Abstract
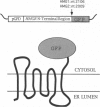
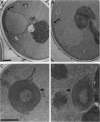
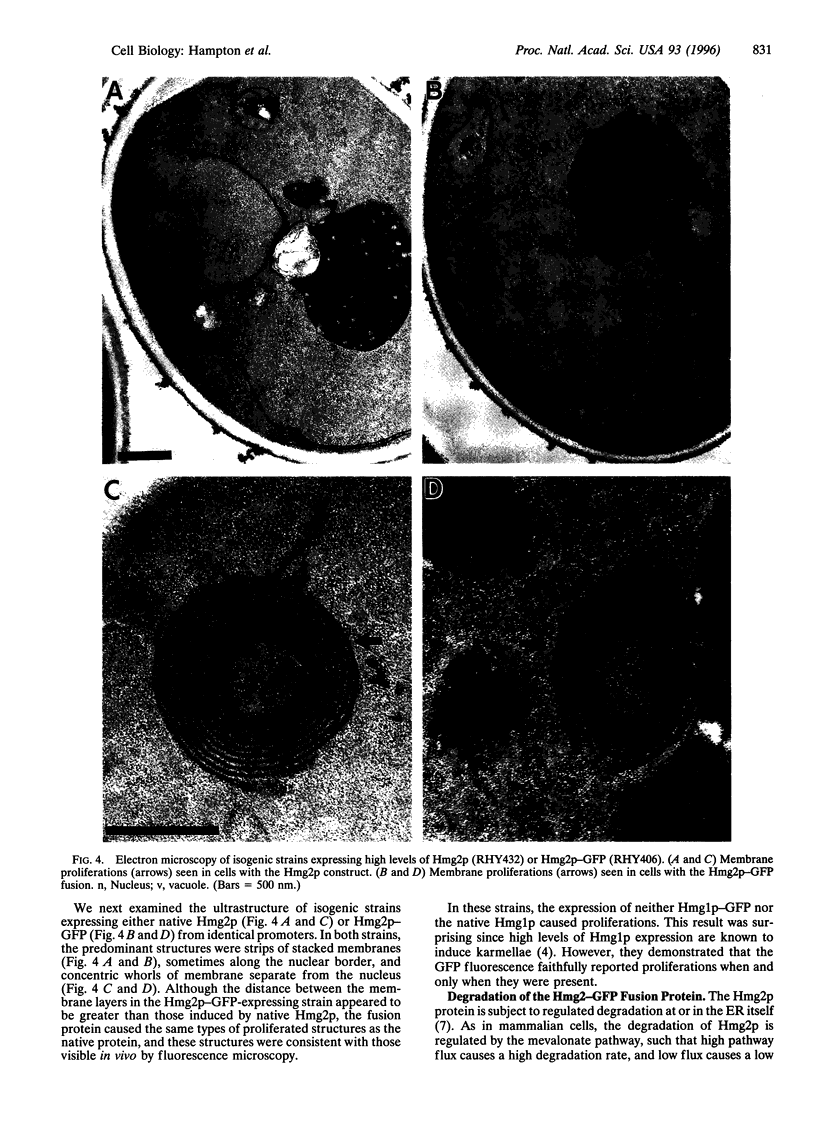
To test the utility of green fluorescent protein (GFP) as an in vivo reporter protein when fused to a membrane domain, we made a fusion protein between yeast hydroxymethylglutaryl-CoA reductase and GFP. Fusion proteins displayed spatial localization and regulated degradation consistent with the native hydroxymethylglutaryl-CoA reductase proteins. Thus, GFP should be useful in the study of both membrane protein localization and protein degradation in vivo.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chalfie M., Tu Y., Euskirchen G., Ward W. W., Prasher D. C. Green fluorescent protein as a marker for gene expression. Science. 1994 Feb 11;263(5148):802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Chun K. T., Bar-Nun S., Simoni R. D. The regulated degradation of 3-hydroxy-3-methylglutaryl-CoA reductase requires a short-lived protein and occurs in the endoplasmic reticulum. J Biol Chem. 1990 Dec 15;265(35):22004–22010. [PubMed] [Google Scholar]
- Cody C. W., Prasher D. C., Westler W. M., Prendergast F. G., Ward W. W. Chemical structure of the hexapeptide chromophore of the Aequorea green-fluorescent protein. Biochemistry. 1993 Feb 9;32(5):1212–1218. doi: 10.1021/bi00056a003. [DOI] [PubMed] [Google Scholar]
- Edwards P. A., Lan S. F., Fogelman A. M. Alterations in the rates of synthesis and degradation of rat liver 3-hydroxy-3-methylglutaryl coenzyme A reductase produced by cholestyramine and mevinolin. J Biol Chem. 1983 Sep 10;258(17):10219–10222. [PubMed] [Google Scholar]
- Hampton R. Y., Rine J. Regulated degradation of HMG-CoA reductase, an integral membrane protein of the endoplasmic reticulum, in yeast. J Cell Biol. 1994 Apr;125(2):299–312. doi: 10.1083/jcb.125.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning A. J., Lum P. Y., Williams J. M., Wright R. DiOC6 staining reveals organelle structure and dynamics in living yeast cells. Cell Motil Cytoskeleton. 1993;25(2):111–128. doi: 10.1002/cm.970250202. [DOI] [PubMed] [Google Scholar]
- Preuss D., Mulholland J., Kaiser C. A., Orlean P., Albright C., Rose M. D., Robbins P. W., Botstein D. Structure of the yeast endoplasmic reticulum: localization of ER proteins using immunofluorescence and immunoelectron microscopy. Yeast. 1991 Dec;7(9):891–911. doi: 10.1002/yea.320070902. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Misra L. M., Vogel J. P. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989 Jun 30;57(7):1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- Terasaki M. Fluorescent labeling of endoplasmic reticulum. Methods Cell Biol. 1989;29:125–135. doi: 10.1016/s0091-679x(08)60191-0. [DOI] [PubMed] [Google Scholar]
- Wright R., Basson M., D'Ari L., Rine J. Increased amounts of HMG-CoA reductase induce "karmellae": a proliferation of stacked membrane pairs surrounding the yeast nucleus. J Cell Biol. 1988 Jul;107(1):101–114. doi: 10.1083/jcb.107.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R., Rine J. Transmission electron microscopy and immunocytochemical studies of yeast: analysis of HMG-CoA reductase overproduction by electron microscopy. Methods Cell Biol. 1989;31:473–512. doi: 10.1016/s0091-679x(08)61624-6. [DOI] [PubMed] [Google Scholar]