Abstract
The microdosimetric-kinetic (MK) model is one of the models that can describe the fraction of cells surviving after exposure to ionizing radiation. In the MK model, there are specific parameters, k and yD, where k is an inherent parameter to represent the number of potentially lethal lesions (PLLs) and yD indicates the dose-mean lineal energy in keV/μm. Assuming the PLLs to be DNA double-strand breaks (DSBs), the rate equations are derived for evaluating the DSB number in the cell nucleus. In this study, we estimated the ratio of DSBs for two types of photon irradiation (6 MV and 200 kVp X-rays) in Chinese hamster ovary (CHO-K1) cells and human non-small cell lung cancer (H1299) cells by observing the surviving fraction. The estimated ratio was then compared with the ratio of γ-H2AX foci using immunofluorescent staining. For making a comparison of the number of DSBs among a variety of radiation energy cases, we next utilized the survival data in the literature for both cells exposed to other photon types, such as 60Co γ-rays, 137Cs γ-rays and 100 kVp X-rays. The ratio of DSBs based on the MK model with conventional data was consistent with the ratio of γ-H2AX foci numbers, confirming that the γ-H2AX focus is indicative of DSBs. It was also shown that the larger yD is, the larger the DSB number is. These results suggest that k and yD represent the characteristics of the surviving fraction and the biological effects for photon irradiation.
Keywords: microdosimetric-kinetic model, γ-H2AX foci, double-strand breaks, RBE37
INTRODUCTION
Photon beams are in widespread use in radiotherapy for eradicating cancers. The biological effect of the photon beams is currently standardized as the relative biological effectiveness (RBE). The RBE has been conventionally regarded as unity in any energy range, but it is also known that the effects of radiation depend on the absorbed dose [Gy], radiation type and dose rate [1–3]. As for the cellular-level response, we are able to evaluate the effects of radiation via cell surviving fraction using the method of colony formation assay reported by several investigators [4, 5]. The primary target for causing cell damage by radiation is the cell nucleus, particularly the DNA [6, 7]. DNA lesions by irradiation may lead to cell death. These lesions are classified into two types: single-strand breaks (SSBs), and double-strand breaks (DSBs). It is known that most SSBs are repairable in a relatively short period of time and do not lead to cell death directly, while DSBs promote lethal damage effectively.
For describing cell survival after irradiation, several models and formulae have been reported [8–11]. Among them, the linear–quadratic (LQ) model is currently the most widely used in radiotherapy. In the LQ model, the surviving fraction is given by S = exp(−αD − βD2), where α and β are the proportionality factors to the absorbed dose (D) [Gy−1] and the dose squared (D2) [Gy−2], respectively. In many cases, the LQ curve fits fairly well to the survival data. However, the implications involved in the terms are not necessarily trivial. The microdosimetric-kinetic (MK) model (presented by R.B. Hawkins) can evaluate the surviving fraction in terms of microdosimetry [12–14]. This model takes into account the spatial distribution of the energy deposition of radiation [which includes the dose mean lineal energy, yD (in keV/μm)] for expressing the difference of radiation energy [15]. The yD value can be measured using the tissue-equivalent proportional counter (TEPC) and calculated by Monte Carlo simulation [16–18]. Another advantage of the MK model is that it contains the inherent parameter (k) that implies the number of potentially lethal lesions (PLLs) per cell nucleus per Gy. k is defined as the characteristic parameter that depends on radiation energy [19]. These parameters (k, yD) provide an explanation of the curving nature in the LQ model and an estimation of damage degree, e.g. the number of DSBs.
From an experimental point of view, we can observe the DSBs visually with immunofluorescent staining using the γ-H2AX antibody, 53BP1 or MDC1 [20, 21]. There have been several investigations concerning the γ-H2AX foci, and the number of γ-H2AX foci has been observed with time-lapse images [22]. Beyreuther et al. suggest that the number of γ-H2AX foci peaks at 30 min after irradiation, and the number of DSBs is larger for lower photon beam energy [23]. As described above, we have quantified the number of foci from a theoretical point of view, where the relation between the foci number obtained from experiment and the k-value in the MK model is illustrated.
The effects on living cells (e.g. surviving fraction and the number of DSBs) depend not only on the type of radiation but also on the energy. For example, it is well known that low-energy electrons such as δ-rays have a larger impact than high-energy electrons [24]. However, the different effects by the same type of radiation with different energies have rarely been discussed in the literature. Although the RBE and radiation weighting factor (WR) are currently regarded as unity for electrons and photons, this energy dependence must be non-negligible for estimating the number of DSBs and the surviving fraction in the same cell line.
In this study, we take the parameter (k) in the MK model and establish a formulation for estimating the number of DSBs by deriving the rate equations for lesions after irradiation. For two types of cell lines, Chinese hamster ovary (CHO-K1) and human non-small cell lung cancer (H1299) cells, we attempt to evaluate the parameter (k) in the MK model by comparing the relative number of DSBs estimated by the β value in the LQ formula with the number of γ-H2AX foci. In parallel, we investigate the relationship between yD and k estimated using the equation derived in the model. We sought to confirm that photon beams with lower energies induce more intensive damage in the cells. We also evaluate the validity of the equation for estimate the number of DSBs.
MATERIALS AND METHODS
Surviving fraction and yD value in the MK model
The MK model was proposed by R.B. Hawkins in 1994 [12]. In this model, each cell nucleus is divided into several domains. The lesions arising from irradiation in a domain are called potentially lethal lesions (PLLs). A PLL can undergo one of four transformations: (i) it may be converted to a lethal and non-repairable lesion via a first-order process (rate constant for transformation is a [h−1]); (ii) it may be converted to a lethal and non-repairable lesion via a second-order process (rate constant for transformation is bd [h−2]); (iii) it may be repaired via a first-order process (rate constant for transformation is c [h−1]); (iv) it may maintain the lesion state for a period of time tr [h], after which time, if it is still present, it becomes lethal and non-repairable [12]. In the MK model, it is assumed that the PLLs are DSBs in DNA. Using the rate constants (a, bd and c) for the transformations above, a rate equation of the number of PLLs per domain for a single-dose irradiation is expressed as:
 |
(1) |
By considering the case of (a + c)P > >2bdP2,
 |
(2) |
and
 |
(3) |
Here, P is the number of PLLs in a domain; kd is the number of PLLs produced on average per dose [Gy−1] just after the irradiation; z is specific energy deposited in the domain [Gy]; t is time after the irradiation to the cell nucleus [h] and satisfies 0 < t < tr. The rate equation of the lethal lesions per domain (L) is expressed by:
 |
(4) |
By solving Eq. (4) after the substitution of Eq. (3) in the terms in the right hand side, we have
 |
(5) |
where
 |
(6) |
and
 |
(7) |
The average number of lethal lesions Ln per cell nucleus and the surviving fraction (S) for the single-dose irradiation can be described by using the expected value (with brackets), assuming the Poisson distribution of the number of lethal lesions in a cell nucleus, as follows:
 |
(8) |
where
 |
(9) |
 |
(10) |
 |
(11) |
 |
(12) |
and N is the number of domains in a cell nucleus;  is the average number of lethal lesions per domain; rd and ρ represent the radius of the domain (= 0.5 µm) and the density of the domain (= 1.0 g/cm3), respectively; D is the absorbed dose in Gy; k is the number of PLLs per cell nucleus; α and β represent LQ (Linear–Quadratic) parameters that are obtained by fitting to the cell surviving fraction data; the yD-value is the dose mean lineal energy (keV/μm). The yD-value is given by
is the average number of lethal lesions per domain; rd and ρ represent the radius of the domain (= 0.5 µm) and the density of the domain (= 1.0 g/cm3), respectively; D is the absorbed dose in Gy; k is the number of PLLs per cell nucleus; α and β represent LQ (Linear–Quadratic) parameters that are obtained by fitting to the cell surviving fraction data; the yD-value is the dose mean lineal energy (keV/μm). The yD-value is given by
 |
(13) |
and
 |
(14) |
where ϵ is the energy deposited in a domain, <l> is the mean chord length expressed as two-thirds times the domain diameter (i.e. 2/3 × 1 µm water-equivalent), y is the lineal energy, f(y) is the probability density of the lineal energy, d(y) is the dose distribution of the lineal energy. Note that Eq. (8) for the surviving fraction in the MK model includes the dose mean lineal energy yD in keV/μm.
Principle for estimating the number of DSBs according to the MK model
Once it is assumed that the PLLs correspond to DSBs in DNA, the number of DSBs per cell nucleus (k) can be obtained by fitting Eq. (8) to the experimental surviving fraction. By substituting Eq. (7) into Eq. (10), β is derived as follows:
 |
(15) |
where b (= bd/N) is the rate constant for transformation to a lethal and non-repairable lesion via a second-order process [h−1]. If we assume that (a + c)tr is > 3 following Hawkins [13], Eq. (15) can be approximated as:
 |
(16) |
Since (a + c) and b are regarded as specific parameters to a certain type of cells, the ratio of β to k2 would be constant as:
 |
Accordingly, if the parameters β and k are given for radiation with a certain energy (e.g. γ-ray), then the parameters β' and k' for radiation with some other energy can be related as:
 |
(17) |
From this relation, we can estimate the relative number of DSBs (k'/k) by the determination of β from the cell survival curves.
Cell line, culture condition and irradiation
The CHO-K1 and H1299 cell lines were obtained from RIKEN Bio Resource Center in Japan and ATCC, respectively. Two cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM, Sigma, St Louis, MO, USA) supplemented with 10% fetal bovine serum (FBS, Nichirei Biosciences Inc., Tokyo, Japan) at 37°C in a humidified 95% air, 5% CO2 incubator. Both cells were kept within vent cap flasks whose area was 25 cm2 (IWAKI, Tokyo, Japan), and were grown to semi-confluence. Each type of cell was irradiated with 200 kVp X-rays (Siemens, Concord, CA) at a dose rate of 1.25 Gy/min, or by 6 MV therapeutic X-rays (Mitsubishi Electric Co., Tokyo, Japan) at a dose rate of 2.5 Gy/min. In the case of kV photon irradiation, we measured the air dose rate at the surface of the cell culture using an ionization chamber (NE2571). In the case of MV photon irradiation, the dish containing the cells was placed on a water-equivalent phantom with a 50 mm-thickness at the side of the gantry head in order to compensate for the build-up effect. The absorbed dose in water (D) was determined according to the dose protocol of TRS 277 [25] (in air method) for 200 kVp X-rays and Japanese standard dosimetry 01 [26] for Linac-6 MV X-rays, respectively. Each irradiation was performed at room temperature.
Clonogenic assay
The cells were grown to semi-confluence, irradiated, trypsinized and counted. 1 × 103, 1 × 104 or 1 × 105 cells were reseeded in plastic dishes. The cells had been cultured in a CO2 incubator for 10–14 d. Then, the DMEM was replaced every 2 d. After the culture period, the cells were fixed with methanol and stained with 2% Giemsa solution (Kanto Chemical Co. Inc., Tokyo, Japan) to determine the number of colonies per dish. The colonies were counted and survival rates were calculated with the plating efficiency of the non-irradiated cells.
Estimation of the DNA damage based on the MK model
To estimate the number of PLLs in a cell nucleus after irradiation of 1.0 Gy, we took the cell survival data from the literature. For CHO-K1 cells, Zellmer et al. (1998) have reported the surviving fraction with 200 kVp X-rays [27]. Using their data and our experimental results for irradiation with 6 MV X-rays for CHO-K1 cells, we deduced the parameters α and β by fitting the LQ formula to the data via the least squares method and confirmed the difference between the effects of two types of photon irradiation. The ratio of parameter (k) for 6 MV X-rays and for 200 kVp X-rays was then obtained. In the same manner, we obtained the ratio of k for H1299 cells using our survival data from the clonogenic assay.
Immunofluorescence staining with γ-H2AX
Two types of cell lines, CHO-K1 and H1299 cells, were grown on the ϕ 35 mm glass cover slips covered with collagen, irradiated with 6 MV and 200 kVp X-rays at 1.0 Gy. At 30 min after the irradiation, the cells on the coverslips were fixed in a 4% paraformaldehyde solution with PBS for 10 min at room temperature and then rinsed three times with PBS. Next, the cells were permeabilized in ice-cold 0.2% Triton X-100 in PBS for 5 min, and blocked with a solution of 1% BSA-containing PBS for 30 min. After that, a primary antibody γ-H2AX (diluted by a solution of 1% BSA-containing PBS) was fed into the glass coverslips, and kept overnight at 4°C. The next day, the primary antibody was removed and rinsed three times with a solution of 1% BSA-containing PBS. Then Alexa Fluor 594-conjugated goat–anti–rabbit (Molecular Probes, Invitrogen, Japan), diluted by a solution of 1% BSA-containing PBS, was put onto coverslips and left for 2 h, then rinsed one time with 1 µg/ml DAPI (4′,6-diamidino-2-phenylindole phenylindole)-containing methanol. The cells on the coverslips were stained with 1 µg/ml DAPI-containing methanol for 15 min in the incubator, and rinsed one time with methanol.
The γ-H2AX foci in the cell nuclei were observed using a High Standard all-in-one fluorescent microscope (model BZ-9000; Keyence, Osaka, Japan). The images of γ-H2AX foci were taken using the Z-stack function, and reconstructed by quick full focus with 3D in the cell nucleus. The number of the γ-H2AX foci was counted using VH-H1A5 software (Keyence, Tokyo, Japan), and presumably corresponds to the number of DSBs. This procedure was conducted for each condition (control and 1.0 Gy in each cell line, for more than 80 cells of CHO-K1 or H1299). In the measurement of the foci, binarized processing was adopted, setting the threshold level of fluorescence intensity corresponding to the visible level.
Comparison between k ratio in the MK model and ratio of γ-H2AX foci number
The γ-H2AX foci number per cell nucleus per Gy obtained in the previous subsection was defined as experimental k, written as kexp in this paper. Using the results of kexp for both cell lines, the ratio of kexp for 6 MV X-rays and for 200 kVp X-rays was calculated. The ratio of kexp was then compared with the ratio of k estimated based on the MK model (Eq. (17)). From the result of ratios, we evaluated the parameter (k) in the MK model.
Analysis of the relationship between the yD-value and the number of DSBs
In order to confirm the dependence of cell survival on photon energy, we collected the cell survival data previously reported in the literature (listed in Table 1) for CHO-K1 cells exposed to 100 kVp X-rays [28], 60Co γ-rays [29–32] and 137Cs γ-rays [27, 33–39], and for H1299 cells exposed to 60Co γ-rays [40] and 137Cs γ-rays [41–43]. Here, the parameters α and β were determined in the same manner as mentioned above. The kexp after irradiation with 200 kVp X-rays was adopted as the standard radiation; then by using Eq. (17), we estimated each k exposed to each photon type (6 MV X-rays, 137Cs γ-rays, 200 kVp X-rays and 100 kVp X-rays). In addition, the RBE value was calculated at the 37% survival level, which is related with the number of DSBs.
Table 1.
A list of references for survival data
| Cell line | Radiation Type | Reference [27–43] |
|---|---|---|
| CHO-K1 | 60Co γ-rays | Mothersill C et al., 1986 [29] |
| Czub J et al., 2008 [30] | ||
| Murakami D et al., 2004 [31] | ||
| Thacker J et al., 1985 [32] | ||
| Linac-6 MV X-rays | present (clonogenic assay) | |
| 137Cs γ-rays | Schneiderman MH et al., 2001 [33] | |
| Schwartz JL et al., 2000 [35] | ||
| Van Putten JWG et al., 2001 [34] | ||
| Woudstra EC et al., 1999 [36] | ||
| Stevens CW et al., 1999 [37] | ||
| Murray D et al., 2000 [38] | ||
| Britten RA et al., 1997 [39] | ||
| 200 kVp X-rays | Zellmer, DL et al., 1998 [27] | |
| 100 kVp X-rays | Wedemeyer N et al., 2001 [28] | |
| H1299 | 60Co γ-rays | Iwasa T et al., 2009 [40] |
| Linac-6 MV X-rays | present (clonogenic assay) | |
| 137Cs γ-rays | Liu S-K et al., 2008 [41] | |
| Chen M-F et al., 2006 [42] | ||
| Shin S-H et al., 2007 [43] | ||
| 200 kVp X-rays | present (clonogenic assay) |
RESULTS
Parameters in the MK model using the surviving fraction
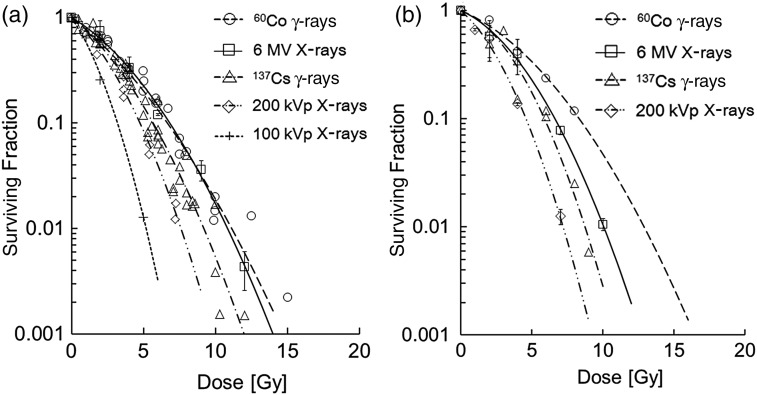
Figure 1a and b shows the cell surviving fractions for CHO-K1 cells and for H1299 cells, respectively, after irradiation with a variety of photon beams. The survival curves in the figures fit well to the experimental data listed in Table 1. The curves exhibit a general tendency that the surviving fraction of cells exposed to lower energy photons was lower than that of cells exposed to higher energy photons, even at the same absorbed dose, i.e., the surviving fraction for 200 kVp X-rays was less than that for 6 MV X-rays. Table 2 presents a list of the parameters in the MK model, which was obtained by fitting Eq. (8) to the surviving fraction. Here, the α and β values were determined as two independent parameters. It should be noted that the β value decreases with photon energy.
Fig. 1.
Surviving fractions as a function of dose after the irradiation of several types of photon beams: (a) for CHO-K1 cells, and (b) for H1299 cells. The lines fitted to the symbol plots were determined based on the MK model. The experimental data were taken from the literature [27–43] listed in Table 1.
Table 2.
MK parameters (α, β, α0, yD, k), experimental k (obtained by counting the number of γ-H2AX foci) and RBE37 for CHO-K1 and H1299 cell lines
| Cell line | Radiation Type | a | α0 | β | yD [keV/ μm] | k | kexp | RBE37 |
|---|---|---|---|---|---|---|---|---|
| CHO-K1 | 60Co γ-rays | 0.246 ± 0.023 | 0.238 ± 0.024 | 0.0152 ± 0.0023 | 2.34 | 30.4 ± 9.3 | 30a | 0.757 |
| Linac-6 MV X-rays | 0.190 ± 0.034 | 0.179 ± 0.035 | 0.0218 ± 0.0033 | 2.36 | 36.5 ± 11.2 | 39.4 ± 13.1 | 0.689 | |
| 137Cs γ-rays | 0.255 ± 0.029 | 0.239 ± 0.032 | 0.0270 ± 0.0036 | 2.90 | 40.6 ± 12.3 | 0.855 | ||
| 200 kVp X-rays | 0.284 ± 0.051 | 0.245 ± 0.070 | 0.0425 ± 0.0083 | 4.51 | 50.9 ± 14.1 | 50.9 ± 14.1 | 1.000 | |
| 100 kVp X-rays | 0.456 ± 0.082 | 0.378 ± 0.135 | 0.0833 ± 0.0179 | 4.70 | 71.3 ± 22.7 | 1.522 | ||
| H1299 | 60Co γ-rays | 0.130 ± 0.033 | 0.121 ± 0.036 | 0.0176 ± 0.0048 | 2.34 | 33.6 ± 9.2 | 0.539 | |
| Linac-6 MV X-rays | 0.137 ± 0.036 | 0.121 ± 0.038 | 0.0319 ± 0.0041 | 2.36 | 45.3 ± 10.8 | 37.9 ± 13.7 | 0.659 | |
| 137Cs γ-rays | 0.113 ± 0.074 | 0.085 ± 0.083 | 0.0474 ± 0.0103 | 2.90 | 55.2 ± 14.2 | 0.713 | ||
| 200 kVp X-rays | 0.277 ± 0.048 | 0.231 ± 0.066 | 0.0526 ± 0.0078 | 4.51 | 58.1 ± 12.5 | 58.1 ± 12.5 | 1.000 |
By using the β values with 200 kVp X-rays and 6 MV X-rays in Table 2, we estimated the ratio of DSBs using Eq. (17). In CHO-K1 cells, the ratio of k for irradiation by 6 MV X-rays to that for irradiation by 200 kVp X-rays was estimated to be 0.716 ± 0.088, while in H1299 cells it was 0.779 ± 0.076. These ratios suggest that the number of DSBs resulting from 200 kVp X-rays is larger than that from 6 MV X-rays, independent of cell type.
The γ-H2AX foci observed in this study are shown in Fig. 2, where the blue area represents a cell nucleus stained with DAPI, and each red point represents a DSB. The number of γ-H2AX foci (kexp) was counted using VH-H1A5 software (Keyence, Tokyo, Japan), and is regarded as the number of DSBs. The γ-H2AX foci numbers per cell nucleus are shown in Fig. 3a for CHO-K1 cells and in Fig. 3b for H1299 cells, respectively. The average numbers of foci per nucleus before irradiation (background) for CHO-K1 and H1299 cells were 18.7 ± 7.69 and 16.5 ± 8.24, respectively. These background numbers of foci were at a comparable level for 6 MV and 200 kVp X-rays. After irradiation of 1.0 Gy, kexp for CHO-K1 cells irradiated with 200 kVp and 6 MV X-rays was 50.9 ± 14.1 and 39.4 ± 13.1, respectively; kexp for H1299 cells similarly irradiated was 58.1 ± 12.5 and 37.9 ± 13.7, respectively. The ratio of kexp for 6 MV X-rays to kexp for 200 kVp X-rays with CHO-K1 cells was 0.774 ± 0.335 and for H1299 cells it was 0.652 ± 0.274. The DSB ratio from γ-H2AX observation (kexp ratio) closes in on the value of the ratio of k estimated by the MK model, convincing us that the parameter (k) in the MK model corresponds to the number of DSBs per cell nucleus.
Fig. 2.
Images in the γ-H2AX antibody fluorescent observation for CHO-K1 and H1299 cells. The blue area represents a cell nucleus, and red points are the γ-H2AX foci indicating DNA double-strand breaks (DSBs).
Fig. 3.
Number of DSBs obtained by counting the γ-H2AX foci: (a) for CHO-K1, and (b) for H1299 cells. The asterisk represents a P < 0.05 significant difference.
Comparison of the parameters with other data, and evaluation of yD
Based on Eq. (17) and the results of γ-H2AX foci observation (kexp) for 200 kVp X-ray irradiation as the standard radiation (i.e. 50.9 ± 14.1 per nucleus per Gy for CHO-K1 cells in Fig. 3a, and 58.1 ± 12.5 per nucleus per Gy for H1299 cells in Fig. 3b), we estimated the numbers of DSBs per cell nucleus (k) for the other cases with 100 kVp X-rays, 137Cs γ-rays, 6 MV X-rays and 60Co γ-rays for CHO-K1 cells, and 6MV X-rays and 60Co γ-rays for H1299 cells, to be as shown in Table 2. In general, the k estimated by the MK model fits well the number of γ-H2AX foci (kexp). The nominal number of DSBs for Chinese hamster ovary cell lines was taken from the reference [12, 44], and we adopted 30 as the expected kexp with 60Co γ-rays for CHO-K1 cells. As is shown in Table 2, the number of DSBs (k) increases with the dose mean lineal energy yD [keV/μm], which suggests that k depends on the energy of photon beams. Table 2 also shows that the RBE37 is generally elevated along with the k value.
DISCUSSION
Parameter k in the MK model and the number of DSBs kexp
We have paid attention primarily to the parameter k in the MK model in this study. Since the PLLs are assumed to be DSBs in the MK model, k can be regarded as the number of DSBs per cell nucleus per Gy. As shown in Table 2, the k estimated in the MK model agreed well with the number of γ-H2AX foci (kexp), supporting this assumption. The k-value was estimated from β in this study. According to the methodology of the MK model, we can deduce a k-value from α0 as well. However, it is expected that the k-value from α0 will have an uncertainty larger than that from β (as shown in Table 2) because α0 is deduced from α and β, while β is directly determined by a curve fitting to survival data. Therefore, it might be reasonable to estimate the k-value from β via Eq. (17).
It is known that the stopping power of electrons has a peak in the low-energy region around 100 eV in liquid water [45], which brings about the peak energy deposition just before the electrons stop. In fact, there are many ionization and excitation events in the vicinity of the track end [46]. This concentration of energy deposition around the track end should have a larger influence on the probability of inducing lesions in a cell nucleus than the other parts of the track. The ratio of the energy depositions around the track end to the other track parts is larger for a short track with a low-energy electron than that for a long track with a high-energy electron. Therefore, even at the same absorbed dose, the damage probability caused by a low-energy electron produced through low-energy photon irradiation is larger than that caused by a high-energy electron produced through high-energy photon irradiation. Further to this, the estimated numbers of DSBs (k) in Table 2 evidently reflect the degree of damage inflicted by secondary electrons produced after several types of photon irradiation.
Evaluation of the effect by photon beams depending on their energies
We also investigated the relationships between the dose mean lineal energy yD [keV/μm], the RBE37 value and the number of DSBs per cell nucleus per Gy (k). As shown in Fig. 4, the number of DSBs per nucleus per Gy increases monotonically as the yD value increases. This result may have arisen from the characteristic that yD depends on the photon energy, as indicated in Table 2. The number of DSBs detected by the γ-H2AX antibody (kexp) shows a clear trend, i.e. that lower energy X-rays induce DSBs more than higher energy X-rays. This is attributable to the energy deposition pattern along the secondary-electron track, which concentrates on the track end, as mentioned in the previous subsection. Since yD represents the energy deposition at every site, including the end part, lower energy electrons arising from low-energy photon irradiations will increase the yD value in a cell nucleus. As we can see in Table 2, the magnitude relationship between the number of DSBs with 60Co γ-rays and that with 6-MV X-rays is opposite to the relationship between the mean energies of both photon beams (the mean energy of 60Co γ-rays is 1.25 MeV, and the mean energy of continuous 6-MV X-rays is roughly estimated to be 1.70 MeV). This result must be explainable on the energy spectra of electrons created by two types of photon beams. We supposed that yD reflects the biological effect along with the microscopic electron processes, which can be verified from the viewpoint of the dose mean lineal energy, yD in keV/μm, not from the mean energy of electrons. Table 2 shows that the value of k (supposedly the number of DSBs) lies in the same order of the value of yD, supporting this presumption.
Fig. 4.
Relationship between the dose mean lineal energy yD (keV/μm) and the number of DSBs (per cell nucleus per Gy): closed diamonds for CHO-K1 cells, and open circles for H1299 cells. The numbers of DSBs were estimated using Eq. (17).
In addition to yD, by replacing the surviving fraction with RBE37, the relationship between RBE37 and k [estimated number of DSBs per nucleus per Gy based on Eq. (17)] is given in Fig. 5 as well. The RBE37 value is likely to be proportional to the number of DSBs. Alternatively the RBE value can be obtained by calculating the relative number of DSBs (when we see the number of DSBs as an endpoint of cell damage) [47]. Since the parameter k is proportional to the number of foci, the ratio of k has to be proportional to the RBE. The proportional relation between k and RBE37 in Fig. 5 suggests the possibility of evaluating the effects on cells and DSB production from the viewpoint of biodosimetry [48, 49].
Fig. 5.
Relationship between RBE37 and the number of DSBs (k). The number of DSBs was estimated using Eq. (17) while the RBE37 value was deduced from the absorbed dose at 37% survival level using Eq. (8).
CONCLUSION
In this study, we evaluated the relationship between k (as the number of DSBs) and yD, and also k and RBE37. It was shown that k deduced using the MK model agrees well with the number of DSBs per cell nucleus that is observed by immunofluorescent staining using γ-H2AX antibody. Further, the surviving fraction at the same dose decreases as the yD value increases, while both the DSB number and the RBE37 value increase with yD. These results suggest that the effects of photons on cell survival and the DNA damage depend on the photon energy in terms of both microdosimetry and biodosimetry. The dependence on the photon energy is explainable from the energy deposition along the track of electrons (created by the photon) passing through the cell nucleus. The present study suggests that the RBE value and the radiation weighting factor (WR) for electrons and photons should not always be treated as unity. It was also demonstrated that a quantitative estimation of DNA damage can be made through the perspective of energy deposition along the electron track, which is promising for evaluating the damage effects on cells exposed to ionizing radiation.
FUNDING
Funding to pay the Open Access publication charges for this article was provided by H. Date (Hokkaido University).
REFERENCES
- 1.Bentzen SM. Dose–response relationships in radiotherapy. In: Michael J, van der Kogel AJ, editors. Basic Clinical Radiobiology. 4th ed. London: Hodder Arnold; 2009. pp. 56–66. [Google Scholar]
- 2.Begg AC, Wouters BG. Irradiation-induced damage and the DNA damage response. In: Michael J, van der Kogel AJ, editors. Basic Clinical Radiobiology. 4th edn. London: Hodder Arnold; 2009. pp. 11–26. [Google Scholar]
- 3.Michael J, van der Kogel AJ. The dose-rate effect. In: Michael J, van der Kogel AJ, editors. Basic Clinical Radiobiology. 4th edn. London: Hodder Arnold; 2009. pp. 158–68. [Google Scholar]
- 4.Puck TT, Marcus PI. Rapid method for viable cell titration and clone production with HeLa cells in tissue culture: the use of X-irradiated cells to supply conditioning factors. Proc Natl Acad Sci U S A. 1955;41:432–7. doi: 10.1073/pnas.41.7.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puck TT, Marcus PI. Action of x-rays on mammalian cells. J Exp Med. 1956;103:653–66. doi: 10.1084/jem.103.5.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munro TR. The relative radiosensitivity of the nucleus and cytoplasm of the Chinese hamster fibroblasts. Radiat Res. 1970;42:451–70. [PubMed] [Google Scholar]
- 7.Warters RL, Hofer KG. Radionuclide toxicity in cultured mammalian cells. Elucidation of the primary site for radiation-induced division delay. Radiat Res. 1977;69:348–58. [PubMed] [Google Scholar]
- 8.Curtis SB. Lethal and potentially lethal lesions induced by radiation—a unified repair model. Radiat Res. 1986;106:252–70. [PubMed] [Google Scholar]
- 9.Tobias C. The repair–misrepair model in radiobiology: comparison to other models. Radiat Res. 1985;8 S77–95. [PubMed] [Google Scholar]
- 10.Park C, Papiez L, Zhang S, et al. Universal survival curve and single fraction equivalent dose: useful tools in understanding potency of ablative radiotherapy. Int J Radiat Oncol Biol Phys. 2008;70(847–52) doi: 10.1016/j.ijrobp.2007.10.059. [DOI] [PubMed] [Google Scholar]
- 11.Sato T, Furusawa Y. Cell survival fraction estimation based on the probability densities of domain and cell nucleus specific energies using improved microdosimetric kinetic models. Radiat Res. 2012;178:341–56. doi: 10.1667/rr2842.1. [DOI] [PubMed] [Google Scholar]
- 12.Hawkins RB. A statistical theory of cell killing by radiation of varying linear energy transfer. Radiat Res. 1994;140:366–74. [PubMed] [Google Scholar]
- 13.Hawkins RB. A microdosimetric-kinetic model of cell death from exposure to ionizing radiation of any LET, with experimental and clinical applications. Radiat Biol. 1996;69:739–55. doi: 10.1080/095530096145481. [DOI] [PubMed] [Google Scholar]
- 14.Hawkins RB. The relationship between the sensitivity of cells to high-energy photons and the RBE of particle radiation used in radiotherapy. Radiat Res. 2009;172:761–76. doi: 10.1667/RR1655.1. [DOI] [PubMed] [Google Scholar]
- 15.Rossi HH, Zaider M, Turner JE. Microdosimetry and its Applications. Berlin: Springer; 1996. [Google Scholar]
- 16.Okamoto H, Kanai T, Kase Y, et al. Relation between lineal energy distribution and relative biological effectiveness for photon beams according to the microdosimetric kinetic model. J Radiat Res. 2011;52:75–81. doi: 10.1269/jrr.10073. [DOI] [PubMed] [Google Scholar]
- 17.McDonald JC, Ma IC, Zeitz L. Microdosimetric properties of encapsulated 125I and other photon sources. Radiat Res. 1979;77:221–32. [PubMed] [Google Scholar]
- 18.ICRU. Microdosimetry. Report 36. Bethesda: MD; 1983. International Commission on Radiation Units and Measurements. [Google Scholar]
- 19.Hawkins RB. Mammalian cell killing by ultrasoft X-rays and high-energy radiation: an extension of the MK model. Radiat Res. 2006;166:431–42. doi: 10.1667/RR3594.1. [DOI] [PubMed] [Google Scholar]
- 20.Franken N, Cate R, Przemek MK, et al. Comparison of RBE values of high-LET α-particles for the induction of DNA-DSBs, chromosome aberrations and cell reproductive death. Radiat Oncol. 2011;6:64. doi: 10.1186/1748-717X-6-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martia M, Ulich G, Frank L, et al. 53BP1 and MDC1 foci formation in HT-1080 cells for low- and high-LET microbeam irradiations. Radiat Environ Biophys. 2011;50:345–52. doi: 10.1007/s00411-011-0366-9. [DOI] [PubMed] [Google Scholar]
- 22.Gerelchuluun A, Hong Z, Sun L, et al. Induction of in situ DNA double-strand breaks and apoptosis by 200 MeV protons and 10 MV X-rays in human tumour cell lines. Int J Radiat Biol. 2011;87:57–70. doi: 10.3109/09553002.2010.518201. [DOI] [PubMed] [Google Scholar]
- 23.Beyreuther E, Lessmann E, Pawelke J, et al. DNA double-strand break signaling: X-ray energy dependence of residual co-localised foci of γ-H2AX and 53BP1. Int J Radiat Biol. 2009;85:1042–50. doi: 10.3109/09553000903232884. [DOI] [PubMed] [Google Scholar]
- 24.Nikjoo H, Lindborg L. RBE of low energy electrons and photons. Phys Med Biol. 2010;55 doi: 10.1088/0031-9155/55/10/R01. R65–109. [DOI] [PubMed] [Google Scholar]
- 25.IAEA. Absorbed dose determination in photon and electron beams (IAEA Technical Reports Series 277) Vienna, 1987. [Google Scholar]
- 26.JSMP. Tokyo: Tsusho Sangyo Kenkyu Sha; 2002. Standard dosimetry of absorbed dose in external beam radiotherapy (Standard Dosimetry 01) [Google Scholar]
- 27.Zellmer DL, Chapman JD, Stobbe CC. Radiation fields backscattered from material interfaces: I. Biological effectiveness. Radiat Res. 1998;150:406–15. [PubMed] [Google Scholar]
- 28.Wedemeyer N, Greve B, Uthe D, et al. Frequency of CD59 mutations induced in human-hamster hybrid AL cells by low-dose X-irradiation. Mutat Res. 2001;473:73–84. doi: 10.1016/s0027-5107(00)00137-8. [DOI] [PubMed] [Google Scholar]
- 29.Mothersill C, Seymour CB. Effect of lactate on the recovery of CHO-KI cells from gamma radiation damage. Acta Radiol Oncol. 1986;25:71–6. doi: 10.3109/02841868609136381. [DOI] [PubMed] [Google Scholar]
- 30.Czub J, Banaś D, Błaszczyk A, et al. Cell survival and chromosomal aberrations in CHO-K1cells irradiated by carbon ions. Int J Radiat Biol. 2008;84:821–9. [Google Scholar]
- 31.Murakami D, Suzuki MF, da Silva Dias M, et al. Genotoxic and cytotoxic effects of 60Co γ-rays and 90Sr/90Y β-rays on Chinese hamster ovary cells (CHO-K1) Radiat Environ Biophys. 2004;43:91–9. doi: 10.1007/s00411-004-0239-6. [DOI] [PubMed] [Google Scholar]
- 32.Thacker J, Stretch A. Responses of 4 X-ray-sensitive CHO cell mutants to different radiations and to irradiation conditions promoting cellular recovery. Mutat Res. 1985;146:99–108. doi: 10.1016/0167-8817(85)90060-4. [DOI] [PubMed] [Google Scholar]
- 33.Schneiderman MH, Hofer KG, Schneiderman GS. Targets for radiation-induced cell death: when dna damage doesn't kill. Radiat Res. 2001;155:529–35. doi: 10.1667/0033-7587(2001)155[0529:tfricd]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 34.Van Putten JWG, Groen HJM, Smid K, et al. End-joining deficiency and radiosensitization induced by gemcitabine end-joining deficiency and radiosensitization induced by gemcitabine. Cancer Res. 2001;61:1585–91. [PubMed] [Google Scholar]
- 35.Schwartz JL, Jordan R, Sun J, et al. Dose-dependent changes in the spectrum of mutations induced by ionizing radiation. Radiat Res. 2000;153:312–7. doi: 10.1667/0033-7587(2000)153[0312:ddcits]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 36.Woudstra EC, Konings AW, Jeggo PA, et al. Role of DNA-PK subunits in radiosensitization by hyperthermia. Radiat Res. 1999;152:214–8. [PubMed] [Google Scholar]
- 37.Stevens CW, Stamato TD, Mauldin SK, et al. Radiation-induced recombination is dependent on Ku80. Radiat Res. 1999;151:408–13. [PubMed] [Google Scholar]
- 38.Murray D, Rosenberg E, Allalunis-Turner MJ. Protection of human tumor cells of differing radiosensitivity by WR-1065. Radiat Res. 2000;154:159–62. doi: 10.1667/0033-7587(2000)154[0159:pohtco]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 39.Britten RA, Murray D. Constancy of the relative biological effectiveness of 42 MeV (p→Be+) neutrons among cell lines with different DNA repair proficiencies. Radiat Res. 1997;148:308–16. [PubMed] [Google Scholar]
- 40.Iwasa T, Okamoto I, Suzuki M. Inhibition of insulin-like growth factor 1 receptor by CP-751,871 radiosensitizes non-small cell lung cancer cells. Clin Cancer Res. 2009;15:5117–25. doi: 10.1158/1078-0432.CCR-09-0478. [DOI] [PubMed] [Google Scholar]
- 41.Liu S-K, Coackleyc C, Kraused M, et al. A novel poly (ADP-ribose) polymerase inhibitor, ABT-888, radiosensitizes malignant human cell lines under hypoxia. Radiother Oncol. 2008;88:258–68. doi: 10.1016/j.radonc.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 42.Chen M-F, Keng P-C. Inhibition of lung tumor growth and augmentation of radiosensitivity by decreasing peroxiredoxin I expression. Int J Radiat Oncol Biol Phys. 2006;64:581–91. doi: 10.1016/j.ijrobp.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 43.Shin S-H, Yoon M-J. Enhanced lung cancer cell killing by the combination of selenium and ionizing radiation. Oncol Rep. 2007;17:209–16. [PubMed] [Google Scholar]
- 44.Ager DD, Dewey WC, Gardiner K, et al. The measurement of radiation-induced DNA double-strand breaks by pulsed-field gel electrophoresis. Radiat Res. 1990;122:181–7. [PubMed] [Google Scholar]
- 45.ICRU. Bethesda: MD: International Commission on Radiation Units and Measurements; 1984. Stopping Powers for Electrons and Positrons. ICRU Report 37. [Google Scholar]
- 46.Date H, Sutherland KL, Hasegawa H, et al. Ionization and excitation collision processes of electrons in liquid water. Nucl Instrum Methods Phys Res B. 2007;265:515–20. [Google Scholar]
- 47.Bernal MA, Almeida CE, David M, et al. Estimation of the RBE of mammography-quality beams using a combination of a Monte Carlo code with a B-DNA geometrical model. Phys Med Biol. 2011;56:7393–403. doi: 10.1088/0031-9155/56/23/004. [DOI] [PubMed] [Google Scholar]
- 48.Redon CE, Nakamura AJ, Martin OA, et al. Recent developments in the use of γ-H2AX as a quantitative DNA double-strand break biomarker. Aging (Albany NY) 2011;3:168–74. doi: 10.18632/aging.100284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Redon CE, Nakamura AJ, Gouliaeva K, et al. The use of gamma-H2AX as a biodosimeter for total-body radiation exposure in non-human primates. PLoS One. 2010;5 doi: 10.1371/journal.pone.0015544. e15544. [DOI] [PMC free article] [PubMed] [Google Scholar]