This work reports crystal structures of trimeric riboflavin synthase from the pathogen B. abortus both as the apo protein and in complex with several ligands of interest. It is shown that ligand binding drives the assembly of the unique active site of the trimer, and these findings are complemented by a detailed kinetic study on this enzyme, in which marked inhibition by substrate and product was observed.
Keywords: enzyme–ligand complex; inhibition by substrate and product; 6,7-dimethyl-8-ribityllumazine; vitamin B2
Abstract
Riboflavin synthase (RS) catalyzes the last step of riboflavin biosynthesis in microorganisms and plants, which corresponds to the dismutation of two molecules of 6,7-dimethyl-8-ribityllumazine to yield one molecule of riboflavin and one molecule of 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione. Owing to the absence of this enzyme in animals and the fact that most pathogenic bacteria show a strict dependence on riboflavin biosynthesis, RS has been proposed as a potential target for antimicrobial drug development. Eubacterial, fungal and plant RSs assemble as homotrimers lacking C 3 symmetry. Each monomer can bind two substrate molecules, yet there is only one active site for the whole enzyme, which is located at the interface between two neighbouring chains. This work reports the crystallographic structure of RS from the pathogenic bacterium Brucella abortus (the aetiological agent of the disease brucellosis) in its apo form, in complex with riboflavin and in complex with two different product analogues, being the first time that the structure of an intact RS trimer with bound ligands has been solved. These crystal models support the hypothesis of enhanced flexibility in the particle and also highlight the role of the ligands in assembling the unique active site. Kinetic and binding studies were also performed to complement these findings. The structural and biochemical information generated may be useful for the rational design of novel RS inhibitors with antimicrobial activity.
1. Introduction
Riboflavin (vitamin B2) is the precursor of FMN and FAD, two essential cofactors for all organisms that are involved in a wide variety of cellular processes, including DNA damage repair, bioluminescence, signal transduction and circadian clock regulation (Müller, 1992 ▶; Massey, 2000 ▶). This vitamin is biosynthesized in microorganisms and plants but not in humans and animals, which must acquire it through their diet. This observation, added to the fact that most pathogenic bacteria are unable to acquire flavins from the environment efficiently, highlight the riboflavin pathway as an interesting target for the development of antimicrobial drugs (Long et al., 2010 ▶).
Riboflavin is biosynthesized from one equivalent of GTP and two equivalents of ribulose 5′-phosphate, as summarized in Fig. 1 ▶ (Fischer & Bacher, 2005 ▶, 2006 ▶). The penultimate step is catalyzed by 6,7-dimethyl-8-ribityllumazine synthase (LS) and involves the condensation of 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione (1) with 3,4-dihydroxy-2-butanone 4-phosphate (2) to yield 6,7-dimethyl-8-ribityllumazine (3). Two molecules of the latter compound then undergo a very unusual dismutation reaction which is catalyzed by riboflavin synthase (RS) and corresponds to the main subject of this work, yielding riboflavin (4) with the regeneration of one molecule of 1, which can be recycled by LS (Fischer & Bacher, 2008 ▶, 2011 ▶; Ladenstein et al., 2013 ▶).
Figure 1.
The main catalytic steps involved in the biosynthesis of riboflavin. 1, 5-Amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione; 2, 3,4-dihydroxy-2-butanone 4-phosphate; 3, 6,7-dimethyl-8-ribityllumazine; 4, riboflavin; DHBPS, 3,4-dihydroxy-2-butanone 4-phosphate synthase; LS, 6,7-dimethyl-8-ribityllumazine synthase; RS, riboflavin synthase (reaction highlighted on a green background).
RS is assembled as a homotrimer in eubacteria, fungi and plants (Eberhardt et al., 1996 ▶; Liao et al., 2001 ▶; Gerhardt, Schott et al., 2002 ▶; Fischer et al., 2005 ▶), whereas it folds as a homopentamer with an unrelated amino-acid sequence in archaea (Fischer et al., 2004 ▶; Ramsperger et al., 2006 ▶). The dismutation reaction can be summarized as a four-carbon unit transfer between two 6,7-dimethyl-8-ribityllumazine molecules, with the donor and acceptor molecules observing an opposite orientation of their 6- and 7-methyl groups at the active site (Supplementary Fig. S11). Briefly, the accepted mechanism involves three key steps: (i) transfer of a hydride ion from the acceptor to the donor molecule, (ii) a [4+2] cycloaddition that yields a pentacyclic intermediate and (iii) a sequence of two β-eliminations that gives rise to the xylene ring of riboflavin (Illarionov, Eisenreich et al., 2001 ▶; Kim et al., 2010 ▶). Interestingly, catalytic studies of selected RS point mutants from Escherichia coli showed that a great number of highly conserved polar residues can be mutated to alanine without a significant change in the catalytic activity (Illarionov, Kemter et al., 2001 ▶). Additionally, the formation of riboflavin can also proceed under relatively mild conditions, i.e. when boiling an aqueous solution of 6,7-dimethyl-8-ribityllumazine at neutral or acidic pH (Beach & Plaut, 1969 ▶). These observations suggest that RS might perform its catalytic activity mainly by assuring the proper orientation of the substrate molecules.
Brucella abortus is a pathogenic Gram-negative bacterium that is responsible for the disease brucellosis, a worldwide zoonosis that affects humans and livestock, and for which a safe and completely reliable vaccine is still unavailable (Godfroid et al., 2005 ▶; Pappas et al., 2005 ▶; Gorvel, 2008 ▶). The enzymes of the riboflavin pathway in B. abortus bear considerable interest as targets against this disease (Bonomi et al., 2010 ▶). Its LS, named BLS and RibH2 in several reports by our group, is a major candidate for the development of a subunit vaccine against brucellosis (Laplagne et al., 2004 ▶; Berguer et al., 2006 ▶; Craig et al., 2012 ▶) and has been described in detail from a structural point of view (Zylberman et al., 2004 ▶; Klinke et al., 2005 ▶). On the other hand, its RS has been recently proposed as another important immunogenic candidate protein for the development of a brucellosis subunit vaccine (Yang et al., 2011 ▶), but structural information is unavailable to date.
In this work, we present the crystallographic structure of RS from B. abortus. In order to describe its substrate-binding sites, we also solved the structure of the enzyme in the presence of bound ligands, namely one of the products of the reaction (riboflavin) and two product analogues [roseoflavin and 5-nitro-6-ribitylamino-2,4(1H,3H)-pyrimidinedione (NRP); see Fig. 7 for their chemical structures]. Previous structural information on eubacterial and fungal RSs includes (i) the trimeric enzyme from Escherichia coli (which was solved only in its apo form; Liao et al., 2001 ▶), (ii) the RS from the yeast Schizosaccharomyces pombe (which was solved in the presence of a substrate analogue but crystallized as a monomer owing to dissociation of the trimer in the crystallization drop; Gerhardt, Schott et al., 2002 ▶) and (iii) the structure of the N-terminal domain of the RS monomer from E. coli in complex with riboflavin (Truffault et al., 2001 ▶; Meining et al., 2003 ▶). Consequently, the crystal models presented in this paper correspond to the second time that a trimeric RS has been crystallized preserving its quaternary organization, and also the first time that an intact RS trimer has been solved in the presence of bound ligands. The models corroborate the existing flexibility in the particle and also highlight the role of the bound ligands in assembling the unique active site of the trimer. To complement these findings, we also performed kinetic and binding studies on the enzyme. Overall, the structural information presented here can be regarded as an excellent starting point for the design of powerful inhibitors against brucellosis.
2. Materials and methods
2.1. Materials
Riboflavin and roseoflavin were purchased from Sigma–Aldrich (St Louis, Missouri, USA) and Toronto Research Chemicals (North York, Canada), respectively. NRP and 6,7-dimethyl-8-ribityllumazine were kindly provided by Professor Markus Fischer, University of Hamburg, Germany.
2.2. Gene cloning
The ribE gene coding for RS (KEGG entry BAB1_0790; http://www.genome.jp/dbget-bin/www_bget?bmf:BAB1_0790) was amplified by PCR using B. abortus strain 2308 chromosomal DNA as template and the following oligonucleotide primers: 5′-TTCATATGTTTACAGGCATAATCACCGAT-3′ (forward) and 5′-GCAAGGCTTGCGCAATATCAGAAACTCGAGAT-3′ (reverse). The resulting PCR product was digested with NdeI and XhoI, and was then ligated into the pET-22b plasmid (Novagen, Billerica, Massachusetts, USA) which had been previously treated with the same restriction enzymes. The final pET-22b-RS construct, the quality of which was checked by DNA sequencing, includes the coding region for the complete RS enzyme (residues 1–202), a two-residue cloning artifact (Leu203 and Glu204) and a C-terminal six-histidine tag (His205–His210). E. coli BL21(DE3) competent cells (Stratagene, La Jolla, California, USA) were transformed with the resulting ligation mixtures.
2.3. Protein expression and purification
Transformed cells with the pET-22b-RS construct were grown overnight in 25 ml LB medium with 150 µg ml−1 ampicillin at 37°C with agitation (200 rev min−1); they were then diluted to 500 ml and grown to an absorbance (at 600 nm) of 0.6. At this point, isopropyl β-d-1-thiogalactopyranoside was added to a final concentration of 1 mM and the culture was incubated for a further 2 h at 37°C with agitation (200 rev min−1). The bacteria were centrifuged at 5000g for 10 min at 4°C. Pellets were resuspended and sonicated in a solution consisting of 20 mM sodium phosphate, 0.5 M sodium chloride, 20 mM imidazole, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM dithiothreitol (DTT) with a final pH of 7.4 (buffer A) and then centrifuged at 160 000g in a Beckman Coulter L7-65 ultracentrifuge (Brea, California, USA) for 45 min at 4°C. The supernatant was filtered through a 0.45 µm membrane and loaded onto a HisTrap HP column (all columns were from GE Healthcare, Little Chalfont, England) in a fast protein liquid chromatography apparatus (Gilson, Luton, England). Elution was performed with a linear gradient of buffer B consisting of 20 mM sodium phosphate, 0.5 M sodium chloride, 0.5 M imidazole, 1 mM PMSF, 1 mM DTT (pH 7.4). A major peak was observed at around 20% buffer B. The appropriate protein fractions were pooled and further purified by gel-filtration chromatography on a Superdex 200 16/60 column with isocratic elution in phosphate-buffered saline with 0.5 M sodium chloride, yielding a major peak at around 90 ml. The final fractions were then concentrated to 20 mg ml−1 by centrifugation in Amicon Ultra-4 devices (Millipore, Billerica, Massachusetts, USA) and simultaneously exchanged into crystallization buffer (10 mM Tris, 25 mM sodium chloride pH 7.4). The protein was aliquoted and stored at −70°C. The quality of the final preparation was checked by SDS–PAGE (15% gel) and UV spectrophotometry.
2.4. Crystallization of the RS apo form
Initial crystallization trials were performed in 96-well plates using a Honeybee 963 robot (Digilab, Marlborough, Massachusetts, USA) and JBScreen Classic (Jena Bioscience, Germany) in a sitting-drop vapour-diffusion configuration. After one week of equilibration at room temperature, ten conditions out of the 240 tested showed promising hits. We were able to reproduce and optimize crystal growth for several of these conditions, and the best diffracting RS crystals (RS-APO) were eventually obtained by the hanging-drop method by mixing 2 µl concentrated protein stock with an equal amount of a crystallization solution consisting of 12%(w/v) PEG 8000, 10%(w/v) glycerol, 0.5 M potassium chloride. Microseeding, which was performed after a 2 d equilibration period, dramatically improved the quality and the size of the crystals. Using microseeding, long bars with maximum dimensions of 0.5 × 0.1 × 0.1 mm were obtained after 3 d. Samples were cryoprotected in mother liquor with an increased glycerol concentration [25%(w/v)] and then cooled in liquid nitrogen in loops (Hampton Research, Aliso Viejo, Califonia, USA).
2.5. Crystallization of RS bound to ligands
Crystals of RS bound to the product analogue roseoflavin (RS-ROS) were grown by adding 1 µl of a roseoflavin solution in a slight molar excess with respect to the binding sites to the crystallization mixture mentioned above. Dark orange bars were obtained after microseeding. In contrast, cocrystallization failed in the case of NRP owing to precipitation of the protein. In this case, 1 µl of a concentrated NRP solution (again in a slight molar excess with respect to the binding sites) was added directly to hanging drops containing fresh RS crystals. Ligand-bound crystals of RS bound to NRP (RS-NRP) were harvested after a 4 d soaking period, observing minor damage to the samples. Finally, riboflavin was incorporated into the RS crystals in a completely different way. For this purpose, solid riboflavin was added directly to the LB medium to a final concentration of 1 mM. The enzyme was then purified and crystallized in the same way as described for the apo form. Crystals of RS bound to riboflavin (RS-RBF) obtained following this procedure showed a marked yellow colour, supporting the presence of bound riboflavin.
2.6. X-ray data collection and processing
Preliminary diffraction data were collected at 100 K at the Institut Pasteur Montevideo, Uruguay on a MicroMax-007 HF rotating-anode diffractometer (Rigaku, The Woodlands, Texas, USA) equipped with a MAR345 image-plate detector (Rayonix, Evanston, Illinois, USA). The final data sets were collected at 100 K on the PROXIMA1 protein crystallography beamline at SOLEIL, France using a PILATUS 6M detector (Dectris, Baden, Switzerland) and on the X6A protein crystallography beamline at the National Synchrotron Light Source (NSLS), USA equipped with an ADSC Quantum 270 CCD detector (Poway, California, USA). Single crystals were used for the collection of each data set. X-ray diffraction data were processed with the programs MOSFLM (Leslie & Powell, 2007 ▶), SCALA (Evans, 2006 ▶) and XDS (Kabsch, 2010 ▶), with the latter running in a semi-automated approach using the xdsme package (http://code.google.com/p/xdsme/). A total of 5% of the recorded reflections were flagged for cross-validation. Details of the data-collection parameters and processing statistics are shown in Table 1 ▶.
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell: RS-APO, 2.01–1.90 Å; RS-RBF, 3.00–2.85 Å; RS-ROS, 1.89–1.78 Å; RS-NRP, 1.96–1.85 Å.
| RS-APO | RS-RBF | RS-ROS | RS-NRP | |
|---|---|---|---|---|
| PDB code | 4fxu | 4e0f | 4g6i | 4gqn |
| Data collection | ||||
| Synchrotron source | SOLEIL | NSLS | SOLEIL | SOLEIL |
| Beamline | PROXIMA1 | X6A | PROXIMA1 | PROXIMA1 |
| No. of frames | 1000 | 220 | 400 | 800 |
| Oscillation step (°) | 0.2 | 1.0 | 0.2 | 0.2 |
| Crystal-to-detector distance (mm) | 320.2 | 310.0 | 295.3 | 344.7 |
| Wavelength (Å) | 0.98011 | 1.0000 | 0.98011 | 0.98011 |
| Exposure per frame (s) | 0.2 | 60 | 0.2 | 0.2 |
| Indexing and scaling | ||||
| Unit-cell parameters | ||||
| a (Å) | 67.42 | 69.98 | 68.04 | 69.85 |
| b (Å) | 93.48 | 91.82 | 92.57 | 92.30 |
| c (Å) | 103.30 | 98.77 | 102.58 | 99.09 |
| α = β = γ (°) | 90 | 90 | 90 | 90 |
| Space group | P212121 | P212121 | P212121 | P212121 |
| Resolution limit (Å) | 1.90 | 2.85 | 1.78 | 1.85 |
| Total No. of reflections | 380981 | 130810 | 178081 | 321086 |
| No. of unique reflections | 52161 | 15167 | 60197 | 55235 |
| Average multiplicity | 7.3 (7.3) | 8.6 (8.7) | 3.0 (3.0) | 5.8 (5.8) |
| 〈I/σ(I)〉 | 21.6 (2.0) | 5.9 (1.9) | 21.2 (2.1) | 15.7 (2.0) |
| R meas | 0.046 (0.971) | 0.115 (0.422) | 0.032 (0.630) | 0.082 (0.850) |
| Completeness (%) | 99.8 (98.7) | 98.7 (98.6) | 96.1 (95.3) | 99.6 (98.7) |
| Trimers per asymmetric unit | 1 | 1 | 1 | 1 |
| Solvent content (%) | 47 | 46 | 47 | 46 |
| B factor (Wilson plot) (Å2) | 41 | 48 | 32 | 26 |
| Refinement | ||||
| Resolution range (Å) | 27.7–1.90 | 67.3–2.85 | 32.3–1.78 | 27.1–1.85 |
| No. of protein atoms | 4143 | 4590 | 4556 | 4637 |
| No. of ligand atoms | — | 27 | 116 | 126 |
| No. of water molecules | 148 | 66 | 240 | 307 |
| R | 0.232 | 0.216 | 0.240 | 0.200 |
| R free | 0.264 | 0.267 | 0.273 | 0.234 |
| R.m.s. deviations from ideal values† | ||||
| Bond lengths (Å) | 0.010 | 0.006 | 0.005 | 0.007 |
| Bond angles (°) | 1.1 | 0.9 | 0.9 | 1.0 |
| B factor (average) (Å2) | 50 | 23 | 35 | 31 |
| MolProbity validation‡ | ||||
| Clashscore | 1.96 | 5.01 | 2.79 | 1.48 |
| Poor rotamers (%) | 5.1 | 5.1 | 2.1 | 3.7 |
| Ramachandran plot | ||||
| Favoured (%) | 97.6 | 96.2 | 97.8 | 96.8 |
| Allowed (%) | 2.2 | 3.6 | 2.2 | 3.2 |
| Disallowed (%) | 0.2 | 0.2 | — | — |
2.7. Structure resolution, model building and refinement
All structures were solved by means of the molecular-replacement method in the resolution range 15–4 Å with AMoRe (Navaza, 1994 ▶). RS-RBF was the first structure to be solved, using the coordinates of the trimeric RS from E. coli (PDB entry 1i8d; Liao et al., 2001 ▶) as a search model. The other structures were then solved using RS-RBF as a search model. A single trimer was located in the asymmetric unit in all cases, and the corresponding crystal packings were successfully checked. The oriented coordinates were subjected to positional and individual B-factor refinement with REFMAC (Murshudov et al., 2011 ▶) and BUSTER (Bricogne et al., 2011 ▶), with the application of noncrystallographic symmetry (NCS) restraints under the local structure similarity restraints (LSSR) approach (Smart et al., 2012 ▶) to take into account the observed differences between individual chains by removing these from the NCS relation. Model building was performed with Coot (Emsley et al., 2010 ▶). In the last steps of the refinement, water and ligand molecules were added to the corresponding models. The coordinates for riboflavin (Gerhardt, Haase et al., 2002 ▶), roseoflavin (Serganov et al., 2009 ▶) and NRP (Klinke et al., 2007 ▶) were obtained from published structures, and their CIF restraint libraries were obtained from the CCP4 database (Winn et al., 2011 ▶). Detailed statistics of the refinement process can be found in Table 1 ▶.
2.8. Validation and analysis of the models
The four crystallographic structures were subjected to validation with MolProbity (Chen et al., 2010 ▶) as well as with the validation module implemented in Coot. The following web servers at the European Bioinformatics Institute (EBI) were used: PDBePISA (study of interface areas; http://www.ebi.ac.uk/msd-srv/prot_int/; Krissinel & Henrick, 2007 ▶), PDBeFold (superpositions and r.m.s.d. calculations; http://www.ebi.ac.uk/msd-srv/ssm/; Krissinel & Henrick, 2004 ▶) and ClustalW2 (sequence alignments; http://www.ebi.ac.uk/Tools/msa/clustalw2/; Larkin et al., 2007 ▶).
2.9. Graphical representation
Molecular structures and their electron densities were represented using PyMOL (Schrödinger, New York, USA). Chemical structures were drawn with Accelrys Draw (Accelrys, San Diego, California, USA).
2.10. Calculation of molecular weights by static light scattering
Molecular weights in solution were calculated using a Precision Detectors PD2080 static light-scattering instrument (Bellingham, Massachusetts) tandemly connected to a fast protein liquid chromatography apparatus and an LKB 2142 differential refractometer (LKB, Bromma, Sweden). In each case, 250 µl of a ∼2 mg ml−1 protein solution was injected into an analytical Superdex 200 column and eluted with a buffer consisting of 20 mM Tris, 0.25 M sodium chloride, 0.5 mM PMSF, 0.5 mM DTT pH 7.8. Both signals corresponding to 90° light scattering (λ = 682 nm) and the refractive index of the eluted peaks were recorded and were then processed with the Discovery32 software supplied by Precision Detectors. Bovine serum albumin (BSA; molecular weight 66 500 Da) was used as standard for light-scattering detector calibration.
2.11. Riboflavin dissociation constant and binding stoichiometry
The dissociation constant for riboflavin (K d) and its binding stoichiometry were determined by fluorescence titration on a Jasco FP-6500 spectrofluorometer (Easton, Maryland, USA) according to published procedures (Fischer et al., 2002 ▶). Briefly, a 45 µM ligand solution was titrated by the sequential addition of 5 µM RS aliquots (subunits) in 50 mM potassium phosphate buffer pH 7.0 at 20°C. Fluorescence emission was measured after each aliquot with excitation and emission wavelengths of 470 and 535 nm, respectively. Control experiments were performed using BSA. Data analysis was performed using GraphPad Prism 5 (GraphPad Software, La Jolla, California, USA).
2.12. Kinetic assay
Catalytic activity experiments were performed fluorometrically as described by Fischer et al. (2005 ▶) with some modifications. Explicitly, reaction mixtures consisted of 50 mM potassium phosphate buffer pH 7.0 and 50 nM RS (subunits). 6,7-Dimethyl-8-ribityllumazine was then added at a broad range of concentrations (0–1000 µM) in a final volume of 1 ml. Riboflavin fluorescence was monitored as described above every 0.5 s for 20 min. Assays were always performed in duplicate. To correct for systematic variations from the spectrofluorometer, riboflavin dilution series were measured in conjunction with each experiment. These measurements were used to define both the linear range and the response of the instrument. Relative fluorescence intensities were converted to absolute riboflavin concentrations using the slope from the linear regression of the fluorescence measurements of a riboflavin dilution series (data not shown). Initial velocities (V i) were then calculated for each substrate concentration. Finally, these points were fitted to various kinetic models derived from the velocity versus substrate-concentration plot using a nonlinear least-squares regression method in Excel 2010 (Microsoft, Redmond, Washington, USA). For the product-inhibition experiments, V i values were determined from reaction mixtures containing a fixed concentration of 6,7-dimethyl-8-ribityllumazine (5 µM) with different riboflavin concentrations.
3. Results and discussion
3.1. Quality of the crystallographic models
In this work, we have solved the structure of four variants of the enzyme RS from B. abortus by X-ray crystallography: (i) unliganded apo protein (RS-APO) and the protein bound to (ii) the reaction product riboflavin (RS-RBF), (iii) the product analogue roseoflavin (RS-ROS) and (iv) the product analogue NRP (RS-NRP). All structures were solved by the molecular-replacement method in space group P212121 as described in §2. The final refined models show very good stereochemistry, with more than 96% of the residues in the favoured region of the Ramachandran plot (Table 1 ▶).
The four structures are isomorphous and present rather similar unit-cell parameters, showing a unique trimer in their asymmetric units. The last 5–13 C-terminal residues lacked electron density and were excluded from the models. It is important to mention that the crystallized construct contains a two-residue cloning artifact plus a six-residue histidine tag at the C-terminus. With the exception of the abovementioned ranges, the structures RS-RBF and RS-NRP show continuous density in their polypeptide chains with no breaks. The RS-ROS complex also lacked density for the loop 55–58 in chain B, which is exposed to the solvent. Finally, the unliganded RS-APO structure presented additional disordered regions, namely residues 55–60 in chain A and residues 16–20, 51–68, 80–85 and 119–121 in chain B, which were excluded from the crystallographic model. The latter observation might be caused by enhanced mobility owing to the absence of bound ligands. In addition to the missing backbone residues, several side chains (most of them corresponding to exposed polar residues) could not be located in the electron density of the four structures. In these cases, the residues were modelled only to the β-carbon. In the complex models, clear electron density for all ligand molecules was observed, which allowed their correct fitting in both position and conformation. Interestingly, a different number of ligand molecules was found in each case. Explicitly, a single riboflavin molecule was found in the RS-RBF trimer, whereas four roseoflavin molecules were modelled in RS-ROS and six NRP molecules in RS-NRP.
When the complete trimeric liganded structures are superimposed against each other, root-mean-square deviation (r.m.s.d.) values of between 0.34 and 0.65 Å for approximately 600 aligned Cα atoms are observed, which stresses the high similarity of the complexes. However, these values rise to an average of 1.30 Å when the liganded structures are superimposed against RS-APO. This is the consequence of a slightly different orientation of chain A in the apo structure, as explained in detail below.
3.2. Structural description of the RS enzyme
RS from B. abortus is arranged as an asymmetric trimer with approximate dimensions of 70 × 65 × 65 Å (Fig. 2 ▶ a). The quaternary structure observed in the crystals is coincident with that in solution. In this regard, molecular-weight calculation performed via static light scattering yielded a value of 66 ± 4 kDa both in the absence and the presence of bound ligands, which is in very good agreement with the value of 69.8 kDa (3 × 23.3 kDa) calculated from its sequence. The overall shape of the particle closely resembles the trimeric RS from E. coli (Liao et al., 2001 ▶), yet there are some differences in the relative orientation of their monomers and in the structures of their C-termini. RS belongs to the α+β protein class, and its monomer shows the following structural elements: (i) an N-terminal six-stranded β-barrel (residues 1–91), (ii) a connecting loop (residues 92–101), (iii) a C-terminal six-stranded β-barrel (residues 102–187) and (iv) a C-terminal α-helix (residues 188–202) (Fig. 2 ▶ b). Both β-barrels, which are capped by two 310-helices and one α-helix, adopt an almost identical fold (Fig. 2 ▶ c), with an average r.m.s.d. of 1.20 Å for 83 aligned Cα atoms. A similar observation was found in the RS structures from E. coli (Liao et al., 2001 ▶) and S. pombe (Gerhardt, Schott et al., 2002 ▶). The structural homology between both barrels is also reflected at the sequence level, with 19 identical residues (23%) and 17 similar residues (20%), as shown in Fig. 3 ▶. Interestingly, the barrels belonging to the same chain are related by pseudo-C 2 symmetry, with an average rotation angle of 177° between each another. An additional unique pseudo-C 2 rotation axis that relates the N-terminal barrel from chain A to the C-terminal barrel from chain B is also present and gives rise to the active site of the trimer, as described in detail below.
Figure 2.
(a) The RS-APO structure. Each chain is depicted in a different colour. N- and C-terminal barrels are indicated for each chain in their respective colours. The C-terminal three-helix bundle can clearly be seen at the centre of the molecule. An arrow marks the approximate location of the unique active site of the enzyme. (b) Structure of the RS monomer depicted in rainbow colours. The major structural elements are identified. The orientation highlights the pseudo-C 2 symmetry observed between the N- and C-terminal barrels. A black oval capped with a tilde marks the approximate location of the pseudo-twofold rotation axis, which is perpendicular to the plane of the paper. The monomer depicted here corresponds to chain A from the RS-NRP complex, which bears two copies of the NRP ligand, as explained in the text. (c) Superposition of the N- and C-terminal barrels. Shown are residues 1–91 (N-terminal barrel, pink) and 102–188 (C-terminal barrel, cyan) belonging to chain A from the RS-NRP complex. Secondary-structure elements and terminal residues are indicated. Bound ligands are also depicted. (d) Interactions between chains in the C-terminal three-helix bundle. The side chains of the interacting residues are represented as sticks. Hydrogen bonds are marked with dotted lines. The colours for each chain are the same as in (a), but the orientation of the figure has been shifted slightly for clarity.
Figure 3.

Sequence alignment of the N- and C-terminal barrels. Secondary-structure elements are indicated. Identical and similar residue pairs are highlighted in dark and light orange, respectively. Residues in direct contact with the ligand molecules, as seen in the complex structures solved in this work, are marked with a star. Gly99 and His101, which belong to the central loop bridging both β-barrels and are not present in the alignment, are also involved in ligand binding.
Finally, when the individual monomers of RS are superimposed, r.m.s.d. values ranging from 0.66 to 1.23 Å are observed (165–200 aligned Cα atoms). The structural elements are very well preserved between chains. However, in the four structures a rather different conformation of the residue range 96–101A is observed (which belongs to the central connecting loop), as well as a deviation of about 20° in the orientation of the C-terminal α-helix in chain C (Supplementary Fig. S2). Additionally, no significant rearrangements are observed within the same chains when the liganded and apo structures are superimposed. This reveals no considerable effect on the architecture of the individual subunits upon ligand binding.
3.3. Comparison with other known RSs at the monomeric level
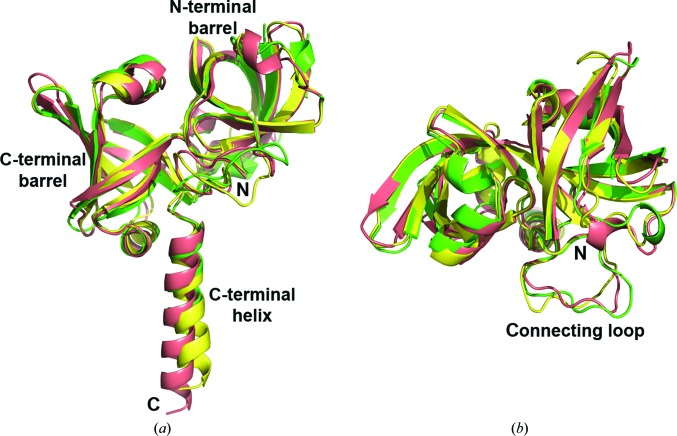
The B. abortus RS monomers can be superposed on those from E. coli (Liao et al., 2001 ▶) and S. pombe (Gerhardt, Schott et al., 2002 ▶) with average r.m.s.d. values of 1.48 and 1.09 Å for over 180 aligned Cα atoms, respectively (Fig. 4 ▶). These small values highlight the existing structural similarity, which extends along the whole polypeptide chains, with moderate divergences in the central connecting loops and in the C-terminal helices. When the corresponding sequences are aligned (Fig. 5 ▶), a total of 43 residues (21%) are coincident in all three species and 79 (39%) are coincident in two. The highly conserved N-terminal sequence MFTG, which is known to be critical in stabilizing the active-site architecture (Illarionov, Kemter et al., 2001 ▶; Liao et al., 2001 ▶), is also present in Brucella RS.
Figure 4.
Superposition of the RS monomers from B. abortus (chain A from RS-APO, green), E. coli (chain A from PDB entry 1i8d, pink) and S. pombe (unique chain from PDB entry 1kzl, yellow). N- and C-termini are indicated, as well as the main structural elements. (a) Front view. (b) Top view.
Figure 5.
Sequence alignment of RSs with known three-dimensional structure. Colours and features are similar to those in Fig. 3 ▶. The location of the secondary-structure elements in the picture refers to the B. abortus enzyme.
3.4. The asymmetric trimeric assembly
The major interactions between chains that are responsible for the trimeric organization of RS from B. abortus can be classified into four different groups. The first interacting group links the N-terminal barrel from chain A with the C-terminal barrel from chain B (eight and seven residues within 3.5 Å distance, respectively). Most of the observed contacts are polar, with the presence of two salt bridges: Glu66A–Arg166B and Glu70A–Lys140B. Interestingly, the interactions between these barrels are only observed in the liganded structures; they are totally absent in RS-APO. This observation may be the consequence of the cementing properties of the bound ligands. The next interacting group involves residues from the central connecting loops. In this way, residues 92–94 from chain A interact with residues 95–98 from chain B, as do residues 93–94 from chain B with residues 95–96 from chain C. These contacts include hydrophobic interactions, several hydrogen bonds and the salt bridge Lys93A–Glu97B. Another important group of contacts are those made by the three C-terminal α-helices, the association of which gives rise to a somewhat open three-helix bundle located in the central region of the protein (Fig. 2 ▶ a). Each α-helix is in contact with its two neighbouring partners, making hydrophilic and hydrophobic interactions (Fig. 2 ▶ d). The most relevant polar contact is the hydrogen bond Asp188–Tyr193, which is present in all chains. Additionally, there is another hydrogen bond between residues Gln189A and Asp188B. Nonpolar interactions are made by the side chains of residues Leu190, Ala194, Ala195 and Leu197. It is important to state that this set of contacts is very well preserved amongst the four structures solved in this work, even in the case of RS-APO. The trimeric RS from E. coli (Liao et al., 2001 ▶) also shows an analogous three-helix bundle that is longer than that in Brucella (a total of 61 versus 41 residues, respectively) and that bears not only hydrophobic contacts but also three salt bridges that are absent in the structures solved here. In this sense, it is important to mention that the sequence of the C-termini in RSs is highly variable amongst species (see Fig. 5 ▶). Finally, the last group of interactions are those observed between the three-helix bundle and other structural elements of the trimer (Supplementary Fig. S3). Explicitly, several C-terminal residues from chain A are in direct contact with the C-terminal barrel from chain B mainly through hydrogen-bond interactions; the C-terminal residues from chain B make the same type of contacts with the C-terminal barrel from chain C, and the C-terminal residues from chain C are linked to the central connecting loop from chain A through polar and hydrophobic contacts.
As also observed in the E. coli enzyme, RS from B. abortus is a homotrimer devoid of trigonal symmetry, with its monomers asymmetrically arranged in the particle with rotation angles that are different from the theoretical symmetric 120° triplet between chains. Interestingly, the rotation angles that relate neighbouring monomers in RS differ not only between species but also between the four structures solved in this work (Table 2 ▶). The complex structures RS-RBF, RS-ROS and RS-NRP bear nearly the same rotation-angle triplets. However, when ligands are absent the rotation angle between chains A and B increases by about 7° (from 86 to 93°). This is accompanied at the same time by a decrease of about 6° in the rotation angle between chains C and A (from 163 to 157°). Since the rotation angle between chains B and C remains similar in all four cases (∼110°), this clearly shows a displacement of monomer A towards monomer C in the absence of bound ligands (Fig. 6 ▶). This molecular opening, which reflects the intrinsic flexibility of the trimer and might be the consequence of crystal-packing optimization, causes a decrease in the interface area between chains A and B from an average value of 1650 Å2 in the complex structures to 1200 Å2 in RS-APO. Remarkably, the interface area between the monomer pairs B–C and C–A remains fairly constant in all four structures, with average values of 1050 and 450 Å2, respectively. At the same time, all contacts between the N-terminal barrel from chain A and the C-terminal barrel from chain B are lost, as mentioned previously. The rest of the contacts made by monomer A (mostly by its C-terminal α-helix and its central connecting loop) are well preserved in all four structures, which identify both β-barrels from chain A as the elements with the highest mobility in the trimeric crystal models. Owing to crystal-packing constraints, we cannot rule out at this point whether the β-barrels from the other two chains might also experience a similar behaviour in solution.
Table 2. Rotation angles (°) between RS monomers in the crystallographic structures.
| A–B | B–C | C–A | Reference | |
|---|---|---|---|---|
| RS-APO | 93 | 110 | 157 | This work |
| RS-RBF | 86 | 110 | 163 | This work |
| RS-ROS | 86 | 113 | 162 | This work |
| RS-NRP | 87 | 111 | 163 | This work |
| E. coli RS† | 85 | 124 | 151 | Liao et al. (2001 ▶) |
Chain IDs in the E. coli enzyme have been modified with respect to its original letters (PDB entry 1i8d) to preserve the location of the active site between chains A and B.
Figure 6.
Superposition of the RS-RBF, RS-ROS and RS-NRP structures (green) with RS-APO (yellow). Only chains B and C are used for the structural alignment in order to stress the displacement observed in chain A from RS-APO (approximately 7°). The relative orientation of chains B and C is fairly similar in the four structures. The calculated r.m.s.d. values with respect to RS-APO for these two chains together (366 superimposed Cα atoms) are only 0.73, 0.92 and 0.70 Å for RS-RBF, RS-ROS and RS-NRP, respectively. Bound ligands are not included for clarity.
From the opposite point of view, we can propose that the binding of any of the different ligands studied in this work to RS promotes the formation of a tight interface between the N-terminal barrel from chain A and the C-terminal barrel from chain B. As described in the next section, this interface holds the unique active site of the trimer, where two molecules of 6,7-dimethyl-8-ribityllumazine bind in an antiparallel manner. As a preliminary conclusion, the ligands may therefore act as the necessary cementing partners required to build up the active site of RS.
It is interesting to note that the rotation angles in E. coli RS show some discrepancy from the values for Brucella RS. This might be the consequence of a slightly different arrangement of the monomers to optimize the unrelated crystal packing in both species, revealing further evidence for the existing flexibility between monomers in the enzyme. Notably, the monomers that build up the active site in RS from E. coli (the structure of which was solved in the absence of bound ligands) are related by a rotation of 85° and give rise to a competent active site at their interface. This value is almost identical to the rotation angles observed between chains A and B only in the liganded forms of RS from B. abortus.
3.5. Substrate-binding sites and the unique active site
Despite the fact that the structure of a complete trimeric RS solved in the presence of bound ligands was not available before this work, previous structural studies (Liao et al., 2001 ▶; Truffault et al., 2001 ▶; Gerhardt, Schott et al., 2002 ▶; Meining et al., 2003 ▶) have identified and described the substrate-binding sites in some detail. Each β-barrel domain is known to bind one molecule of 6,7-dimethyl-8-ribityllumazine, yielding two binding sites per monomer and a total of six binding sites per RS trimer. The binding site in each of the two barrels involves different residues but its architecture is essentially the same in both cases, corresponding to a shallow surface cavity located adjacent to the β-barrel between helix α1 and strands β4 and β5 (see Figs. 2 ▶ b and 2 ▶ c for the location of the NRP molecules, which occupy all of the 6,7-dimethyl-8-ribityllumazine binding sites). For the dismutation reaction to occur, it is mandatory that two binding sites come close to each other in space. In the conformation observed in the crystal structure, this situation can only happen at the interface between chains A and B, yielding a unique active site for the whole trimer (see Fig. 2 ▶ a). In addition to the two binding sites that build up the catalytic site of RS, the other four substrate-binding sites (one for chain A, one for chain B and two for chain C) are open to the solvent and are therefore not competent for the dismutation reaction, at least in the conformation observed in the crystal models.
As mentioned previously, the formation of the active site gives rise to a pseudo-twofold rotation axis that relates the N-terminal barrel from chain A to the C-terminal barrel from chain B. This symmetry element, which is unrelated to the pseudo-twofold rotation axes between β-barrels in the same chains, is particularly seen in the three liganded structures solved in this work. In these cases, a rotation angle of about 175° is observed between both barrels forming the active site, but this value decreases to 171° in RS-APO owing to the displacement experimented by chain A, altering the architecture of the active site and precluding the dismutation reaction, as shown in detail later. Interestingly, the value for the same angle in the unliganded E. coli RS is 178°, stressing again that this enzyme is able to assemble a competent active site in the crystal model even without bound ligands.
The following sections will describe the molecular interactions between B. abortus RS and the introduced ligands. It is important to reiterate that the binding of these molecules does not modify the structure of the monomers significantly, but instead alters the structure of the trimer by shifting the relative location of chain A in the crystals.
3.6. Binding of the reaction product riboflavin
The RS-RBF structure solved in this work bears a single riboflavin molecule, which is located at the substrate-binding site in the N-terminal barrel in chain A (Figs. 7 ▶ a and 8 ▶ a). The recognition of the bound ligand occurs mainly through hydrogen-bond interactions involving (i) the amide groups of residues Thr49A, Trp68A and Val107A, (ii) the carbonyl groups of residues Cys47A, Glu66A, Gly105A and Val107A, and (iii) the polar side chains of residues Thr49A and Lys140B. Additionally, residues Gly99B and Thr151B are at van der Waals distance from the riboflavin molecule, and the side chains of residues Leu48A and Ile165B are located on both sides of the pyrimidinedione ring of the ligand, giving rise to a hydrophobic stabilizing environment. The fact that four residues from the neighbouring chain B are involved in riboflavin binding clearly indicates that this molecule facilitates the formation of the tight interface between the N-terminal barrel from chain A and the C-terminal barrel from chain B, building up the unique active site.
Figure 7.
Representation of the ligands found at the substrate-binding sites. The left panels highlight the locations of the different molecules observed, together with some of the most representative residues involved in binding (drawn in sticks). Monomers A and B are depicted in green and pink, respectively. 2mF o − DF c Fourier maps are represented at the 1.0σ level. The right panels summarize the hydrogen-bond interactions between the individual ligands and the RS enzyme, indicating residue numbers and atoms, distances (in Å, with a maximum value of 3.5) and ligand atom numbers. (a) RS-RBF. (b) RS-ROS (roseoflavin molecule bound at the N-terminal barrel from chain A). (c) RS-ROS (roseoflavin molecule bound at the C-terminal barrel from chain A). (d) RS-NRP [pair of NRP molecules bound at the N-terminal barrel from chain A (i) and at the C-terminal barrel from chain B (ii)]. Absolute contour levels for the electron-density maps (in e Å−3) are as follows: 0.196 (a), 0.288 (b), 0.274 (c) and 0.284 (d).
Figure 8.
Cartoon representation of the complex RS structures highlighting the different ligand-binding stoichiometries in each case. Each monomer is depicted as a violet drop. N- and C-terminal barrels are identified inside the monomers with the letters N and C. Individual chains are labelled outside the drops. Ligands are represented with their aromatic groups in orange (riboflavin), pink (roseoflavin) and yellow (NRP). Ribityl moieties are represented as tails. (a) RS-RBF. (b) RS-ROS. (c) RS-NRP.
The RS-RBF complex represents the first time that an intact trimeric RS has been crystallized in the presence of bound ligands. However, when riboflavin is modelled in the active site of the trimeric E. coli RS (Liao et al., 2001 ▶) using the model of monomeric S. pombe RS bound to 6-carboxymethyl-7-oxo-8-ribityllumazine as a template (Gerhardt, Schott et al., 2002 ▶), it can clearly be seen that the contacts made by the riboflavin molecule are very well conserved between these species (Supplementary Fig. S4). Only one residue whose side chain makes a hydrogen bond to the pyrimidinedione ring of the ligand in S. pombe and E. coli RS (Ser67 and Thr67, respectively) is replaced by an alanine in Brucella RS (Ala71), therefore losing this particular contact. It is important to mention that the interactions of riboflavin with the neighbouring chain B are also very well conserved between these three species.
During the reaction course, it has been proposed that the substrate molecule that acts as the four-carbon unit donor may be the one that binds to the C-terminal barrel in chain B, whereas the acceptor molecule (which is ultimately converted into riboflavin) may be that bound to the N-terminal barrel in chain A. However, there was no direct structural evidence for this observation. In this work, the single riboflavin molecule found in the RS-RBF complex at the N-terminal barrel in chain A gives a straight corroboration of this hypothesis.
In principle, and owing to the structural similarity between the product riboflavin and the substrate 6,7-dimethyl-8-ribityllumazine, up to six riboflavin molecules (two per chain) could bind simultaneously to the RS trimer. However, when modelling riboflavin into all substrate-binding sites, only four sites can be filled without steric clashes: all three binding sites associated with the N-terminal barrels plus the binding site associated with the C-terminal barrel in chain A. The C-terminal barrel from chain B (which builds up the ‘second half’ of the active site) is unable to bind riboflavin because the methyl group linked to the C8 atom would clash with residues Gly99 and Gly100 from the same chain, which are part of the central connecting loop between β-barrels. Additionally, the whole xylene ring of this newly placed molecule would be in steric conflict with the riboflavin molecule already bound in the active site. Next, the hypothetical binding of riboflavin to the C-terminal barrel from chain C (which is open to the solvent) would result in the same kind of clashes with residues Gly99 and Gly100 from the same chain. Interestingly, the C-terminal barrel from chain A (which is also open to the solvent) may be fully capable of binding this ligand, since the loop formed by residues 96–101 in chain A is shifted about 5 Å away from the substrate-binding site in comparison with the other two chains (see Supplementary Fig. S2 for a superposition between monomers). The analysis presented above suggests that up to four riboflavin molecules may bind simultaneously to the protein trimer. This observation is in contrast to previous findings for RS from B. subtilis (Otto & Bacher, 1981 ▶) and A. thaliana (Fischer et al., 2005 ▶), where binding studies indicated the presence of a maximum of three riboflavin molecules per trimer.
It is important to mention that the RS-RBF complex was not obtained by cocrystallization or by soaking, but by the addition of solid riboflavin to the culture medium. At the end of the purification process, the crystal structure revealed that only one molecule of the ligand remained bound per RS trimer, stressing that the acceptor site from chain A (N-terminal barrel) corresponds to the site with the highest affinity for riboflavin.
3.7. Binding of the product analogue roseoflavin
The RS-ROS structure shows the presence of four bound roseoflavin molecules, which are located at the same four predicted riboflavin sites as mentioned in the previous section, namely all three substrate-binding sites associated with the N-terminal barrels plus the binding site associated with the C-terminal barrel in chain A (Fig. 8 ▶ b). In this particular case, the structure was obtained by cocrystallization, adding an excess of the ligand, thus filling all available binding sites that do not give rise to steric clashes. The 8-dimethylamino group in roseoflavin that replaces the original 8-methyl group in riboflavin is not involved in protein–ligand contacts, and for this reason these molecules can be considered to be similar from a structural point of view in this analysis. As a preliminary conclusion, these observations again support the fact that B. abortus RS might be able to bind up to four riboflavin molecules per trimer.
The binding mode and contacts of the roseoflavin molecules found in the N-terminal barrels are essentially similar as in the case of RS-RBF (Fig. 7 ▶ b). The unique roseoflavin bound to the C-terminal barrel in chain A (Fig. 7 ▶ c) exhibits a comparable binding mode with respect to those in the N-terminal barrels. Explicitly, there is a series of hydrogen-bond interactions that involve (i) the amide groups of residues Ile6A, Thr151A and Ile165A, (ii) the carbonyl groups of residues Gly4A, Ile6A, Ser149A and Leu163A and (iii) the polar side chains of the residues Thr151A and Ser168A. Additionally, the side chain of Leu150A faces the pyrimidinedione ring of the ligand. All of these residues are coincident in the alignment shown in Fig. 3 ▶ with respect to the residues contacting the roseoflavin molecules bound in the N-terminal barrels. The only exception is Ser168A, whose related residue in the N-terminal barrel (Ala71A) is unable to make the contact mentioned above. Since this binding site is open to the solvent, there are no contacts with the neighbouring chain for the unique roseoflavin molecule bound to the C-terminal barrel in chain A. Finally, modelling of roseoflavin molecules into the two empty binding sites (C-terminal barrels from chains B and C) results in severe clashes of their xylene moieties, in a similar way to that observed for RS-RBF.
3.8. Binding of the product analogue NRP
NRP differs chemically from the subproduct 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione only by a nitro group at position 5. This compound is of considerable interest since it is also a known substrate-analogue inhibitor of LS (which catalyzes the previous step in the riboflavin-biosynthetic pathway) and several published structures describe its binding mode to this enzyme in detail (Ritsert et al., 1995 ▶; Gerhardt, Haase et al., 2002 ▶; Klinke et al., 2005 ▶, 2007 ▶).
The RS-NRP structure was obtained by soaking RS crystals in the presence of an excess of NRP. The crystal model reveals that all six substrate-binding sites are occupied by NRP molecules (Fig. 8 ▶ c). This observation is not surprising, since the latter molecule is a structural analogue of 6,7-dimethyl-8-ribityllumazine, which bears a comparable yet smaller heteroaromatic system. The series of contacts between this analogue and the enzyme are summarized in Fig. 7 ▶(d) and are essentially equivalent to those observed for the ligands described previously. Interestingly, RS-NRP is the only structure in this work in which the substrate-binding site associated with the C-terminal barrel from chain B is occupied with a bound ligand, therefore ‘filling’ the whole active site. This particular NRP molecule practically does not interact with the neighbouring chain A. Only residues His106A and Thr49A are located about 4 Å away from the ligand molecule and no hydrogen bonds are observed. This leads to the proposition that the only NRP molecule that favours the assembly of the unique active site of the trimer is that bound to the N-terminal barrel in chain A. Fig. 7 ▶(d) also reveals that the ribityl moieties from both NRP molecules bound to the active site adopt an antiparallel orientation, which is the result of the pseudo-twofold symmetry observed between the two β-barrels that build up the catalytic site.
Based on the location of the ligand molecules, RS-NRP allows the modelling of the substrate 6,7-dimethyl-8-ribityllumazine into the active site, which is presented in Fig. 9 ▶(a). As expected, both substrate molecules are properly oriented for the dismutation reaction, with a rotation angle of approximately 40° between the planes containing both aromatic systems. According to the latest postulated reaction mechanism (Kim et al., 2010 ▶), one of its key steps corresponds to a cyclo-addition that involves both xylene methyl groups from the acceptor molecule and atoms C6 and C7 from the donor molecule. The modelled distances between these reacting pairs are 2.5 and 2.6 Å, confirming the favourable geometric arrangement of both 6,7-dimethyl-8-ribityllumazine molecules. A comparable behaviour was observed for S. pombe RS (Meining et al., 2003 ▶). Interestingly, when this modelling analysis is repeated for RS-APO (Fig. 9 ▶ b), much longer distances are observed between the reacting atoms, namely 4.2 and 3.6 Å, owing to the displacement of chain A towards chain C in the unliganded structure (see Fig. 6 ▶). These longer distances may preclude the dismutation reaction, again supporting the fact that the absence of bound substrates may avoid the assembly of a competent active site in the RS-APO crystal model.
Figure 9.
Modelling of 6,7-dimethyl-8-ribityllumazine molecules into the active site of RS-NRP (a) and RS-APO (b). Monomers are depicted in green (chain A) and pink (chain B). Substrate molecules are identified with the letters a and d, which correspond to the acceptor and donor molecules, respectively. Dashed lines represent the distances between the C6α methyl group from the acceptor and the C7 atom from the donor (top) and between the C7α methyl group from the acceptor and the C6 atom from the donor (bottom). Substrate molecules were placed by superposition taking into account the location of the aromatic systems and the ribityl moieties of the ligands in RS-NRP. For (b), the individual subunits from RS-APO were superimposed on the RS-NRP structure first.
The presence of two molecules of NRP at the interface between chains A and B in RS-NRP allows an interesting comparison with the only pentameric archaeal RS with known three-dimensional structure: that of the thermophilic microorganism Methanocaldococcus jannaschii (Ramsperger et al., 2006 ▶). This enzyme was crystallized bound to the substrate-analogue inhibitor 6,7-dioxo-8-ribityllumazine and bears five active sites located at the interface between monomers, each of them accommodating two inhibitor molecules in an antiparallel orientation. The M. jannaschii and B. abortus RSs share the following features: (i) two substrate molecules bind at the interface between monomers that form the active site, (ii) each of the substrate molecules in the active site interacts mainly with a single monomer and (iii) the substrate molecules are oriented in an antiparallel fashion. However, there are also marked divergences, namely (i) the quaternary structure and number of active sites are different in each case, (ii) the active-site architecture is completely different for both enzymes as a consequence of nonhomologous amino-acid sequences and (iii) the substrate pairs at the active site cannot be superimposed as a whole between the enzymes because they give rise to different diastereomeric pentacyclic intermediates during the reaction course.
3.9. Kinetic and binding studies
In order to complement the structural study on RS, we performed a detailed kinetic study of the enzyme. The recombinant RS protein shows clear activity in vitro, with a turnover number of 1.02 min−1 at 20°C and pH 7.0. Fig. 10 ▶ presents an analysis of the initial catalytic rates for a broad range of substrate concentrations. It can be observed that RS does not follow a simple Michaelis–Menten model (dashed curve, inset). In contrast, the enzymatic activity decays progressively as the 6,7-dimethyl-8-ribityllumazine concentration increases (solid curve). A similar behaviour has been reported for Arabidopsis thaliana RS (Fischer et al., 2005 ▶). Taking into account that the Michaelis–Menten model does not fit the experimental curve, and therefore the kinetic constants determined by this model are inadequate to describe the catalytic nature of the enzyme, we propose the substrate-inhibition model shown in Fig. 11 ▶ to describe the kinetics of RS. This model, derived from the Haldane equation (Haldane, 1930 ▶), is the one that best fits our experimental data. Briefly, it assumes that RS bears a catalytic site that binds two substrate S molecules to generate one molecule of product P, and two noncatalytic (or allosteric) sites that cannot generate product and reduce the catalytic rate of the enzyme E. The enzymatic species with two substrate molecules bound to the catalytic site, which can generate a product molecule, is indicated as ES2. Likewise, species with one and two substrate molecules bound to the noncatalytic sites simultaneously with the other two substrate molecules in the active site are indicated as SES2 and SSES2, respectively. In this inhibition model, we hypothesize that the substrate molecules may first bind to the catalytic site and then, as the substrate concentration increases, they may occupy the rest of the binding sites, for which their affinity may be lower. We also assume that the complex intermediates SES2 and SSES2 are not able to generate product. Then, considering a rapid equilibrium, the following kinetic equation can be derived,
 |
where [S] refers to the substrate concentration, V i is the initial reaction rate, [E]o is the total enzyme concentration, k cat is the turnover number, K m is the classic Michaelis–Menten constant (which is approximately equal to K S2 in Fig. 11 ▶ considering that k cat is significantly smaller than the substrate-dissociation constant) and K I1 and K I2 are the dissociation constants for the binding reactions of one substrate molecule to a different site rather than the catalytic site. Table 3 ▶ shows the kinetic parameters calculated by fitting the experimental points to (1) applying a nonlinear regression method. As seen in Fig. 10 ▶, the velocity curve reaches a maximum and then decreases towards zero at higher substrate concentrations. This is owing to an increasing amount of enzyme that is held in the noncatalytic SES2 and SSES2 species. Interestingly, a sigmoidal response in the activity curve should be observed since the binding of two substrate molecules is required for catalysis. However, the curve shows a Michaelian response at low substrate concentration (<5 µM). This behaviour could be owing to the fact that the first substrate molecule binds to the enzyme with very high affinity and the increase in the velocity rate may be caused solely by the binding of the second substrate molecule. This implies that K S1 << K S2 and therefore we excluded K S1 from the calculations in the kinetic model.
Figure 10.
Kinetic study of RS. The initial velocities of riboflavin formation (V i) for different 6,7-dimethyl-8-ribityllumazine concentrations are represented by a solid line. The inset highlights the substrate inhibition observed at high substrate concentrations. For comparison, the curve representing the classic Michaelis–Menten model is drawn as a dashed line.
Figure 11.

Kinetic scheme used for the calculations in the substrate-inhibition model. E, enzyme; S, substrate; P, product (riboflavin).
Table 3. Kinetic constants calculated for the substrate-inhibition model presented in Fig. 11 ▶ .
| Value | Error | |
|---|---|---|
| k cat (min−1) | 1.02 | 0.02 |
| K d (µM) | 20.6 | 1.0 |
| K S2 (µM) | 0.459 | 0.011 |
| K I1 (µM) | 3780 | 70 |
| K I2 (µM) | 59.7 | 1.3 |
The substrate-inhibition model in which the enzymatic activity is inhibited by two substrate molecules exhibits the best fit to our experimental data (Fig. 11 ▶) according to the Akaike information criterion (AIC; Akaike, 1974 ▶). Nevertheless, a greater number of inhibition sites may be involved. The model may underestimate the number of these sites owing to two possible reasons: (i) a lack of precision in the activity measurements in the inhibitory zone and (ii) an insufficient difference between K I values that could hide a larger number of substrate molecules involved in inhibition.
To test the structural analysis presented previously, in which up to four riboflavin molecules were proposed to bind simultaneously to the RS trimer, we determined the binding stoichiometry of this complex in solution based on the binding curve presented in Supplementary Fig. S5. We found that only one molecule of riboflavin binds per RS monomer, with a K d value of 20.6 µM. This difference may be explained by considering that the fourth binding site might only be occupied after the addition of an excess of the ligand. Finally, we studied the effect of riboflavin on the catalytic activity. Similarly to A. thaliana RS (Fischer et al., 2005 ▶), the initial velocity decays as the riboflavin concentration increases (Supplementary Fig. S6), indicating a product-inhibition mechanism as well. Assuming that the 6,7-dimethyl-8-ribityllumazine concentration is lower than that of riboflavin and taking into account that RS exhibits an unusually high affinity for its product, it is likely that product inhibition is the major mechanism by which RS regulates its enzymatic activity in vivo. Moreover, substrate inhibition is only observed at very high nonphysiological substrate concentrations.
4. Conclusions
The crystal structures presented in this work reveal a flexible organization of the monomers in the RS trimer, with an enhanced mobility for the β-barrel regions. On the other hand, the central three-helix bundle preserves its architecture both in the presence and absence of bound ligands, playing a key role in the assembly of the particle. From a pictorial point of view, RS can be regarded as a bunch of three flowers in which the stems are held together but where each of the flowers are able to move with respect to the others. In solution, this relative movement may be rather more pronounced, bringing together other adjacent pairs of substrate-binding sites that are exposed to the solvent in the crystal picture, namely the C-terminal barrel from chain A with the N-terminal barrel from chain C, or the N-terminal barrel from chain B with the C-terminal barrel from chain C (see Fig. 8 ▶). In any case, only two out of the six substrate-binding sites may be able to perform as a catalytic site at a given time. The crystal packing may indeed represent an obstacle for the proposed relative movement of the individual subunits, thus limiting their ability to assemble a different active site. Additionally, in comparison with the crystal state, the binding of riboflavin and substrate analogues in solution may be more dynamic and the disruption of an active site to form a different one might help in the release of the catalysis products.
The stability of the trimer seems to be strongly affected by the chemical environment. For example, RS from S. pombe crystallized as a monomer owing to dissociation of the trimer in the sitting drops after equilibration against a reservoir solution containing, amongst other chemicals, 65% 2-methyl-2,4-pentanediol (MPD; Gerhardt, Schott et al., 2002 ▶). Interestingly, we were also able to obtain good diffracting B. abortus RS crystals by using only ∼20% MPD as the precipitating agent that showed a similar unit cell and space group to those presented in this work. However, in the molecular-replacement step only two of the three expected monomers could be located with success. Forcing the presence of a third monomer affected the statistics negatively. The fact that one chain could not be found reflects that this particular monomer is highly mobile within the crystal packing when MPD is used for crystallization, once more corroborating the intrinsic flexibility of the whole particle.
The structures solved here also demonstrate that ligand binding drives the assembly of the unique active site. More explicitly, binding of any of the three ligands studied in this work (and consequently also 6,7-dimethyl-8-ribityllumazine) to the N-terminal barrel in chain A gives rise to new contacts between these molecules and chain B, bringing both monomers together. Overall, the structural characterization of RS may be useful for the rational design of novel inhibitors with antimicrobial activity against Brucella. It is important to state that some inhibitors of Mycobacterium tuberculosis and E. coli RS have already been discovered (Zhang et al., 2008 ▶; Zhao et al., 2009 ▶), yet their antibiotic activity was only moderate.
The kinetic analysis performed on RS demonstrated a marked inhibition by substrate and product, as also observed for the A. thaliana enzyme (Fischer et al., 2005 ▶). The trimeric organization may be a key factor in the regulation of the catalytic activity of RSs in eubacteria, fungi and plants. RS, in common with many of the enzymes involved in the riboflavin-biosynthetic pathway, has evolved to be a low-rate catalyst. This might justify why evolution has preserved an enzyme that is not using two thirds of its catalytic sites at any given moment, and might open the possibility for yet unknown functions in addition to riboflavin biosynthesis. In this regard, it has recently been reported that 6,7-dimethyl-8-ribityllumazine, as well as reduced and hydroxylated 8-ribityllumazine derivatives, are strong and specific activators of mucosal-associated invariant T (MAIT) cells, which play a key role in immunity regulation and detection of microbial infection (Kjer-Nielsen et al., 2012 ▶; Patel et al., 2013 ▶). However, there is no information about potential carriers or reservoirs for these molecules prior to their recognition by the MAIT cells. We hypothesize that RS might be a strong candidate for this function, which could also explain why this enzyme is a strong immunogen in brucellosis.
Supplementary Material
PDB reference: RS-APO, 4fxu
PDB reference: RS-RBF, 4e0f
PDB reference: RS-ROS, 4g6i
PDB reference: RS-NRP, 4gqn
Supporting Information.. DOI: 10.1107/S1399004714005161/kw5091sup1.pdf
Acknowledgments
This work was supported by the Argentinian Research Council (CONICET) and the Argentinian Agency for Scientific and Technological Development (ANPCyT). We acknowledge access to the X-ray facilities at the Institut Pasteur Montevideo, Uruguay, the SOLEIL synchrotron, France and the NSLS, USA. The X6A beamline at the NSLS is funded by the National Institute of General Medical Sciences, National Institutes of Health under agreement GM-0080. The NSLS, Brookhaven National Laboratory is supported by the US Department of Energy under contract No. DE-AC02-98CH10886. We are grateful to the Center for Structural Biology of the Mercosur (CEBEM) and the Argentinian Ministry of Science (MINCyT) for travel support.
Footnotes
Supporting information has been deposited in the IUCr electronic archive (Reference: KW5091).
References
- Akaike, H. (1974). IEEE Trans. Automat. Contr. 19, 716–723.
- Beach, R. & Plaut, G. W. (1969). Tetrahedron Lett., pp. 3489–3492. [DOI] [PubMed]
- Berguer, P. M., Mundiñano, J., Piazzon, I. & Goldbaum, F. A. (2006). J. Immunol. 176, 2366–2372. [DOI] [PubMed]
- Bonomi, H. R., Marchesini, M. I., Klinke, S., Ugalde, J. E., Zylberman, V., Ugalde, R. A., Comerci, D. J. & Goldbaum, F. A. (2010). PLoS One, 5, e9435. [DOI] [PMC free article] [PubMed]
- Bricogne, G., Blanc, E., Brandl, M., Flensburg, C., Keller, P., Paciorek, W., Roversi, P., Sharff, A., Smart, O. S., Vonrhein, C. & Womack, T. O. (2011). BUSTER v.2.10.0. Cambridge: Global Phasing Ltd.
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Craig, P. O., Alzogaray, V. & Goldbaum, F. A. (2012). Biomacromolecules, 13, 1112–1121. [DOI] [PubMed]
- Eberhardt, S., Richter, G., Gimbel, W., Werner, T. & Bacher, A. (1996). Eur. J. Biochem. 242, 712–719. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Engh, R. A. & Huber, R. (1991). Acta Cryst. A47, 392–400.
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Fischer, M. & Bacher, A. (2005). Nat. Prod. Rep. 22, 324–350. [DOI] [PubMed]
- Fischer, M. & Bacher, A. (2006). Physiol. Plant. 126, 304–318.
- Fischer, M. & Bacher, A. (2008). Arch. Biochem. Biophys. 474, 252–265. [DOI] [PubMed]
- Fischer, M. & Bacher, A. (2011). Chembiochem, 12, 670–680. [DOI] [PubMed]
- Fischer, M., Haase, I., Feicht, R., Richter, G., Gerhardt, S., Changeux, J.-P., Huber, R. & Bacher, A. (2002). Eur. J. Biochem. 269, 519–526. [DOI] [PubMed]
- Fischer, M., Haase, I., Feicht, R., Schramek, N., Köhler, P., Schieberle, P. & Bacher, A. (2005). Biol. Chem. 386, 417–428. [DOI] [PubMed]
- Fischer, M., Schott, A. K., Römisch, W., Ramsperger, A., Augustin, M., Fidler, A., Bacher, A., Richter, G., Huber, R. & Eisenreich, W. (2004). J. Mol. Biol. 343, 267–278. [DOI] [PubMed]
- Gerhardt, S., Haase, I., Steinbacher, S., Kaiser, J. T., Cushman, M., Bacher, A., Huber, R. & Fischer, M. (2002). J. Mol. Biol. 318, 1317–1329. [DOI] [PubMed]
- Gerhardt, S., Schott, A. K., Kairies, N., Cushman, M., Illarionov, B., Eisenreich, W., Bacher, A., Huber, R., Steinbacher, S. & Fischer, M. (2002). Structure, 10, 1371–1381. [DOI] [PubMed]
- Godfroid, J., Cloeckaert, A., Liautard, J.-P., Kohler, S., Fretin, D., Walravens, K., Garin-Bastuji, B. & Letesson, J.-J. (2005). Vet. Res. 36, 313–326. [DOI] [PubMed]
- Gorvel, J.-P. (2008). Microbes Infect. 10, 1010–1013. [DOI] [PubMed]
- Haldane, J. B. S. (1930). Enzymes London: Longmans, Green & Co.
- Illarionov, B., Eisenreich, W. & Bacher, A. (2001). Proc. Natl Acad. Sci. USA, 98, 7224–7229. [DOI] [PMC free article] [PubMed]
- Illarionov, B., Kemter, K., Eberhardt, S., Richter, G., Cushman, M. & Bacher, A. (2001). J. Biol. Chem. 276, 11524–11530. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kim, R., Illarionov, B., Joshi, M., Cushman, M., Lee, C. Y., Eisenreich, W., Fischer, M. & Bacher, A. (2010). J. Am. Chem. Soc. 132, 2983–2990. [DOI] [PMC free article] [PubMed]
- Kjer-Nielsen, L. et al. (2012). Nature (London), 491, 717–723.
- Klinke, S., Zylberman, V., Bonomi, H. R., Haase, I., Guimarães, B. G., Braden, B. C., Bacher, A., Fischer, M. & Goldbaum, F. A. (2007). J. Mol. Biol. 373, 664–680. [DOI] [PubMed]
- Klinke, S., Zylberman, V., Vega, D. R., Guimarães, B. G., Braden, B. C. & Goldbaum, F. A. (2005). J. Mol. Biol. 353, 124–137. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2004). Acta Cryst. D60, 2256–2268. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Ladenstein, R., Fischer, M. & Bacher, A. (2013). FEBS J. 280, 2537–2563. [DOI] [PubMed]
- Laplagne, D. A., Zylberman, V., Ainciart, N., Steward, M. W., Sciutto, E., Fossati, C. A. & Goldbaum, F. A. (2004). Proteins, 57, 820–828. [DOI] [PubMed]
- Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J. & Higgins, D. G. (2007). Bioinformatics, 23, 2947–2948. [DOI] [PubMed]
- Leslie, A. G. W. & Powell, H. R. (2007). Evolving Methods for Macromolecular Crystallography, edited by R. J. Read & J. L. Sussman, pp. 41–51. Dordrecht: Springer.
- Liao, D.-I., Wawrzak, Z., Calabrese, J. C., Viitanen, P. V. & Jordan, D. B. (2001). Structure, 9, 399–408. [DOI] [PubMed]
- Long, Q., Ji, L., Wang, H. & Xie, J. (2010). Chem. Biol. Drug Des. 75, 339–347. [DOI] [PubMed]
- Massey, V. (2000). Biochem. Soc. Trans. 28, 283–296. [PubMed]
- Meining, W., Eberhardt, S., Bacher, A. & Ladenstein, R. (2003). J. Mol. Biol. 331, 1053–1063. [DOI] [PubMed]
- Müller, F. (1992). Chemistry and Biochemistry of Flavoenzymes Boca Raton: CRC Press.
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Navaza, J. (1994). Acta Cryst. A50, 157–163.
- Otto, M. K. & Bacher, A. (1981). Eur. J. Biochem. 115, 511–517. [DOI] [PubMed]
- Pappas, G., Akritidis, N., Bosilkovski, M. & Tsianos, E. (2005). N. Engl. J. Med. 352, 2325–2336. [DOI] [PubMed]
- Patel, O. et al. (2013). Nature Commun. 4, 2142. [DOI] [PubMed]
- Ramsperger, A., Augustin, M., Schott, A. K., Gerhardt, S., Krojer, T., Eisenreich, W., Illarionov, B., Cushman, M., Bacher, A., Huber, R. & Fischer, M. (2006). J. Biol. Chem. 281, 1224–1232. [DOI] [PubMed]
- Ritsert, K., Huber, R., Turk, D., Ladenstein, R., Schmidt-Bäse, K. & Bacher, A. (1995). J. Mol. Biol. 253, 151–167. [DOI] [PubMed]
- Serganov, A., Huang, L. & Patel, D. J. (2009). Nature (London), 458, 233–237. [DOI] [PMC free article] [PubMed]
- Smart, O. S., Womack, T. O., Flensburg, C., Keller, P., Paciorek, W., Sharff, A., Vonrhein, C. & Bricogne, G. (2012). Acta Cryst. D68, 368–380. [DOI] [PMC free article] [PubMed]
- Truffault, V., Coles, M., Diercks, T., Abelmann, K., Eberhardt, S., Lüttgen, H., Bacher, A. & Kessler, H. (2001). J. Mol. Biol. 309, 949–960. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Yang, Y., Wang, L., Yin, J., Wang, X., Cheng, S., Lang, X., Wang, X., Qu, H., Sun, C., Wang, J. & Zhang, R. (2011). Mol. Immunol. 49, 175–184. [DOI] [PubMed]
- Zhang, Y., Illarionov, B., Morgunova, E., Jin, G., Bacher, A., Fischer, M., Ladenstein, R. & Cushman, M. (2008). J. Org. Chem. 73, 2715–2724. [DOI] [PubMed]
- Zhao, Y., Bacher, A., Illarionov, B., Fischer, M., Georg, G., Ye, Q.-Z., Fanwick, P. E., Franzblau, S. G., Wan, B. & Cushman, M. (2009). J. Org. Chem. 74, 5297–5303. [DOI] [PubMed]
- Zylberman, V., Craig, P. O., Klinke, S., Braden, B. C., Cauerhff, A. & Goldbaum, F. A. (2004). J. Biol. Chem. 279, 8093–8101. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: RS-APO, 4fxu
PDB reference: RS-RBF, 4e0f
PDB reference: RS-ROS, 4g6i
PDB reference: RS-NRP, 4gqn
Supporting Information.. DOI: 10.1107/S1399004714005161/kw5091sup1.pdf