Summary
Metastasis is a major clinical challenge for cancer treatment. Emerging evidence suggests that epigenetic aberrations contribute significantly to tumor formation and progression. However, the drivers and roles of such epigenetic changes in tumor metastasis are still poorly understood. Using bioinformatic analysis of human breast cancer gene expression datasets, we identified histone demethylase RBP2 as a putative mediator of metastatic progression. By using both human breast cancer cells and genetically engineered mice, we demonstrated that RBP2 is critical for breast cancer metastasis to the lung in multiple in vivo models. Mechanistically, RBP2 promotes metastasis as a pleiotropic positive regulator of many metastasis genes. In addition, RBP2 loss suppresses tumor formation in the MMTV-neu transgenic mice. These results suggest that therapeutically targeting RBP2 is a potential strategy to inhibit tumor progression and metastasis.
Introduction
In the United States, breast cancer is the most common cancer and the second leading cause of cancer death in women (Desantis et al., 2011). Advanced breast cancer is associated with significant mortality because it metastasizes to vital organs, primarily to lung, brain and bone (Bos et al., 2009; Kang et al., 2003; Minn et al., 2007; Minn et al., 2005). There are still limited treatment options for patients with metastatic breast cancer. Thus, it is critical to identify and validate novel drug targets for the development of effective therapies.
Tumor metastasis is a multistage process that includes local invasion, intravasation, survival in the circulation, extravasation, and colonization in distant organs (Nguyen et al., 2009a; Sethi and Kang, 2011). During this process, cancer cells must overcome various physiological barriers and adapt to foreign environments. This requires the coordinate modulation of pleiotropic genetic programs at different stages of tumor progression (Brabletz, 2012; Peinado et al., 2012). To achieve this kind of plasticity, it is conceivable that reversible transcription programs may be required in addition to somatic genetic alterations. Consistent with this idea, many epigenetic regulators were reported to play critical roles in this process (Nguyen and Massague, 2007). For example, histone H3K27 methyltransferase EZH2 and histone demethylase JMJD2C (also known as KDM4C) were shown to promote tumor progression and metastasis (Luo et al., 2012; Min et al., 2010; Varambally et al., 2002). In contrast, histone H3K4 demethylase LSD1 was reported to inhibit breast cancer metastasis (Wang et al., 2009b).
Breast cancer metastasis to different tissues is mediated in part by organ-specific metastasis genes, some of which are also highly expressed in the primary tumors. Some of these genes, such as MMP1 and CXCL1, provide growth advantages in both the primary and metastatic sites, while others like TNC and MMP2 promote aggressive growth only in the metastatic niche (Minn et al., 2007). Despite their known functions, the mechanisms by which these genes are up-regulated remain unknown. These gene products can be modulated individually or more broadly by pleiotropic regulators. Among such potential pleiotropic regulators, transcription factors are difficult to target, while epigenetic regulators are very attractive targets for cancer therapies, in part because epigenetic changes are reversible (Blair and Yan, 2012; Rodriguez-Paredes and Esteller, 2011).
To identify novel epigenetic regulators that can be targeted in breast cancer metastasis, we conduct an unbiased bioinformatic analysis of human breast cancer datasets. We identify a strong association between the expression of histone demethylase RBP2 (also known as JARID1A and KDM5A) with breast cancer metastasis. RBP2 is a member of the JARID1 family histone demethylases, which catalyze the removal of methyl-groups from tri- or di-methylated lysine 4 in histone H3 (Blair et al., 2011; Christensen et al., 2007; Iwase et al., 2007; Klose et al., 2007; Lee et al., 2007; Secombe et al., 2007; Tahiliani et al., 2007; Yamane et al., 2007). We show that RBP2 positively regulates many metastasis related genes, including TNC, which is required for metastatic colonization in the lungs. These findings are further validated in the MMTV-neu transgenic mouse model. Importantly, RBP2 promotes TNC expression and malignant invasion through a demethylase-independent mechanism. In summary, our findings suggest that RBP2 regulates a critical epigenetic switch that sets the stage for tumor metastasis and can be targeted to inhibit breast cancer progression and metastasis.
Results
RBP2 Expression Is Strongly Associated with Breast Cancer Metastasis
To identify novel epigenetic regulators of breast cancer metastasis, we conducted an unbiased bioinformatic analysis of gene expression profiles of mammary tumors from 533 breast cancer patients using Kaplan-Meier Plotter, a meta-analysis based biomarker assessment tool (Gyorffy et al., 2010). This analysis tool utilizes Affymetrix gene expression profiling data, which have multiple probe sets for most genes. We examined the correlation between increased incidence of distant tumor metastasis with the gene expression levels of a comprehensive list of targetable histone methylation and acetylation enzymes, including histone lysine methyltransferases (KMTs), histone lysine demethylases (KDMs), histone acetyltransferases (KATs), and histone deacetylases (HDACs). This analysis revealed that high mRNA levels of two enzymes, EZH2 and RBP2, correlated significantly with early and high incidence of tumor metastasis (Figure 1A and Table S1). Our approach was validated by the fact that EZH2 (only one probe available) was previously shown to promote invasion and metastasis of breast cancer cells (Kleer et al., 2003; Moore et al., 2013). Two probes of RBP2 are present on the microarray platform, and both probes showed similar results (Figure 1B and S1A). RBP2 was found to promote tumorigenesis (Lin et al., 2011; Zeng et al., 2010) and drug resistance (Sharma et al., 2010). However, the role of RBP2 in breast cancer metastasis has not been studied. Therefore, we focused on the role of RBP2 in breast cancer metastasis.
Figure 1.

High RBP2 expression level is associated with breast cancer metastasis. (A) Correlation of the mRNA levels of histone modifying enzymes with breast cancer metastasis. The patients were divided into two groups with either higher or lower expression based on each probe. Plotted were hazard ratio (HR) with 95% confidence and Bonferroni multiple testing corrected p-value (MTCPV). (B) Kaplan-Meier analysis of metastasis-free survival of lymph node negative patients with breast cancer, stratified by RBP2 expression level based on the 202040_s_at probe. (C) Summary of Kaplan-Meier analysis of metastasis-free survival of all patients, ER+ or ER− breast cancer patients in the EMC286 cohort. (D) Western blot analysis of RBP2 and tubulin in the indicated cells. 231, MDA-MB-231 cells.
We validated the association of RBP2 expression with breast cancer metastasis using a large clinical dataset, EMC286 (Figure S1B). To determine whether the association of RBP2 expression with metastasis is subtype specific, we performed independent analysis of estrogen receptor (ER) positive (ER+) and negative (ER−) patients. This revealed that RBP2 expression was strongly associated with metastasis in ER− patients in both the EMC286 and the combined datasets, while it was more weakly associated with metastasis in ER+ patients only in the combined dataset (Figure 1C and S1C–I). To further determine whether RBP2 can be an independent clinical indicator of metastasis, we conducted a Cox multivariate analysis of EMC286. Our analysis indicated that RBP2 was associated with tumor metastasis independent of ER, PR, and HER2 status (Table S2).
We then extended our analysis to two matched breast cancer cell line series, including the MDA-MB-231and 4T1 cell series. LM2 and 1833 cells were derived from the ER− MDA-MB-231 human breast cancer cells by in vivo selection, and have increased metastatic activity to the lungs or the bones, respectively, when compared to their parental cells (Kang et al., 2003; Minn et al., 2005). Mouse mammary tumor cell lines 67NR, 168FARN, 4TO7, 66cl4, and 4T1 were isolated from a single spontaneous mammary tumor and have varying metastatic capabilities to the lungs (Aslakson and Miller, 1992). Western blot analysis showed that RBP2 protein was expressed at a higher level in the more lung metastatic LM2 cells, compared to the bone metastatic 1833 cells or the less metastatic parental MDA-MB-231 cells (Figure 1D). Furthermore, the expression levels of RBP2 strongly correlated with the metastatic capabilities of the 4T1 series, with RBP2 levels being the highest in the most aggressive 4T1 cell line (Figure 1D). These results are consistent with our observation in patient samples that RBP2 expression level correlated with increased metastatic potential.
RBP2 Is Critical for Pro-Metastatic Gene Expression
To determine the roles of RBP2 in breast cancer, we transfected MDA-MB-231 cells with siRNA against RBP2 or siRNA control. We found that knockdown of RBP2 led to global increase of H3K4me3 levels, suggesting that RBP2 is the major H3K4me3 demethylase in MDA-MB-231 cells (Figure S2A–B). To determine the effects of RBP2 depletion on gene expression, we conducted microarray analysis of these cells and identified 176 up-regulated and 212 down-regulated genes after RBP2 knockdown (Table S3). Next, we performed gene set enrichment analysis (GSEA) (Subramanian et al., 2005) to determine whether RBP2 loss causes alterations of known breast cancer organotropic metastasis gene signatures, including the lung metastasis signature (LMS), bone metastasis signature (BoMS), and brain metastasis signature (BrMS) (Bos et al., 2009; Kang et al., 2003; Minn et al., 2007; Minn et al., 2005). These analyses revealed that RBP2 knockdown significantly decreased expression of genes linked to breast cancer metastasis to the lungs (Figure 2A). In contrast, there were no significant changes of genes involved in breast cancer metastasis to the bone or brain in RBP2 knockdown cells (Figure 2A, right panel). This result is consistent with our observations that RBP2 is highly expressed in the lung metastatic LM2 cells but not in the bone metastatic 1833 cells.
Figure 2.
RBP2 regulates the expression of lung metastasis genes. (A) Gene-set enrichment analyses of MDA-MB-231 cells with RBP2 siRNAs versus control siRNA using organ-specific metastasis gene signatures. Left panel, enrichment plot showing decreased enrichment of the lung metastasis gene signature in MDA-MB-231 cells transfected with RBP2 siRNA compared with those with control siRNA. Right Panel, summary of enrichment analyses. Shown are normalized enrichment scores (NES), nominal p-value (p-val), and false discovery rate q-value (q-val). RBP2 KD, RBP2 siRNA knockdown; Ctrl KD, control siRNA knockdown. LMS, lung metastasis signature; BoMS, bone metastasis signature; BrMS, brain metastasis signature; Up, upregulated genes; Down, downregulated genes. (B) Real time RT-PCR analysis of the indicated mRNAs in MDA-MB-231 (231) or LM2 cells transfected with scrambled control or RBP2 siRNA. Scr si, scrambled siRNA; RBP2 si-1, RBP2 siRNA-1; RBP2 si-2, RBP2 siRNA-2. Error bars represent SD.
We selected several genes, including TNC, SOX4, FSCN1, PDGFA, SERPINE2, and PLCB1, all of which were implicated in breast cancer metastasis to the lungs (Minn et al., 2007; Minn et al., 2005; Tavazoie et al., 2008; Tiwari et al., 2013), for confirmation via real time RT-PCR analysis. As expected, all these genes were expressed at higher levels in LM2 cells compared to MDA-MB-231 cells (Figure 2B). Consistent with our GSEA results, RBP2 knockdown led to significantly down-regulation of all these genes in MDA-MB-231 cells and most of these genes in LM2 cells (Figure 2B).
RBP2 Regulates Breast Cancer Cell Invasion through TNC
Invasion of tissue basement membranes is one of the key steps of metastasis. To dissect the roles of RBP2 in tumor metastasis, we first examined its role in invasion using trans-well invasion assays. LM2 cells, which expressed higher levels of RBP2, showed increased invasion ability compared to MDA-MB-231 cells (Figure S3A). Furthermore, we found that the LM2 cells transfected with siRNAs against RBP2 showed a dramatic decrease of their invasion ability, compared with the cells transfected with the control siRNAs (Figure 3A–B).
Figure 3.
Knockdown of RBP2 reduces cell invasion in vitro. (A) Representative image of DAPI staining and (B) Quantification of LM2 cells invaded through Matrigel coated membrane inserts after treatment with the indicated siRNA. (C) Western blot analysis of the indicated proteins in whole cell lysates or conditioned media of MDA-MB-231 (231) and LM2 cells transfected with the indicated siRNAs. (D) Quantification of LM2 cells invaded through Matrigel coated membrane inserts after transfection with the indicated siRNAs and treatment with the indicated concentration of recombinant TNC protein. (B and D) 4 random fields of each insert were quantified and normalized to the control. Error bars represent SEM of three inserts. **, p<0.01; ***, p<0.001. Luc, luciferase siRNA; Scr, scrambled siRNA, RBP2 si-1, RBP2 siRNA-1; RBP2 si-2, RBP2 siRNA-2.
TNC, which encodes tenascin C, was the most significantly decreased metastasis genes in LM2 cells with RBP2 knockdown. TNC is an extracellular matrix protein that enhances cancer cell migration and invasion (Hancox et al., 2009; Nagaharu et al., 2011). TNC also promotes colonization of breast cancer cells to the lungs and is a critical component of the metastatic niche in vivo (Oskarsson et al., 2011). Therefore we further examined the regulation of TNC as an output for epigenetic modulation of invasion by RBP2. We analyzed protein levels in the media of LM2 cells and confirmed that extracellular TNC protein production was also regulated by RBP2 (Figure 3C). Next, we examined the correlation of RBP2 and TNC expression in human primary breast tumors. Consistent with our data from the cell lines, ER− tumors with high levels of RBP2 expressed high levels of TNC (Figure S3B).
Since we showed that TNC expression is regulated by RBP2, we next tested whether TNC is required for the pro-invasive function of RBP2. To this end, recombinant TNC protein was added into the chambers in trans-well assays. The addition of TNC partially rescued the decreased invasion phenotype in RBP2 knockdown cells (Figure 3D), suggesting that regulation of invasion by RBP2 was at least partially due to the decrease of TNC expression in RBP2 knockdown cells.
RBP2 Promotes TNC Expression and Invasion in a Demethylase-independent Manner
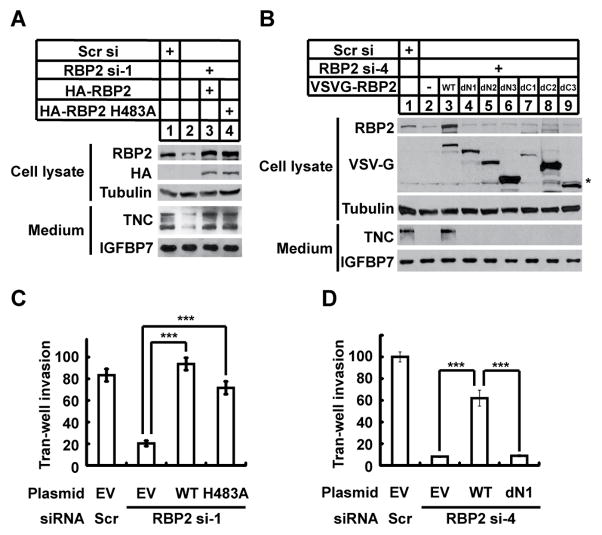
To dissect the mechanism by which RBP regulates gene expression and invasion, we reintroduced siRNA-resistant forms of RBP2 into RBP2 knockdown cells. Consistent with the idea that RBP2 is critical for TNC expression, restoration of wild type RBP2 expression rescued the TNC level in RBP2 knockdown cells (Figure 4A). Interestingly, re-introduction of a histone demethylase-defective form of RBP2 (RBP2 H483A) also rescued the decrease of TNC (Figure 4A). Moreover, chromatin-immuneprecipataion (ChIP) assays showed that knockdown of RBP2 in LM2 cells only led to a slightly decrease of H3K4me3 near the TNC transcriptional start site (Figure S4A–B). These results suggest that the demethylase activity of RBP2 is not required for TNC expression in LM2 cells.
Figure 4.
RBP2 promotes TNC expression and cell invasion in a demethylase-independent manner. (A and B) Western blot analysis of the indicated proteins in whole cell lysates or conditioned media of LM2 cells co-transfected with the indicated siRNAs and plasmids. (C and D) Quantification of LM2 cells invaded through Matrigel coated membrane inserts after transfection with the indicated siRNAs and plasmids. Scr si or Scr, scrambled siRNA, RBP2 si-1, RBP2 siRNA-1; RBP2 si-4, RBP2 siRNA-4. EV, empty vector plasmid; WT, wild type RBP2 plasmid; HA-RBP2: HA-RBP2 plasmid; HA-RBP2 H483A: HA-RBP2 plasmid with H483A mutation; VSVG-RBP2: VSVG-RBP2 plasmids. A schematic map of VSVG-RBP2 plasmids is shown in Figure S4C. Error bars represent SEM of three inserts.
To further investigate whether other RBP2 domains are required for TNC expression, we tested the effects of expressing RBP2 truncation mutants in LM2 cells transfected with an siRNA targeting 3′ untranslated region of RBP2. We found that RBP2 mutants lacking either the N- or C-terminal domains failed to rescue TNC expression (Figure 4B and S4C), suggesting that the N- and C-terminal domains are required for TNC expression in LM2 cells. To determine if TNC is a direct target of RBP2, we performed ChIP assays using an anti-RBP2 antibody. Despite strong enrichment of RBP2 at the promoter of a known RBP2 target gene NDUFA9, we did not detect RBP2 binding at the TNC promoter (Figure S4D). Hence, RBP2 does not regulate expression of TNC by modulating its promoter activity.
Next, we determined the abilities of wild type and mutant RBP2 to promote invasion. Reintroduction of either wild type or a demethylase-defective RBP2 in LM2 cells rescued the diminished cell invasion induced by RBP2 knockdown (Figure 4C). In contrast, an N-terminal truncated form of RBP2 was unable to rescue the invasion ability of LM2 cells (Figure 4D), suggesting that the N-terminal domains, but not the histone demethylase activity of RBP2, are required to promote cell invasion.
RBP2 is Critical for Breast Cancer Metastasis to the Lungs in vivo
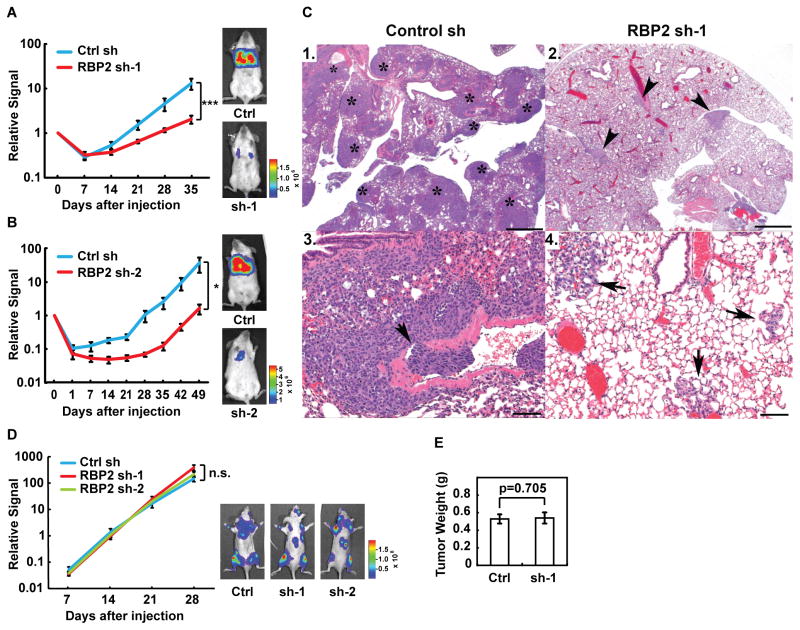
To investigate the roles of RBP2 in metastasis in vivo, we generated LM2 cells with scrambled shRNA control hairpin or stable knockdown of RBP2 using two independent shRNA hairpins. LM2 cells were engineered to express firefly luciferase (Minn et al., 2005), which allows for live imaging of metastasis in vivo. Similar to our results from siRNA mediated knockdown, LM2 cells with RBP2 stable knockdown secreted lower levels of TNC compared to the control cells (Figure S5A). Cells with stable RBP2 knockdown and their respective control cell line were then injected into the tail vein of NOD.SCID mice. Lung metastatic activity was assayed by bioluminescence imaging weekly as well as by examination of the lungs at necropsy. RBP2 knockdown led to an approximately 10-fold decrease in the lung metastasis abilities of LM2 cells (Figure 5A–B). The effect was observed even at the early time points (Figure 5A–B), suggesting that RBP2 controls extravasation and/or early seeding of lung metastasis in vivo. Histological analysis of the lungs isolated at necropsy confirmed that mice injected with RBP2 knockdown cells had fewer and smaller lesions in the lungs (Figure 5C).
Figure 5.
Knockdown of RBP2 decreases tumor metastasis in vivo. (A, B) Normalized bioluminescence signals of lung metastasis and representative bioluminescence images of mice injected intravenously with LM2 cells stably expressing control or RBP2 shRNA. The data represent average ± SEM. *, p<0.05; ***, p<0.001. (C) Representative H&E-stained lung sections. Mice injected with LM2 cells carrying control shRNA (Control sh) have more tumors [1 *(not all tumor foci marked), 3] than mice with LM2 cells carrying RBP2 shRNA (RBP2 sh-1) (2 arrowheads, 4) visible at low power. Vascular invasion (3, arrow) and small foci of metastatic nodules (4 arrows) are observed at increased magnification. Scale bars for panels 1, 2 = 500 μm and for panels 3,4 = 100 μm. (D) Normalized bioluminescence signals of bone metastasis and representative bioluminescence images of mice injected intracardially with 1833 cells stably expressing control or RBP2 shRNA. The data represent average ± SEM. n.s., non-significant. (E) The average weight of primary tumors at the endpoint in mice implanted in mammary fat pads with LM2 cells stably expressing control or RBP2 shRNA. Ctrl sh or Ctrl, control shRNA. Sh-1, RBP2 sh-1. Error bars represent SEM.
To determine whether RBP2 more broadly affects tumor progression or metastasis to multiple organs in vivo, we performed stable RBP2 knockdown in the bone selective 1833 breast cancer cell lines. Knockdown of RBP2 in 1833 cells had no significant effect on the ability of these cells to colonize the bone (Figure 5D and S5B). Knockdown of RBP2 also showed no effect on mammary tumor formation following orthotopic transplantation of LM2 cells into the mammary gland (Figure 5E). Altogether, these in vivo results are consistent with the observation that RBP2 regulates the expression of genes specific for lung metastasis.
To further validate our findings in a genetically engineered mouse model, we examined the effects of RBP2 loss on breast cancer progression and metastasis using the MMTV-neu transgenic mice, a breast cancer mouse model wherein more than 70% of mice with mammary tumors developed lung metastases (Guy et al., 1992). These mice carry wild type neu (the rat HER2 gene) under the control of the mouse mammary tumor virus (MMTV) promoter. We crossed the RBP2 knockout mice with the MMTV-neu transgenic mice and monitored mammary tumor formation and lung metastasis. We found that RBP2 knockout delayed mammary tumor formation in the MMTV-neu mice (Figure 6A), suggesting that RBP2 can contribute to mammary tumor development in this model. Lungs were obtained from mice when their primary mammary tumors were of similar size and reached approximately 1 cm3 (Figure S6). Similar to the results obtained from the experimental lung colonization model, RBP2 knockout mice showed dramatic decrease in the number of lung metastasis nodules and the incidence of lung metastasis (Figure 6B–D). Interestingly, in this model, most of the lung metastasis nodules were located inside the blood vessels, suggesting that RBP2 can also mediate the survival and proliferation of breast cancer cells in the pulmonary vasculature. Taken together, our findings from the human cell line models, preclinical mouse models and patient tumor samples strongly support a role for RBP2 as an epigenetic driver of metastasis.
Figure 6.
Loss of RBP2 decreases tumor progression and metastasis in the MMTV-neu transgenic mice. (A) Kaplan-Meier tumor-free survival curves of the MMTV-neu transgenic mice with the indicated genotypes of Rbp2. N, animal number in each group; M, days of medium survival in each group. p < 0.0001 based on Log-rank (Mental-Cox) test. (B) Column scatter plot showing the number of lung metastatic nodules in the MMTV-neu transgenic mice with the indicated Rbp2 genotypes. (C) Incidence of lung metastasis of the MMTV-neu transgenic mice with the indicated Rbp2 genotypes. (D) Representative H&E-stained lung sections. Rbp2+/+:MMTV-neu mice (1, 3, 5) showed greater numbers of tumors in the lungs than Rbp2−/−:MMTV-neu mice (2, 4, 6), and the morphology of the tumor cells are similar (5, 6). Scale bars for panels 1–4 = 500 μm and for panels 5–6 = 100 μm.
Discussion
Through mining the gene expression profiles of human mammary tumors, we identified a novel and strong association of RBP2 expression with breast cancer metastasis. Consistent with this finding, we found that RBP2 is overexpressed in highly metastatic cell lines that were generated from both human and mouse breast tumors. We revealed that RBP2 has pleiotropic roles in invasion and metastasis and we have linked RBP2 function to the regulation of several well-known genetic mediators of metastasis. Although recent studies highlighted the connection of epigenetic regulators to tumor metastasis, these studies were mostly limited to experiments using cancer cell lines or mouse xenograft models (Cao and Yan, 2013). Our study is bolstered by functional data from several preclinical mouse models, which demonstrate RBP2 as a critical epigenetic mediator of metastasis and an ideal cancer target. Combined with our analysis of human patient data, these results provide a strong rationale to target RBP2 to treat invasive and metastatic breast cancer.
We showed with two independent bioinformatic pipelines that RBP2 level correlated with increased incidence of breast cancer metastasis. However, a recent report using GOBO, a different breast cancer online tool (Ringner et al., 2011), showed that RBP2 level correlated with increased relapse free survival (Paolicchi et al., 2013), although no functional experiments were performed to validate these results. The discrepancy could be due to the cohort sizes and method of analysis. Kaplan-Meier Plotter for our analysis included more comprehensive gene expression profiles of 2,977 non-redundant breast tumors (Gyorffy et al., 2010), while GOBO included 1,881 breast tumors (Ringner et al., 2011). Compared to the one data normalization step used by GOBO, our analysis also incorporates two data normalization steps and was independently validated. Therefore, the results from this prior study may be more sensitive to dataset inclusion and sample stratification. The inconsistency between these datasets begs further functional validation using cell based assays and mouse models. Our extensive functional studies clearly validated our bioinformatic analysis, highlighting the importance of functional validation.
RBP2 has been implicated as an oncoprotein in various cancer types (Blair et al., 2011; Sharma et al., 2010). For example, RBP2 amplification was reported in approximately 15% of breast cancers (Hou et al., 2012). We showed previously that RBP2 is critical for endocrine tumor formation in mice lacking Rb and menin tumor suppressors (Lin et al., 2011). In this study, we demonstrate that knockdown or loss of RBP2 inhibits breast cancer metastasis to the lungs using two in vivo metastasis models. In contrast, RBP2 is not required for bone metastasis by 1833 cells. These results highlight the critical role of a pleiotropic epigenetic regulator in directing transcription program of tissue-specific metastasis. Although RBP2 knockdown did not affect the rate of primary tumor growth by LM2 cells in orthotopic xenograft model, RBP2 loss inhibited primary tumor formation in the MMTV-neu model. These results suggest that RBP2 likely regulates early stages of tumor progression when overexpressed early on in the mammary gland, or promotes a more amenable mammary tumor microenvironment for early tumorigenesis. Therefore, our studies highlight the importance of targeting RBP2 both in early tumorigenesis and tumor metastasis.
ER− breast cancer is an aggressive subtype of breast cancer that is often associated with rapid and frequent metastasis and poor patient outcome (Carey et al., 2010). However, only a limited number of targeted therapeutic methods for these patients are currently under investigation in the clinic. Here, we demonstrated that RBP2 is critical for invasion and metastasis of ER− breast cancer cells. Suppression of tumor formation and metastasis by RBP2 loss in the MMTV-neu model suggests that RBP2 is also critical for breast cancer metastasis of HER2 positive (HER2+) patients, the majority of whom are also ER−. These findings are consistent with our results from human patient samples, where RBP2 is strongly associated with increased metastasis in ER− patients. Taken together, these results support the requirement of RBP2 in aggressive breast cancers and provide the rationale for novel treatment methods targeting RBP2 in ER− and HER2+ patients.
Our gene expression analyses showed that RBP2 is a pleiotropic positive regulator of lung metastasis genes, suggesting that RBP2 is a critical epigenetic switch that enables tumor cells to metastasize through activating a constellation of genes. Contrary to the prevailing notion that RBP2 can only repress gene expression through histone demethylation, RBP2 has diverse activities in gene regulation and can activate gene expression (Benevolenskaya et al., 2005; Wang et al., 2009a). Consistent with its role in gene activation, our mechanistic studies indicated that RBP2 promotes TNC expression and cell invasion via a demethylase-independent mechanism. Similar to our findings, Lid, the fly homolog of RBP2, has demethylase-independent function in dMyc-mediated cell growth and development (Li et al., 2010). Furthermore, our results also indicated that the N- and C-terminal domains of RBP2 are required for regulation of TNC expression and the N-terminal domains of RBP2 are required for cell invasion. The ARID domain located in the N-terminus of RBP2 is involved in DNA binding (Tu et al., 2008), while the LXCXE motif located in the C-terminus of RBP2 interacts with Rb and p107 (Defeo-Jones et al., 1991; Kim et al., 1994). Thus, our results suggest that DNA binding and recruitment of Rb or p107 by RBP2 may be required for activation of RBP2 target genes, such as TNC. In contrast, RBP2 demethylase activity was reported to promote proliferation of mouse embryonic fibroblasts (Lin et al., 2011) and growth and invasion of lung cancer cells (Teng et al., 2013). Even though demethylase inhibitors such as the small molecules we identified recently (Sayegh et al., 2013) could be used to suppress cancer progression in general, our results suggest that alternative targeting strategy is needed to inhibit invasion and metastasis by breast cancer cells.
Experimental Procedures
Cell Culture and Trans-well Invasion Assays
MDA-MB-231 and 4T1 breast cancer cell series were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin and streptomycin. Retroviruses were generated by co-transfection of pLKO.1 plasmids carrying the indicated shRNAs with packaging plasmids into 293T cells as described previously (Klose et al., 2007). To generate RBP2 stable knockdown cell lines, LM2 cells were infected with the indicated viruses and selected with 1 μg/ml puromycin for two weeks. siRNA transfections were performed using RNAiMAX (Invitrogen). Plasmid transfections and plasmid/siRNA cotransfections were performed using Lipofectamine 2000 (Invitrogen). The shRNA sequences targeting RBP2 were: sh-1, ccagacttacagggacactta; sh-2, ccttgaaagaagccttacaaa. Scrambled control shRNA was described previously (Yang et al., 2007). The siRNA targeting sequences of RBP2 were: siRNA-1, gctgtacgagagtatacac; siRNA-2: cttctgtactgctgactgg; siRNA-3: gccaagaacattccagcct; and siRNA-4: gatagtagttagaggctta, which targets 3′ untranslated region of RBP2. Scrambled siRNA was described previously (Beshiri et al., 2012) and luciferase siRNA was obtained from Dharmacon. pcDNA3/HA-RBP2 construct was described previously (Benevolenskaya et al., 2005) and the H483A mutation was introduced by site-directed mutagenesis as described previously (Klose et al., 2007). pCI-neo/VSVG-RBP2 constructs were described previously (Chan and Hong, 2001). Trans-well invasion assay were performed as described previously (Cheung et al., 2013). Pictures of four random fields from each well were taken under the microscope at 10 × magnification, and the number of cells on the basal side was counted. In the TNC rescue experiment, BSA and/or recombinant TNC protein (CC065, Millipore) were pre-incubated with the insert for 2 hours and added to the media when seeding cells at the final concentration of 100 ng/ml.
Animal Studies
6–8 weeks old female NOD.SCID mice were used for lung metastasis and mammary fat pad tumor growth experiments. 4–6 weeks old male athymic nude mice were used for bone metastasis experiments. For lung metastasis assay, 2 × 105 viable cells were re-suspended in 0.1 ml saline, and injected into the lateral tail vein. For bone metastasis assay, 1 × 105 viable cells were re-suspended in 0.1 ml saline, and injected into the left ventricle of the heart. Lung or bone metastatic colonization was monitored and quantified immediately after the injection and at the indicated time points using non-invasive bioluminescence, and validated at the end point by histological analysis, as previously described (Bos et al., 2009; Lu et al., 2011; Nguyen et al., 2009b). All values of luminescence photon flux were normalized to the value of the same mouse obtained immediately after injection of the cells. For mammary fat pad tumor growth assay, 1 × 106 viable cells were re-suspended in 0.1 ml of 1:1 mixture of saline and Matrigel, and injected into the fat pads of 4th mammary glands. Tumor weight was determined at the end point.
The MMTV-neu (FVB/N-Tg(MMTVneu)202Mul/J) mice were obtained from The Jackson Laboratory. Rbp2−/− mice (Klose et al., 2007) were backcrossed to the FVB background for at least eight generations, and then crossed with the MMTV-neu transgenic mice for two generations. The copy numbers of the MMTV-neu transgene were determined by genotyping at least 12 offsprings from the crossing of Rbp2+/− mice carrying the MMTV-neu transgene with wild type mice. Rbp2+/− mice with two copies of the MMTV-neu transgenes were selected and intercrossed. Breast tumor formation of Rbp2+/+:MMTV-neu and Rbp2−/−:MMTV-neu mice were monitored weekly and mice were euthanized when primary tumors reached approximately 1 cm 3.
Bioinformatic and Statistical Analysis
Kaplan Meier-plotter analysis of histone modifying enzymes in metastasis-free survival of breast cancer patients were performed by using the tool generated by the Szallasi group (Gyorffy et al., 2010). The settings for the analysis were: DMFS, auto select best cutoff, 10 years follow up threshold, ER status all, PR status all, lymph node negative, grade all, and molecular subtype all. Multiple testing corrected p-values of each probe were calculated by timing original p-value with the number of genes (69) analyzed. The results of these experiments were described in Figure 1A–B and S1A, C–E, and G–H, and Table S1.
EMC286 cohort gene expression data was downloaded from GSE2034 (Wang et al., 2005) and normalized using robust multi-array average method within R Affy package. For this cohort, RBP2 probe 202040_s_at was selected for Kaplan Meier metastasis-free survival analysis, and correlation analysis. Samples with high, medium and low RBP2 expression levels were clustered using quartile or k-means as indicated in the figure legends. The metastasis-free survival plots, Cox univariate and multivariate metastasis-free survival analyses were performed via R survival package. Pearson correlation test was performed using the cor.test R function. The results of these experiments were described in Figure 1C, S1B, F and I, S3C, and Table S2.
Triplicate total RNA samples were isolated from MDA-MB-231 cells 72 hours after transfection with siRNAs against RBP2 or luciferase control. Gene expression profiling was performed using HumanHT-12 V3.0 Expression Beadchip Kit (BD-103-0203, Illumina) by Yale Center for Genome Analysis. Non-normalized gene expression profiling data exported from Illumina Genome Studio were processed using Lumi R package. The differential expressed gene list by comparing the average of two RBP2 knockdown (RBP2 siRNA-1 and siRNA-2) samples with the control knockdown (luciferase siRNA) samples was generated using Limma R package. Using adjusted p value < 0.05 and > 1.5 fold difference as the cutoff, 176 genes were up-regulated and 212 were down-regulated after RBP2 knockdown. The raw data were deposited in the NCBI Gene Expression Omnibus database under accession number GSE49119. Gene expression profiles of the control knockdown and the average of two RBP2 knockdown cells were used for gene set enrichment analysis using GSEA v2.0 software. Gene sets were generated from published gene signatures (Bos et al., 2009; Kang et al., 2003; Minn et al., 2007; Minn et al., 2005). Statistical significance was assessed by comparing the enrichment score to enrichment results generated from 10,000 random permutations of the gene set. Log-rank (Mantel-Cox) test were used for analysis of tumor-free survival curves of the MMTV-neu transgenic mice. Comparison of number of metastatic nodules in lungs from wild type or RBP2 knockout mice was performed using negative binomial model. T-test was used for other statistical analysis.
Additional methods can be found in Supplemental Experimental Procedures.
Supplementary Material
Highlights.
RBP2 expression correlates with metastatic relapse in breast cancer patients.
RBP2 is a pleiotropic positive regulator of a tissue-specific metastasis gene set.
RBP2 activates TNC and promotes invasion in a demethylase-independent manner.
RBP2 is critical for breast cancer progression and lung metastasis in vivo.
Acknowledgments
We thank members of the Nguyen, Stern, Wajapeyee and Yan laboratories for their kind help and valuable discussions. We thank Dr. Joan Massague for providing MDA-MB-231, LM2, 1833, 67NR and 4T1 cells, Dr. Yibin Kang for providing 168FARN, 4TO7, and 66cl4 cells, Drs. Lee Cheng-Feng and Li-Jung Juan for sharing RBP2 shRNA constructs, Dr. Wanjin Hong for sharing the VSVG-RBP2 constructs, Dr. Narendra Wajapeyee for providing anti-IGFBP7 antibody, and Dr. Tian Xu for sharing his equipment. This work was supported by the Alexander and Margaret Stewart Trust Fellowship (to Q.Y.), Connecticut Department of Public Health Biomedical Research grant 2013-0201 (to Q.Y.), American Cancer Society Research Scholar Grant RSG-13-384-01-DMC (to Q.Y.), NIH grants R01CA166376 (to D.X.N.) and P30CA16359 (to the Yale Comprehensive Cancer Center). J.C., D.X.N. and Q.Y. designed the research. J.C. performed most of the experiments. J.C., Z. L. and D.X.N. performed the bioinformatic analysis. J.C., W.K.C.C., and M.Z. performed the in vivo imaging experiments. C.J.B. performed the histological analysis. S.Y.Z. performed some cell culture work. S.W.C. provided VSVG-RBP2 constructs. J.C., Z.L., C.J.B., D.X.N. and Q.Y. analyzed the data. J.C., D.X.N., and Q.Y. wrote the paper.
Footnotes
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer research. 1992;52:1399–1405. [PubMed] [Google Scholar]
- Benevolenskaya EV, Murray HL, Branton P, Young RA, Kaelin WG., Jr Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Molecular cell. 2005;18:623–635. doi: 10.1016/j.molcel.2005.05.012. [DOI] [PubMed] [Google Scholar]
- Beshiri ML, Holmes KB, Richter WF, Hess S, Islam AB, Yan Q, Plante L, Litovchick L, Gevry N, Lopez-Bigas N, et al. Coordinated repression of cell cycle genes by KDM5A and E2F4 during differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:18499–18504. doi: 10.1073/pnas.1216724109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair LP, Cao J, Zou MR, Sayegh J, Yan Q. Epigenetic Regulation by Lysine Demethylase 5 (KDM5) Enzymes in Cancer. Cancers. 2011;3:1383–1404. doi: 10.3390/cancers3011383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair LP, Yan Q. Epigenetic mechanisms in commonly occurring cancers. DNA and Cell Biology. 2012;31(Suppl 1):S49–61. doi: 10.1089/dna.2012.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA, Massague J. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabletz T. EMT and MET in metastasis: where are the cancer stem cells? Cancer cell. 2012;22:699–701. doi: 10.1016/j.ccr.2012.11.009. [DOI] [PubMed] [Google Scholar]
- Cao J, Yan Q. Histone Demethylases Set the Stage for Cancer Metastasis. Science Signaling. 2013;6:pe15. doi: 10.1126/scisignal.2004188. [DOI] [PubMed] [Google Scholar]
- Carey L, Winer E, Viale G, Cameron D, Gianni L. Triple-negative breast cancer: disease entity or title of convenience? Nature reviews Clinical oncology. 2010;7:683–692. doi: 10.1038/nrclinonc.2010.154. [DOI] [PubMed] [Google Scholar]
- Chan SW, Hong W. Retinoblastoma-binding protein 2 (Rbp2) potentiates nuclear hormone receptor-mediated transcription. J Biol Chem. 2001;276:28402–28412. doi: 10.1074/jbc.M100313200. [DOI] [PubMed] [Google Scholar]
- Cheung William KC, Zhao M, Liu Z, Stevens Laura E, Cao Paul D, Fang Justin E, Westbrook Thomas F, Nguyen Don X. Control of Alveolar Differentiation by the Lineage Transcription Factors GATA6 and HOPX Inhibits Lung Adenocarcinoma Metastasis. Cancer cell. 2013;23:725–738. doi: 10.1016/j.ccr.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J, Agger K, Cloos PAC, Pasini D, Rose S, Sennels L, Rappsilber J, Hansen KH, Salcini AE, Helin K. RBP2 belongs to a family of demethylases, specific for tri- and dimethylated lysine 4 on histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Defeo-Jones D, Huang PS, Jones RE, Haskell KM, Vuocolo GA, Hanobik MG, Huber HE, Oliff A. Cloning of cDNAs for cellular proteins that bind to the retinoblastoma gene product. Nature. 1991;352:251–254. doi: 10.1038/352251a0. [DOI] [PubMed] [Google Scholar]
- Desantis C, Siegel R, Bandi P, Jemal A. Breast cancer statistics, 2011. CA: a cancer journal for clinicians. 2011;61:409–418. doi: 10.3322/caac.20134. [DOI] [PubMed] [Google Scholar]
- Guy CT, Webster MA, Schaller M, Parsons TJ, Cardiff RD, Muller WJ. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:10578–10582. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast cancer research and treatment. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- Hancox RA, Allen MD, Holliday DL, Edwards DR, Pennington CJ, Guttery DS, Shaw JA, Walker RA, Pringle JH, Jones JL. Tumour-associated tenascin-C isoforms promote breast cancer cell invasion and growth by matrix metalloproteinase-dependent and independent mechanisms. Breast Cancer Res. 2009;11:R24. doi: 10.1186/bcr2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J, Wu J, Dombkowski A, Zhang K, Holowatyj A, Boerner JL, Yang ZQ. Genomic amplification and a role in drug-resistance for the KDM5A histone demethylase in breast cancer. Am J Transl Res. 2012;4:247–256. [PMC free article] [PubMed] [Google Scholar]
- Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J. A multigenic program mediating breast cancer metastasis to bone. Cancer cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- Kim YW, Otterson GA, Kratzke RA, Coxon AB, Kaye FJ. Differential specificity for binding of retinoblastoma binding protein 2 to RB, p107, and TATA-binding protein. Mol Cell Biol. 1994;14:7256–7264. doi: 10.1128/mcb.14.11.7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Yan Q, Tothova Z, Yamane K, Erdjument-Bromage H, Tempst P, Gilliland DG, Zhang Y, Kaelin WG., Jr The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell. 2007;128:889–900. doi: 10.1016/j.cell.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Lee MG, Norman J, Shilatifard A, Shiekhattar R. Physical and functional association of a trimethyl H3K4 demethylase and Ring6a/MBLR, a polycomb-like protein. Cell. 2007;128:877–887. doi: 10.1016/j.cell.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Li L, Greer C, Eisenman RN, Secombe J. Essential functions of the histone demethylase lid. PLoS Genetics. 2010;6:e1001221. doi: 10.1371/journal.pgen.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Cao J, Liu J, Beshiri ML, Fujiwara Y, Francis J, Cherniack AD, Geisen C, Blair LP, Zou MR, et al. Loss of the retinoblastoma binding protein 2 (RBP2) histone demethylase suppresses tumorigenesis in mice lacking Rb1 or Men1. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:13379–13386. doi: 10.1073/pnas.1110104108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Mu E, Wei Y, Riethdorf S, Yang Q, Yuan M, Yan J, Hua Y, Tiede BJ, Lu X, et al. VCAM-1 promotes osteolytic expansion of indolent bone micrometastasis of breast cancer by engaging alpha4beta1-positive osteoclast progenitors. Cancer cell. 2011;20:701–714. doi: 10.1016/j.ccr.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo WB, Chang R, Zhong J, Pandey A, Semenza GL. Histone demethylase JMJD2C is a coactivator for hypoxia-inducible factor 1 that is required for breast cancer progression. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E3367–E3376. doi: 10.1073/pnas.1217394109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Zaslavsky A, Fedele G, McLaughlin SK, Reczek EE, De Raedt T, Guney I, Strochlic DE, MacConaill LE, Beroukhim R, et al. An oncogene-tumor suppressor cascade drives metastatic prostate cancer by coordinately activating Ras and nuclear factor-[kappa]B. Nature Medicine. 2010;16:286–294. doi: 10.1038/nm.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Padua D, Bos P, Nguyen DX, Nuyten D, Kreike B, Zhang Y, Wang Y, Ishwaran H, et al. Lung metastasis genes couple breast tumor size and metastatic spread. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:6740–6745. doi: 10.1073/pnas.0701138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore HM, Gonzalez ME, Toy KA, Cimino-Mathews A, Argani P, Kleer CG. EZH2 inhibition decreases p38 signaling and suppresses breast cancer motility and metastasis. Breast cancer research and treatment. 2013;138:741–752. doi: 10.1007/s10549-013-2498-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaharu K, Zhang X, Yoshida T, Katoh D, Hanamura N, Kozuka Y, Ogawa T, Shiraishi T, Imanaka-Yoshida K. Tenascin C induces epithelial-mesenchymal transition-like change accompanied by SRC activation and focal adhesion kinase phosphorylation in human breast cancer cells. Am J Pathol. 2011;178:754–763. doi: 10.1016/j.ajpath.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DX, Bos PD, Massague J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 2009a;9:274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- Nguyen DX, Chiang AC, Zhang XHF, Kim JY, Kris MG, Ladanyi M, Gerald WL, Massagué J. WNT/TCF Signaling through LEF1 and HOXB9 Mediates Lung Adenocarcinoma Metastasis. Cell. 2009b;138:51–62. doi: 10.1016/j.cell.2009.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DX, Massague J. Genetic determinants of cancer metastasis. Nat Rev Genet. 2007;8:341–352. doi: 10.1038/nrg2101. [DOI] [PubMed] [Google Scholar]
- Oskarsson T, Acharyya S, Zhang XH, Vanharanta S, Tavazoie SF, Morris PG, Downey RJ, Manova-Todorova K, Brogi E, Massague J. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nature Medicine. 2011;17:867–874. doi: 10.1038/nm.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolicchi E, Crea F, Farrar WL, Green JE, Danesi R. Histone lysine demethylases in breast cancer. Crit Rev Oncol Hematol. 2013;86:97–103. doi: 10.1016/j.critrevonc.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, Garcia-Santos G, Ghajar C, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringner M, Fredlund E, Hakkinen J, Borg A, Staaf J. GOBO: gene expression-based outcome for breast cancer online. PLoS One. 2011;6:e17911. doi: 10.1371/journal.pone.0017911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nature Medicine. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- Sayegh J, Cao J, Zou MR, Morales A, Blair LP, Norcia M, Hoyer D, Tackett AJ, Merkel JS, Yan Q. Identification of Small Molecule Inhibitors of Jumonji AT-Rich Interactive Domain 1B (JARID1B) Histone Demethylase by a Sensitive High-throughput Screen. J Biol Chem. 2013;288:9408–9417. doi: 10.1074/jbc.M112.419861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secombe J, Li L, Carlos L, Eisenman RN. The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes Dev. 2007;21:537–551. doi: 10.1101/gad.1523007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi N, Kang Y. Unravelling the complexity of metastasis - molecular understanding and targeted therapies. Nat Rev Cancer. 2011;11:735–748. doi: 10.1038/nrc3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Mei P, Fang R, Leonor T, Rutenberg M, Shimizu F, Li J, Rao A, Shi Y. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature. 2007;447:601–605. doi: 10.1038/nature05823. [DOI] [PubMed] [Google Scholar]
- Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng YC, Lee CF, Li YS, Chen YR, Hsiao PW, Chan MY, Lin FM, Huang HD, Chen YT, Jeng YM, et al. Histone demethylase RBP2 promotes lung tumorigenesis and cancer metastasis. Cancer research. 2013;73:4711–4721. doi: 10.1158/0008-5472.CAN-12-3165. [DOI] [PubMed] [Google Scholar]
- Tiwari N, Tiwari VK, Waldmeier L, Balwierz PJ, Arnold P, Pachkov M, Meyer-Schaller N, Schubeler D, van Nimwegen E, Christofori G. Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer cell. 2013;23:768–783. doi: 10.1016/j.ccr.2013.04.020. [DOI] [PubMed] [Google Scholar]
- Tu S, Teng YC, Yuan C, Wu YT, Chan MY, Cheng AN, Lin PH, Juan LJ, Tsai MD. The ARID domain of the H3K4 demethylase RBP2 binds to a DNA CCGCCC motif. Nat Struct Mol Biol. 2008;15:419–421. doi: 10.1038/nsmb.1400. [DOI] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt R, Otte AP, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Wang GG, Song J, Wang Z, Dormann HL, Casadio F, Li H, Luo JL, Patel DJ, Allis CD. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009a;459:847–851. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang H, Chen YP, Sun YM, Yang F, Yu WH, Liang J, Sun LY, Yang XH, Shi L, et al. LSD1 Is a Subunit of the NuRD Complex and Targets the Metastasis Programs in Breast Cancer. Cell. 2009b;138:660–672. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H, Taylor-Papadimitriou J, Tempst P, Zhang Y. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Molecular cell. 2007;25:801–812. doi: 10.1016/j.molcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Yang H, Minamishima YA, Yan Q, Schlisio S, Ebert BL, Zhang X, Zhang L, Kim WY, Olumi AF, Kaelin WG., Jr pVHL acts as an adaptor to promote the inhibitory phosphorylation of the NF-kappaB agonist Card9 by CK2. Molecular cell. 2007;28:15–27. doi: 10.1016/j.molcel.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng JP, Ge Z, Wang LX, Li Q, Wang N, Bjorkholm M, Jia JH, Xu DW. The Histone Demethylase RBP2 Is Overexpressed in Gastric Cancer and Its Inhibition Triggers Senescence of Cancer Cells. Gastroenterology. 2010;138:981–992. doi: 10.1053/j.gastro.2009.10.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.