GG, Woldegiorgis G, Kier AB, Schroeder F. Acyl-CoA binding proteins interact with the acyl-CoA binding domain of mitochondrial carnitine palmitoyltransferase I. Mol Cell Biochem. 2011;355:135–148. doi: 10.1007/s11010-011-0847-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Martin GG, McIntosh AL, Huang H, Gupta S, Atshaves BP, Kier AB, Schroeder F. Human liver fatty acid binding protein (L-FABP) T94A variant alters structure, stability, and interaction with fibrates. Biochemistry. 2013;52:9347–9357. doi: 10.1021/bi401014k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Farruggia B, Pico GA. Thermodynamic features of the chemical and thermal denaturations of human serum albumin. Int J Biol Macromolecules. 1999;26:317–323. doi: 10.1016/s0141-8130(99)00054-9. [DOI] [PubMed] [Google Scholar]
- 89.Wang Q, Christiansen A, Samiotakis A, Wittung-Stafshede P, Cheung MS. Comparison of chemical and thermal protein denaturation by combination of computational and experimental approaches. II J Chem Phys. 2011;135:175102. doi: 10.1063/1.3656692. [DOI] [PubMed] [Google Scholar]
- 90.Veerkamp JH, van Moerkerk HT, Prinsen CF, Van Kuppevelt TH. Structural and functional studies on different human FABP types. Mol Cell Biochem. 1999;192:137–142. [PubMed] [Google Scholar]
- 91.Long D, Yang D. Millisecond timescale dynamics of human liver fatty acid binding protein: testing of its relevance to the ligand entry process. Biophys J. 2010;98:3054–3061. doi: 10.1016/j.bpj.2010.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ray SS, Balaram P. 1-Anilino-8-naphthalene-sulfonate (ANS) binding to proteins investigated by electrospray ionization mass spectrometry: correlation of gas-hase dye binding to population of molten globule states in solution. J Phys Chem. 1999;103:7068–7072. [Google Scholar]
- 93.Di Pietro SM, Santome JA. Isolation, characterization, and binding properties of two rat liver fatty acid binding protein isoforms. Biochim Biophys Acta. 2000;1478:186–200. doi: 10.1016/s0167-4838(00)00042-x. [DOI] [PubMed] [Google Scholar]
- 94.Maatman RG, van Moerkerk HT, Nooren IM, van Zoelen EJ, Veerkamp JH. Expression of human liver fatty acid-binding protein in Escherichia coli and comparative analysis of its binding characteristics with muscle fatty acid-binding protein. Biochim Biophys Acta. 1994;1214:1–10. doi: 10.1016/0005-2760(94)90002-7. [DOI] [PubMed] [Google Scholar]
- 95.Lagakos WS, Guan X, Ho S-Y, Sawicki LR, Corsico B, Murota K, Stark RE, Storch J. L-FABP binds monoacylglycerol in vitro and in mouse liver cytosol. J Biol Chem. 2013;288:19805–19815. doi: 10.1074/jbc.M113.473579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Storch J. Diversity of fatty acid-binding protein structure and function: studies with fluorescent ligands [Review] Mol Cell Biochem. 1993;123:45–53. doi: 10.1007/BF01076474. [DOI] [PubMed] [Google Scholar]
- 97.Hall AM, Kou K, Chen Z, Pietka TA, et al. Evidence for regulated monoacylglycerol acyltransferase expression and activity in human liver. J Lipid Res. 2012;53:990–999. doi: 10.1194/jlr.P025536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jolly CA, Hubbell T, Behnke WD, Schroeder F. Fatty acid binding protein: Stimulation of microsomal phosphatidic acid formation. Arch Biochem Biophys. 1997;341:112–121. doi: 10.1006/abbi.1997.9957. [DOI] [PubMed] [Google Scholar]
- 99.Schroeder F, Huang H, Hostetler HA, Petrescu AD, Hertz R, Bar-Tana J, Kier AB. Stability of fatty acyl CoA thioester ligands of hepatocyte nuclear factor -4alpha and peroxisome proliferator-activated receptor alpha. Lipids. 2005;40:559–568. doi: 10.1007/s11745-005-1416-y. [DOI] [PubMed] [Google Scholar]
- 100.Atshaves BP, Storey SM, Petrescu AD, Greenberg CC, Lyuksyutova OI, Smith R, Schroeder F. Expression of fatty acid binding proteins inhibits lipid accumulation and alters toxicity in L-cell fibroblasts. Am J Physiol. 2002;283:C688–C703. doi: 10.1152/ajpcell.00586.2001. [DOI] [PubMed] [Google Scholar]
- 101.Weisiger RA. Cytosolic fatty acid binding proteins catalyze two distinct steps in intracellular transport of their ligands. Mol Cell Biochem. 2005;239:35–42. [PubMed] [Google Scholar]
- 102.Jefferson JR, Powell DM, Rymaszewski Z, Kukowska-Latallo J, Schroeder F. Altered membrane structure in transfected mouse L-Cell fibroblasts expressing rat liver fatty acid-binding protein. J Biol Chem. 1990;265:11062–11068. [PubMed] [Google Scholar]
- 103.Schroeder F, Atshaves BP, Starodub O, Boedeker AL, Smith R, Roths JB, Foxworth WB, Kier AB. Expression of liver fatty acid binding protein alters growth and differentiation of embryonic stem cells. Mol Cell Biochem. 2001;219:127–138. doi: 10.1023/a:1010851130136. [DOI] [PubMed] [Google Scholar]
- 104.Chao H, Zhou M, McIntosh A, Schroeder F, Kier AB. Acyl CoA binding protein and cholesterol differentially alter fatty acyl CoA utilization by microsomal acyl CoA: cholesterol transferase. J Lipid Res. 2003;44:72–83. doi: 10.1194/jlr.m200191-jlr200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. L-FABP secondary structural changes (%) upon binding stearic acid (SA), arachidonic acid (AA), eicosapentaenoic acid (EPA), docosahexaenoic acid (DHA).
Table S2. L-FABP secondary structure changes (%) upon binding to intermediates in TG synthesis.